Abstract
Precursor mRNA splicing is one of the most highly regulated processes in metazoan species. In addition to generating vast repertoires of RNAs and proteins, splicing has a profound impact on other gene regulatory layers, including mRNA transcription, turnover, transport and translation. Conversely, factors regulating chromatin and transcription complexes impact the splicing process. This extensive cross-talk between gene regulatory layers takes advantage of dynamic spatial, physical and temporal organizational properties of the cell nucleus, and further emphasizes the importance of developing a multidimensional understanding of splicing control.
Introduction
The splicing of messenger RNA precursors (pre-mRNA) to mature mRNAs is a highly dynamic and flexible process that impacts almost every aspect of eukaryotic cell biology. The formation of active splicing complexes – or “spliceosomes” – occurs via step-wise assembly pathways on pre-mRNAs. Small nuclear ribonucleoprotein particles (snRNPs): U1, U2, U4/U6 and U5 in the case of the major spliceosome, and U11, U12, U4atac/U6atac and U5 in the case of the minor spliceosome, together with an additional ~150 proteins associate with pre-mRNAs, initially through direct recognition of short sequences at the exon/intron boundaries. Key features of spliceosome formation are shown in Figure 1 and have been reviewed in detail elsewhere (Hoskins and Moore, 2012; Wahl et al., 2009).
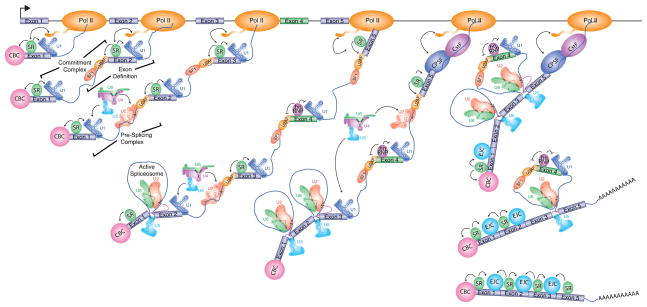
Figure 1. Co-transcriptional and post-transcriptional aspects of pre-mRNA splicing.
Co-transcriptional spliceosome assembly initiates with the binding of U1 snRNP to the 5′ splice site, which is enhanced by exon-bound SR proteins and, for the first exon, the Cap Binding Complex (CBC). A cross-intron commitment complex is formed upon association of U2 snRNP auxiliary factor (U2AF) with the 3′ splice site and adjacent intronic polypyrimidine tract, and branch point binding protein (BBP/SF1) with the branch site. Bridging interactions between these factors across internal exons, or “exon-definition”, occurs within the commitment complex. Transition from a commitment complex to a pre-splicing complex entails communication between 5′ and 3′ splice sites, and the addition of U2 snRNP to the branch site along with numerous additional proteins (not shown). Subsequent association of U4/U6/U5 tri-snRNP, together with still more protein factors, and dynamic remodeling of RNA-protein, protein-protein, and RNA-RNA interactions, ultimately leads to formation of the catalytically-active spliceosome. The two trans-esterification steps of splicing yield the excised intron in the form of the characteristic branched “lariat” structure, and the ligated exons that form mature mRNA. The assembly of most splicing factors and splicing of constitutive introns is thought to occur co-transcriptionally, while splicing of regulated alternative introns often occurs post-transcriptionally. In the example shown, exon 4 is a regulated alternative exon controlled by an hnRNP protein, which prevents the splicing factors bound to flanking splice sites from engaging in productive interactions and therefore promotes exon skipping. At terminal exons (Exon 5), interactions between the splicing factors bound to the upstream 3' splice site and the exon interact with components of the cleavage and polyadenlyation machinery (CPSF and CstF are shown; see also Figure 4A). The association of the splicing factors with the pre-mRNA is enhanced throughout the transcription process by interactions with the C-terminal domain of RNA polymerase II. The Exon-Junction complex (EJC) is recruited upstream of splice junctions upon splicing. The EJC and SR proteins mutually stabilize one another to generate the mature mRNP, which is then exported to the cytoplasm.
Spliceosome assembly can be regulated in extraordinarily diverse ways, particularly in metazoans. The major steps involve formation of the commitment complex followed by the pre-splicing complex and culminating with assembly of the active spliceosome. These steps appear to be reversible and potential points of regulation (Hoskins et al., 2011), and accumulating evidence indicates that formation of the commitment and pre-splicing complexes may be the most often subject to control (Chen and Manley, 2009).
Analysis of human genome architecture emphasizes a major challenge for accurate recognition and regulation of splice sites by the splicing machinery, namely that exons represent only 3% of the human genome (ENCODE Project Consortium, 2012). Accumulating evidence indicates that the high-fidelity process of splice site selection is not simply governed by the interaction of snRNPs and non-snRNP protein factors with pre-mRNA, but that factors associated with chromatin and the transcriptional machinery are also important (Luco et al., 2011). Moreover, splicing can “reach back” to impact chromatin composition and transcriptional activity, as well as influence parallel or downstream steps in gene expression including 3′-end processing, mRNA turnover and translation (de Almeida and Carmo-Fonseca, 2012; Moore and Proudfoot, 2009). Therefore, understanding fundamental biological processes such as cell differentiation, development, as well as disease mechanisms, will require knowledge of the cross-talk between splicing and other regulatory layers in cells. A major facet of developing such knowledge is to understand how splicing is physically, spatially and temporally integrated with other gene expression processes in the cell nucleus. This review focuses on these topics, with an emphasis on knowledge that has been gained from the application of genome-wide strategies, together with focused molecular, biochemical and cell biological approaches.
Regulation of splicing at the level of RNA
Regulatory RNA sequences
Alternative splicing (AS) is the process by which different pairs of splice sites are selected in a pre-mRNA transcript to produce distinct mRNA and protein isoforms. The importance of understanding AS regulation is underscored by its widespread nature and its numerous defined roles in critical biological processes including cell growth, cell death, pluripotency, cell differentiation, development, circadian rhythms, responses to environmental challenge, pathogen exposure, and disease (Irimia and Blencowe, 2012; Kalsotra and Cooper, 2011). Analysis of data from high-throughput RNA sequencing (RNA-Seq) of organ transcriptomes has indicated that at least 95% of human multi-exon genes produce alternatively spliced transcripts (Pan et al., 2008; Wang et al., 2008), and that the frequency of AS scales with cell type and species complexity (Barbosa-Morais et al., 2012; Nilsen and Graveley, 2010). The main types of AS found in eukaryotes are “cassette” exon skipping, alternative 5′ and 3′ splice site selection, alternative retained introns, and mutually exclusive exons. The vast majority of AS events have not been functionally characterized on any level, and this represents a major challenge for biological research. However, large-scale studies of splice variants employing a mix of computational and experimental approaches have provided evidence for widespread roles of regulated alternative exons in the control of protein interaction networks, and in cell signalling (Buljan et al., 2012; Ellis et al., 2012; Weatheritt and Gibson, 2012).
The selection of correct pairs of 5′ and 3′ splice sites in pre-mRNA is governed in part by cis-acting RNA sequences that collectively comprise the “splicing code” (Wang and Burge, 2008). The code utilizes a surprisingly minimal set of highly conserved features; these are the intronic dinucleotides GU and AG (with variations used by the minor spliceosome) at the 5′ and 3′ splice sites, respectively, and the intronic adenosine residue that forms the branched lariat structure. Additional nucleotides surrounding these positions display sequence preferences that reflect requirements for base-pairing interactions with the snRNA components of snRNPs during spliceosome formation (Wahl et al., 2009). While these minimal core elements delineate sites of splicing, they lack sufficient information to discriminate correct from incorrect splice sites and to regulate AS.
Combinations of additional sequence elements referred to as exonic/intronic splicing enhancers (E/ISEs) and silencers (E/ISSs) serve to promote and repress splice site selection. They operate in the context of achieving fidelity and in the regulation of this process (Wang and Burge, 2008). The majority of the code elements comprise short and degenerate linear motifs, although interesting examples of structured RNA elements have been discovered that function in splice site selection (Graveley, 2005; McManus and Graveley, 2011). The major contribution of linear motifs to splicing regulation is reflected by the ability of increasingly sophisticated computer algorithms to predict splicing outcomes from genomic sequence alone (Barash et al., 2010; Zhang et al., 2010). The emerging picture, supported by site-directed mutagenesis of cis-elements, is that splice site selection involves the concerted action of multiple enhancer and silencer elements that are concentrated in regions proximal (typically within ~300 nts) to splice sites (Barash et al., 2010). In particular, enhancers that support constitutive exon splicing are typically concentrated in exons, whereas enhancers and silencers that function in the regulation of AS can be located in alternative exons, although they are most often are concentrated in the immediate flanking intronic regions (Barash et al., 2010). Additionally, silencer elements are enriched in sequences surrounding cryptic splice sites – sequences that resemble splice sites, but are not functional splice sites (Wang and Burge, 2008).
Regulatory proteins
Two major classes of widely expressed trans-acting factors that control splice site recognition are the SR proteins and heterogeneous ribonucleoproteins (hnRNPs) (Long and Caceres, 2009; Martinez-Contreras et al., 2007). Depending on their binding location and the surrounding sequence context, members of each class can promote or repress splice site selection through associating with enhancers or silencers, respectively. For example, members of the SR family of proteins contain one or two RNA recognition motifs that bind ESEs and are thought to promote splicing by facilitating exon-spanning interactions that occur between splice sites (referred to as “exon definition”), but also by forging interactions with core spliceosomal proteins (Figure 1). In addition to widely expressed trans-acting factors, several tissue-specific RNA binding splicing regulators have been characterized (Irimia and Blencowe, 2012; Licatalosi and Darnell, 2010). These include the neural-specific factors Nova, PTBP2/nPTB/brPTB and nSR100/SRRM4, and factors such as RBFOX, MBNL, CELF, TIA and STAR family proteins that are differentially expressed between a variety of cell and tissue types. Through the use of splicing sensitive microarrays and RNA-Seq to detect exons affected by the knockout or knockdown of these factors, in combination with splicing code predictions and in vivo cross-linking coupled to immunoprecipitation and sequencing (HITS-Seq or CLIP-Seq), “maps” of several of these proteins have been generated that correlate their binding location (i.e. within alternative exons and/or the flanking introns) with functions in promoting exon inclusion or skipping (Licatalosi and Darnell, 2010; Witten and Ule, 2011). As mentioned earlier, where studied, these proteins appear to act primarily at the earliest stages of spliceosome formation to control splice site selection.
Integration of splicing with chromatin and transcription
Despite major progress in the characterization of factors that control splicing at the level of RNA, the impact of linked steps in gene regulation and of nuclear organization on the splicing process is less well understood. The fact that synthetic pre-mRNAs can be efficiently spliced in nuclear extracts demonstrates that splicing can be uncoupled from other nuclear processes in vitro. However, mounting evidence indicates that splicing, transcription, and chromatin modification are highly integrated in the cell. Thus, key to understanding the role of chromatin and transcription in the control of splicing is knowing which aspects of the splicing process occur co- or post-transcriptionally.
Some of the first mechanistic insights into the co-transcriptional nature of splicing came from chromatin immunoprecipitation studies in yeast. These experiments revealed that splicing factors fail to associate with intronless genes, but are recruited to intron-containing genes concomitant with the transcription of the splice sites they recognize (Gornemann et al., 2005; Lacadie and Rosbash, 2005). The main exceptions were genes containing short last exons, in which case U1 snRNP was recruited co-transcriptionally, but U2 snRNP was recruited post-transcriptionally (Tardiff et al., 2006). Similar approaches have been used in human cells, with similar results (Listerman et al., 2006). These data paint a general picture in which the splicing machinery is typically recruited to pre-mRNA in a co-transcriptional manner.
Although splicing factors are co-transcriptionally recruited, it does not necessarily follow that the splicing reaction itself occurs co-transcriptionally. Recently, Vargas et al. used in situ hybridization methods with single-molecule resolution, and found that constitutively spliced introns, which typically are efficiently spliced, were removed co-transcriptionally (Vargas et al., 2011). However, mutations that decreased the splicing efficiency, for instance by sequestering splicing signals in RNA secondary structures, caused introns to be post-transcriptionally spliced. More interestingly, two alternatively spliced introns examined were found to be post-transcriptionally spliced. This study suggested that introns could be either co-transcriptionally or post-transcriptionally spliced, in part depending on the strength and type of surrounding cis-regulatory elements.
The extent to which specific classes of splicing events occur co- or post-transcriptionally has since been examined on a genome-wide level. Several groups have analyzed RNA-Seq data generated from total cellular RNA, total nuclear RNA, nucleoplasmic RNA, or chromatin-associated RNA (Ameur et al., 2011; Bhatt et al., 2012; Khodor et al., 2012; Khodor et al., 2011; Tilgner et al., 2012). Each group used a different method to assess the extent of co-transcriptional splicing. Though the precise frequency differed in each study, most introns appeared to be co-transcriptionally spliced. The likelihood of co-transcriptional splicing increases with increased distance of introns from the 3′ ends of genes (Khodor et al., 2012). Strikingly, the set of post-transcriptionally spliced introns are strongly enriched for alternatively spliced introns. Moreover, it was observed that most human transcripts are cleaved and polyadenylated before splicing of all introns is complete, yet these transcripts remain associated with the chromatin until splicing is finished (Bhatt et al., 2012).
Because most splicing events (constitutive and alternative) occur co-transcriptionally, an important goal is to determine the extent to which chromatin and transcription factors impact them. Understanding such links necessitates considering the possible contribution of each step in transcription, through initiation, elongation and termination, and therefore also how transcription is impacted by different chromatin states.
Promoter-directed control of splicing
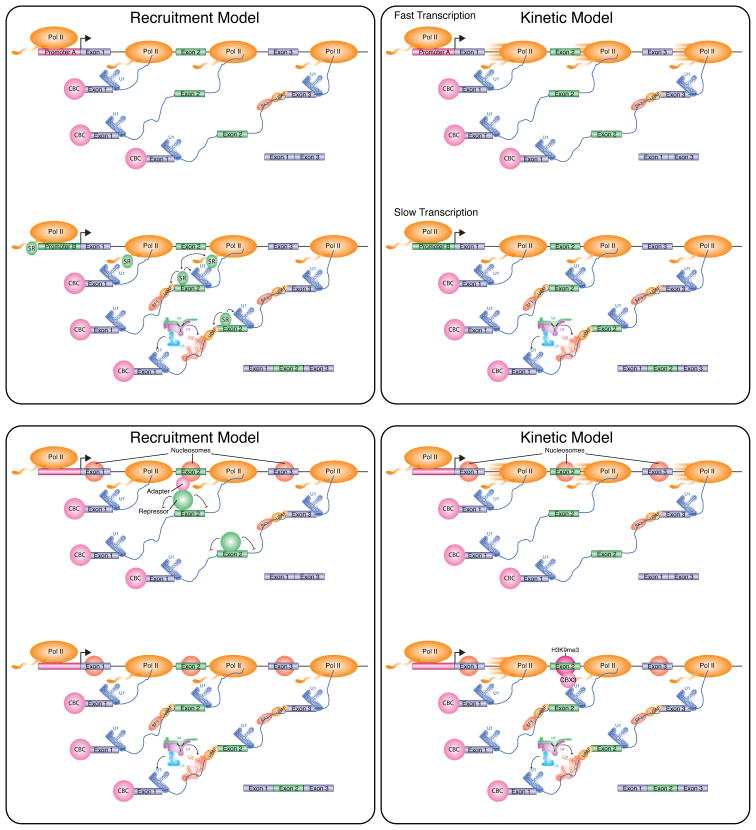
Pioneering studies performed in the late 90’s employing transfected mini-gene reporter experiments demonstrated that the type of promoter used to drive transcription by RNA polymerase II (Pol II) can impact the level of AS of a downstream exon (Cramer et al., 1997). Two non-exclusive models were proposed to explain this effect (Figure 2). In the “recruitment model”, a change in promoter architecture results in the recruitment of one or more splicing factors to the transcription machinery that in turn impact splicing of the nascent RNA. In the “kinetic model”, the change in promoter architecture affects the elongation rate of Pol II, such that there is more or less time for splice sites or other splicing signals flanking the alternative exon to be recognized by trans-acting factors (Kornblihtt, 2007). For example, if these splice sites are weak (i.e. they deviate from consensus splice site sequences associated with efficient recognition by the splicing machinery), rapid elongation will expose distal, stronger splice sites such that exon skipping occurs, as productive splicing complexes will associate with the stronger splice sites first. If elongation is slow, there is increased time for splicing factors to bind to the weak sites in the nascent RNA, and promote exon inclusion. Conversely, reduced Pol II elongation kinetics can also favour the recognition of splicing silencer elements surrounding an alternative exon, resulting in increased exon skipping.
Figure 2. Models for Chromatin and Transcription Elongation-mediated Modulation of Alternative Splicing.
(Top Left) Promoter recruitment model. Different promoters differentially recruit splicing factors to the transcription complex. At promoters which fail to recruit a key splicing factor (shown as an SR protein), the regulated alternative exon (Exon 2) will be skipped, while genes containing promoters that recruit the splicing factor will include exon 2.
(Top Right) Promoter-directed kinetic model. Different promoters assemble transcription complexes capable of different transcription elongation rates. At promoters that assemble fast transcription elongation complexes, the regulated alternative exon (Exon 2) will be skipped, while genes containing promoters that assemble slow elongation complexes will include exon 2. This model requires that the alternative exon contains weak 3′ and/or 5′ splice sites in order to be skipped when the gene is rapidly transcribed.
(Bottom Left) Chromatin-mediated recruitment model. The splicing of an alternative exon can be regulated by the chromatin-mediated recruitment of a splicing repressor. In cells that skip the exon, an adapter protein associates with the nucleosome assembled at the alternative exon, which in turn recruits a splicing repressor. In cells that include the alternative exon, the adaptor protein and/or repressor are not expressed, or the nucleosome at the regulated alternative exon is not modified and therefore cannot recruit the repressor. Similar to this model, a nucleosome-associated adaptor protein may also function to recruit a splicing activator, as proposed for Psip1/Ledgf (Pradeepa et al., 2012)
(Bottom Right) Chromatin-mediated kinetic model. The splicing of an alternative exon can be regulated by a chromatin-mediated change in the rate of transcription elongation. Unmodified nucleosomes can be transcribed rapidly, resulting in skipping of the regulated alternative exon. In cells where the nucleosome assembled on exon 2 has an H3K9me3 mark, CBX3 interacts with the modified nucleosome, slows down the transcription elongation complex, and enhances splicing of the regulated alternative exon.
While the mechanistic basis of promoter-dependent effects on AS has been investigated using model splicing reporters (see below), it is unclear to what extent and under what conditions natural switching of promoters may function in the regulation of downstream AS events in vivo. The analysis of large collections of full length transcript sequences has revealed weak correlations between the use of alternative transcript start sites and the splicing of downstream cassette exons (Chern et al., 2008), although it was not determined whether such correlations may reflect tissue-dependent effects that independently result in the increased complexity of transcription start site usage, and the increased complexity of AS. With the accumulation of datasets from the modENCODE/ENCODE projects and other studies that have yielded parallel genome-wide surveys of multiple aspects of gene regulation, including transcription factor occupancy, epigenetic modifications, long-range chromatin interactions and transcriptome profiles, it should in principle be possible to obtain higher resolution predictions of causative promoter-dependent effects on splicing and other RNA processing steps.
Despite our incomplete understanding of promoter-dependent effects on RNA processing in vivo, evidence from numerous model systems indicates that the strength and composition of a promoter can impact splicing outcomes. For example, the recruitment of the multifunctional proteins PSF/p54nrb by promoter-bound activators stimulates splicing of first introns (Rosonina et al., 2005). Activation of hormone receptors by cognate ligands has been linked to specific splicing outcomes (Auboeuf et al., 2002), and the association of PGC-1, a transcriptional coactivator that plays a major role in the regulation of adaptive thermogenesis, alters splicing activity when it is bound to a gene (Monsalve et al., 2000). Interestingly, PGC-1 contains an RS domain that may function to recruit splicing factors to PCG-1-activated promoters. In the above and additional examples, the type of promoter-bound activator may influence splicing outcomes, in part by altering the composition and/or the processivity of Pol II (David and Manley, 2011). Understanding such effects therefore entails knowledge of factors that bridge activators and Pol II, and of components of Pol II that in turn transmit information to the nascent RNA to impact splicing.
A recent study suggests that the Mediator complex may be involved in integrating and relaying information to direct splicing decisions (Huang et al., 2012). Mediator is a large multi-subunit complex that functions as a general factor at the interface between promoter-bound transcriptional activators and Pol II (Malik and Roeder, 2010). In addition to its general role, locus-specific functions have been ascribed to Mediator, where changes in its composition can lead to differential outcomes in transcription, and possibly RNA processing. Huang and colleagues showed that the MED23 subunit of Mediator physically interacts with several splicing and polyadenylation factors, most notably hnRNP L (Huang et al., 2012). Indeed, MED23 was required for regulating the AS of a subset of hnRNP L targets. It will be of interest to determine how and to what extent Mediator relays information to impact the splicing machinery on hnRNP L-regulated targets, and whether it acts similarly to regulate RNA processing through other RNA binding proteins.
The RNA Polymerase II CTD in splicing control
The C-terminal domain (CTD) of Pol II’s largest subunit impacts different stages of mRNA biogenesis, including addition of a protective cap structure on the 5′-end, splicing and formation of the mature 3′-end. The CTD consists of a repeating heptad amino acid sequence with the consensus Y1S2P3T4S5P6S7, and is predicted to be unstructured in isolation of other factors (Hsin and Manley, 2012). The CTD can be post-translationally modified by phosphorylation on each of the residues Y1S2T4S5S7, and these changes play important and distinct roles in transcription and RNA processing (Hsin and Manley, 2012). Initial evidence for a role of the CTD in RNA processing came from experiments employing expression of an alpha-amanitin resistant mutant of Pol II that harbors a truncated CTD. Truncation to five repeats led to defects in capping, splicing, and 3′-end processing of model pre-mRNA reporters (McCracken et al., 1997b; McCracken et al., 1997a), and the CTD was later found to affect AS outcomes (de la Mata and Kornblihtt, 2006; Rosonina and Blencowe, 2004). The CTD promotes capping and 3′-end formation through direct interactions with sets of factors dedicated to these processes, and increasing evidence indicates that it also serves as a platform to recruit splicing factors that may participate in commitment complex formation and the regulation of AS (David and Manley, 2011; Hsin and Manley, 2012).
Affinity chromatography identified splicing and dual splicing/transcription-associated factors as CTD binding proteins. These include yeast Prp40, human TCERG1/CA150, p54nrb/PSF proteins, SR proteins, and U2AF (reviewed in: (Hsin and Manley, 2012). Recent work supports an RNA-dependent interaction of U2AF with the phosphorylated CTD to stimulate splicing in vitro through an association with the core spliceosomal factor PRP19C (David et al., 2011). Taken together with previous work showing that a phosphorylated CTD polypeptide can stimulate splicing in vitro (Hirose et al., 1999), and that the CTD is more active in promoting splicing of a substrate that has the capacity to form exon-definition interactions compared to a substrate that cannot (Zeng and Berget, 2000), it is interesting to consider that the CTD might function as a platform to facilitate exon definition and commitment complex formation (Figure 2). In this manner, the CTD may also serve to tether exons separated by great intronic distances to promote co-transcriptional splicing (Dye et al., 2006). It will be important to determine whether the CTD plays such roles in vivo in future work.
RNA polymerase II elongation and the control of alternative splicing
Numerous studies employing model experimental systems designed to alter the rate of Pol II elongation have provided evidence supporting the aforementioned kinetic model (Kornblihtt, 2007; Luco et al., 2011). More recent work has applied genome-wide approaches to understand the extent and functional relevance of this mode of regulation. In one study, UV-induced DNA damage was found to result in a hyperphosphorylated form of the CTD and reduced Pol II elongation kinetics, and these changes were proposed to cause changes in AS of genes that function in cell cycle control and apoptosis (Munoz et al., 2009). Another study globally monitored AS changes following treatment of cells with camptothecin and 5,6-dichloro-1-β-D-ribofuranosyl-benzimidazole (DRB), which act through different mechanisms to inhibit Pol II elongation (Ip et al., 2011). Concentrations of these drugs that partially inhibit Pol II elongation preferentially affected AS and transcript levels of genes encoding RNA splicing factors and other RBP genes. Many of the induced AS changes introduced premature termination codons (PTCs) that elicited nonsense-mediated mRNA decay (NMD; see below), which further contributed to reductions in transcript levels. These results suggest that conditions globally impacting elongation rates can lead to the AS-mediated down-regulation of RNA processing factors, such that the levels of these factors are calibrated with the overall RNA processing “needs” of the cell. This type of Pol II-coupled AS network appears to be highly conserved, since amino acid starvation, which causes reduced elongation and/or increased Pol II pausing, was also found to affect the AS of transcripts from splicing factor genes, including several that can elicit NMD, in C. elegans (Ip et al., 2011).
Chromatin structure distinguishes exons from introns
Although recognition of splice sites fundamentally has to occur through direct interactions with pre-mRNA, chromatin features can shape decisions about splice site usage and exon selection. The basic unit of chromatin structure is the nucleosome, which comprises 147 base-pairs of DNA wrapped around a histone octamer consisting of two copies each of histones H2A, H2B, H3 and H4 (Luger et al., 1997). Chromatin function can be regulated by substituting canonical histones with non-allelic variants and through post-translational modification of histone tail residues most notably by methylation and acetylation (Kouzarides, 2007; Talbert and Henikoff, 2010). These “histone “marks” and direct modifications of DNA, including the addition of 5-methylcytosine, 5-hydroxymethylcytosine, and other derivatives (Wu and Zhang, 2011) affect the functional state of chromatin by altering its compaction and by modulating the binding of effector proteins. It is well established that these features have nonuniform distribution along genes with unique signatures marking promoters and gene bodies in a transcription-dependent manner (Smolle and Workman, 2012). More recently, it has become apparent that these chromatin features are also differentially distributed with respect to exon-intron boundaries, and that this differential marking participates in exon recognition.
Analysis of datasets from chromatin immunoprecipitation- high-throughput sequencing (ChIP-Seq), and from micrococcal nuclease digestion followed by sequencing revealed that nucleosomes in a range of organisms display increased occupancy over exons relative to neighboring intronic sequence (Andersson et al., 2009; Chodavarapu et al., 2010; Schwartz et al., 2009; Spies et al., 2009; Tilgner et al., 2009; Wilhelm et al., 2011). Suggesting a possible role in facilitating splicing, exons that have weak splice sites and that are surrounded by relatively long introns have greater levels of nucleosome occupancy than do exons with strong splice sites or that are flanked by short introns (Spies et al., 2009; Tilgner et al., 2009). To assess whether exon-enriched nucleosomes might be compositionally – and therefore functionally – distinct, a number of studies examined global distributions of specific histone modifications with respect to exon-intron boundaries (Andersson et al., 2009; Dhami et al., 2010; Hon et al., 2009; Huff et al., 2010; Kolasinska-Zwierz et al., 2009; Schwartz et al., 2009; Spies et al., 2009). Some of these studies reached different conclusions as to which modifications show enrichment over exons, and to what extent such enrichment is a consequence of increased nucleosome occupancy. Nevertheless, tri-methylation of lysine 36 on histone H3 (H3K36me3) was shown in multiple studies to be enriched over exons above background nucleosome levels (Andersson et al., 2009; Huff et al., 2010; Spies et al., 2009). Exon-enriched nucleosomes may also differ in their histone variant composition. The H2A variant, H2A.Bbd, which is associated with active, intron-containing genes, is enriched in positioned nucleosomes flanking both 5′ and 3′splice sites (Tolstorukov et al., 2012). Such specific histone variants could therefore play a widespread role in splicing (see below).
Base pair composition affects physical properties of the DNA and is not uniform across the genome. Exons are in general associated with higher GC content, which is an important feature governing nucleosome occupancy (Tillo and Hughes, 2009). A recent study found differences in relative GC content between exons and introns that may have evolved to contribute to splicing (Amit et al., 2012). In a reconstructed “ancestral” state, genes contained exons with a low GC content that were flanked by short introns of an even lower GC content. These subsequently diverged to yield two different types of gene architecture in animal species. In one architectural state, genes retained low exonic GC content with lower GC content in introns, but experienced an increase in intron length. In the other state, genes retained short intron length, but saw an overall increase in GC content that eliminated differential exon-intron composition (Amit et al., 2012). Bioinformatic and experimental evidence supports a role for differential GC content in promoting exon recognition in the context of the first type of architecture (Amit et al., 2012). However, to what extent differential GC content between exons and introns influences exon recognition through possible mechanisms associated with (modified) nucleosome deposition is unclear.
Studies employing genome-wide bisulphite sequencing have suggested a role for modified cytosines at exonic CpG dinucleotides in exon recognition and the regulation of AS. Modified CpG dinucleotides are enriched within exons relative to introns in both plants and animals (Chodavarapu et al., 2010; Feng et al., 2010; Laurent et al., 2010) with characteristic patterns at the 5′ and 3′ splice sites (Laurent et al., 2010). Moreover, widespread differences in CpG methylation have been detected between worker and queen bee genomes and, intriguingly, some of these differential methylation patterns appear to correlate with differential AS (Lyko et al., 2010). Highlighting a possible role of DNA epigenetic marks in mediating tissue-specific differences, in mammalian neuronal tissues hydroxymethylation rather than methylation was found to have significant exonic enrichment (Khare et al., 2012). The possible mechanisms by which such modifications affect splicing await future work.
Chromatin-dependent recruitment of the splicing machinery
Analogous to roles of promoter architecture and the Pol II CTD, accumulating evidence suggests that chromatin structure throughout a gene facilitates splicing factor recruitment to nascent transcripts. It has been proposed that splicing factors interact with chromatin directly, or indirectly through intermediate “adaptor” proteins (Figure 2). H3K4me3, which marks the promoters of actively-transcribed genes, binds specifically to CHD1, a protein that associates with U2 snRNP. Indeed, this interaction was shown to increase splicing efficiency (Sims et al., 2007). Similarly, H3K36me3, which is enriched over exons, was recently reported to interact with a short splice isoform of Psip1/Ledgf, which in turn associates with several splicing factors including the SR protein SRSF1 (Pradeepa et al., 2012). Supporting a possible role as a recruitment “adapter”, knockdown of Psip1 led to a change in SRSF1 localization and affected AS.
The aforementioned H2A.Bbd histone variant appears to function in splicing through the recruitment of splicing components (Tolstorukov et al., 2012). Mass spectrometry data revealed that H2A.Bbd interacts with numerous components of the spliceosome, and depletion of this histone variant led to the widespread disruption of constitutive and alternative splicing. Another recent study suggests that recruitment of splicing components by chromatin may be effected through global changes in histone hyperacetylation, or changes in the levels of the heterochromatin-associated protein HP1α (Schor et al., 2012). These alterations result in the global redistribution of numerous splicing factors from chromatin to nuclear speckle domains, which are thought to predominantly represent sites of splicing factor storage (Schor et al., 2012) (see below). Collectively, these studies point to characteristic patterns of chromatin structure associated with active gene expression that may have a widespread impact on the nuclear localization of the splicing machinery, which in turn can impact splicing of nascent transcripts.
Chromatin structure can be altered in highly specific ways within genes, for example, in response to environmental and developmental cues. Such “local” changes are thought to also impact AS of proximal exons on nascent RNA through the action of adapter proteins that bridge chromatin marks and splicing factors. The first example of this type of proposed mechanism involves the mutually exclusive exons IIIb and IIIc in the FGFR2 gene. Switching from exon IIIb to exon IIIc alters the ligand affinity of this receptor and represents an important step in the epithelial to mesenchymal transition. In mesenchymal cells, the region encompassing these exons is characterized by elevated levels of H3K36me3 and low levels of H3K4me3 and H3K27me3(Luco et al., 2010). H3K36me3 modifications favour the binding of MRG15, which promotes the recruitment of the splicing regulator PTBP1 to nascent RNA, and as a consequence represses the use of exon IIIb in these cells (Luco et al., 2010). Consistent with a more widespread role for an MRG15-adapter mechanism to control AS, significantly overlapping subsets of cassette exons were affected by individual knockdown of MRG15 and PTBP1 (Luco et al., 2010). However, the affected exons generally displayed modest changes in inclusion level and were found to be surrounded by relatively weak PTBP1 binding sites, suggesting that this adapter mechanism may be more important for augmenting or stabilizing patterns of AS achieved by direct action of RNA-based regulators, rather than acting to promote pronounced cell type-dependent, switch-like regulation of AS.
Chromatin structure affects splicing by influencing Pol II elongation
Specific features of chromatin structure, as well as chromatin-associated regulators, can influence splice site choice by impacting transcription elongation (Figure 2). SWI/SNF chromatin remodelling factors interact directly with Pol II (Neish et al., 1998; Wilson et al., 1996), and with splicing factors (Batsché et al., 2006), suggesting that these factors might impact splicing in an elongation-dependent manner. Supporting this view, the association of the ATP-dependent SWI/SNF-type chromatin remodelling factor BRM with the human CD44 gene coincides with a change in inclusion levels of alternative exons in CD44 transcripts (Batsché et al., 2006). Increased occupancy of Pol II with elevated S5 phosphorylation of the CTD (which is associated with a paused form of Pol II), was detected specifically over CD44 alternative exons, indicating that a reduced elongation rate or increased pausing of Pol II might be responsible for the change in AS. The Brm ATPase activity required for chromatin remodelling was, however, not required for the change in AS (Batsché et al., 2006).
Recent studies analyzing BRM in Drosophila suggests that it acts together with other members of the SWI/SNF complex to regulate AS and polyadenylation in a locus-specific manner (Waldholm et al., 2011; Zraly and Dingwall, 2012). Developmentally-regulated intron retention of the Eig71Eh pre-mRNA required the SNR1/SNF5 subunit, which suppresses BRM ATPase, and reduced elongation was correlated with more efficient intron splicing (Zraly and Dingwall, 2012).
Covalent modifications of histones impinge on Pol II elongation in ways that impact AS (Figure 2). The heterochromatin protein HP1γ/CBX3, which binds di- and trimethylated histone H3K9 (Bannister et al., 2001; Lachner et al., 2001), mediates inclusion of alternative exons in CD44 transcripts in human cells upon stimulation of the PKC pathway, concomitantly with an increase in Pol II occupancy over the alternatively spliced region (Saint-Andre et al., 2011). However, CBX3 may also play a more direct role in splicing factor recruitment. Depletion of CBX3 in human cells resulted in the accumulation of unspliced transcripts and loss of recruitment of the U1 snRNP-70 KDa (SNRNP70) protein and other splicing factors to active chromatin (Smallwood et al., 2012).
Intriguingly, components of the RNAi machinery in association with CBX3 were recently shown to also regulate AS of CD44 transcripts. Specifically, the Argonaute proteins AGO1 and AGO2 were found by ChIP-Seq analysis to bind the alternative exon-containing region of CD44, and were loaded onto this region by short RNAs derived from CD44 antisense transcripts (Ameyar-Zazoua et al., 2012). Recruitment of AGO1 and AGO2 to CD44 required Dicer and CBX3, and resulted in increased histone H3K9 methylation over the variant exons. Recruitment of AGO proteins to the CD44 gene thus appears to locally induce a chromatin state that affects Pol II elongation and AS.
RNA binding proteins bound to nascent RNA may also alter chromatin composition in ways that impact elongation and splicing (Figure 3). Hu-family proteins, which have well defined roles in the control of mRNA stability, were recently shown to regulate AS by binding to nascent RNA proximal to alternative exons in a manner that induced local histone hyperacetylation and increased Pol II elongation (Mukherjee et al., 2011; Zhou et al., 2011). This activity was linked to the direct inhibition of histone deacetylase 2 (HDAC2) by Hu proteins (Zhou et al., 2011).
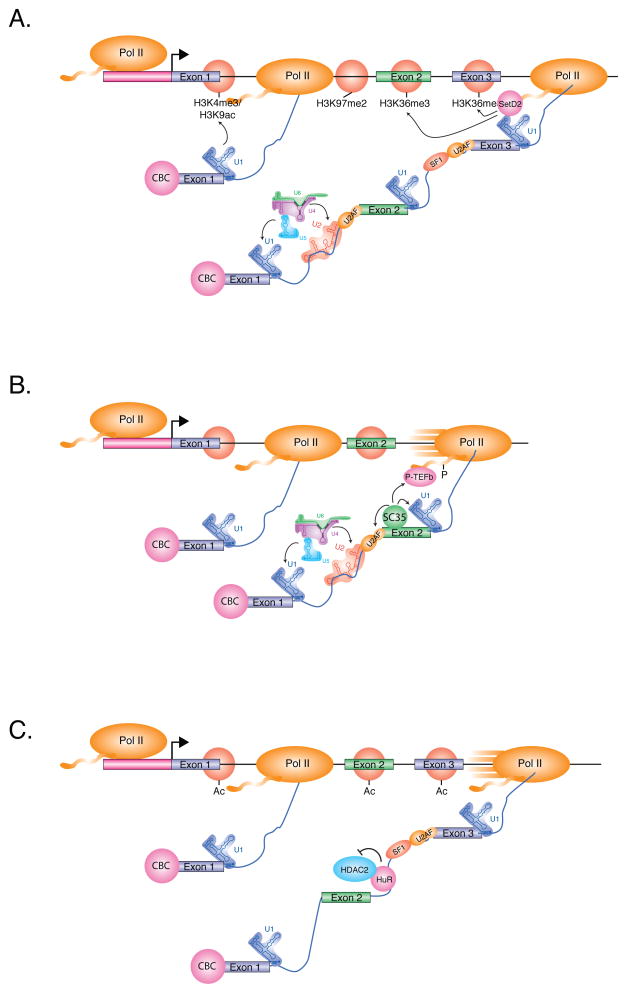
Figure 3. Reverse-Coupling Mechanisms.
(A) Splicing enhances transcription-associated histone modification. Splicing of the first intron enhances transcription initiation and stabilizes promoter-associated marks, including H3K4me3 and H3K9ac, near the 5' splice site of exon 1. Splicing may also facilitate a transition between the elongation-associated marks H3K79me2 and H3K36me3 at the 3' splice site of the first intron. Internal exons are particularly enriched for H3K36me3-modified nucleosomes, due in part to splicing-increased nucleosome occupancy and action of the histone methyltransferase SETD2 associated with elongating Pol II. These marks may also serve to reinforce splicing patterns of nascent pre-mRNA.
(B) The SR protein SRSF2/SC35, which regulates splicing of alternative exons, also enhances transcription elongation by recruiting P-TEFb. P-TEFb phosphorylates the Pol II CTD at Serine 2, which enhances the rate of transcription elongation.
(C) The Hu family of splicing regulators bind to AU-rich sequences within introns and repress the splicing of regulated alternative exons. Shown here, HuR interacts with and represses the activity of the histone deacetylase, HDAC2, which stabilizes nearby acetylated nucleosomes. Acetylated nucleosomes may enhance the rate of transcription elongation, and consequently, promote the skipping of exons with weak splice sites.
RNA Pol II elongation rates are also impacted by nucleotide sequence composition. A/T-rich sequences, in particular, are more difficult for Pol II to transcribe. A novel complex found to be associated with human mRNPs, termed DBIRD, facilitates Pol II elongation across A/T rich sequences (Close et al., 2012). Depletion of this complex resulted in reduced Pol II elongation and changes in the splicing of exons proximal to A/T-rich sequences. It was therefore proposed that DBIRD acts at the interface of RNA Pol II and mRNP complexes to control AS (Close et al., 2012).
Finally, the zinc finger DNA-binding transcription factor and chromatin organizer CTCF has been linked to the regulation of AS of exon 5 of the receptor-linked protein tyrosine phosphatase CD45, and of other transcripts, by locally affecting Pol II elongation (Shukla et al., 2011). Variable inclusion of CD45 exon 5 is controlled by RNA binding proteins during peripheral lymphocyte maturation (Motta-Mena et al., 2010). Intriguingly, CTCF appears to maintain the inclusion of exon 5 at the terminal stages of lymphocyte development by causing Pol II pausing proximal to this exon (Shukla et al., 2011). CTCF binding is inhibited by CpG methylation. Accordingly, increased methylation proximal to CD45 exon 5 led to reduced CTCF occupancy and reduced exon inclusion (Shukla et al., 2011). Analysis of AS changes genome-wide using RNA-Seq following depletion of CTCF further revealed that this factor is likely to have a more widespread role in regulating AS through altering pol II elongation kinetics. However, CTCF is known to mediate intrachromosomal interactions (Ohlsson et al., 2010), and it therefore remains to be determined whether the changes in AS caused by CTCF reflect a direct inhibition of Pol II elongation, or whether these effects are a consequence of more complex topological changes to chromatin architecture.
In the examples described above and others (Luco et al., 2011), changes in AS can be achieved through a variety of mechanisms that perturb Pol II elongation in a widespread or locus-specific manner. In other cases, AS is affected through mechanisms involving the differential recruitment of splicing factors to transcription or chromatin components. It is currently unclear to what extent these mechanisms are distinct or overlap as the recruitment of splicing factors to a transcript in some cases appears to affect elongation kinetics, and in other cases altered elongation kinetics may affect the recruitment of splicing components to chromatin or transcription factors associated with nascent transcripts. For example, as summarized earlier, regulation of variable exon inclusion in CD44 transcripts appears to involve the concerted action of chromatin remodeling, inhibition of Pol II elongation, and the recruitment of splicing factors and the RNAi machinery. Individual genes may therefore possess a unique set of mechanistic principles that are governed by the specific combinatorial interplay between cis-elements of the splicing code and genomic features, which together determine the formation and activity of chromatin features and transcription complexes. The increased use of comparative analyses of parallel datasets interrogating transcriptomic, genomic and chromatin features should nevertheless facilitate a more detailed mechanistic understanding of common principles by which chromatin, transcription and splicing are coupled to coordinate the regulation of subsets of genes.
Regulation of chromatin and transcription by the splicing machinery
In addition to the extensive set of interactions and mechanisms by which chromatin and transcription components can impact splicing, increasing evidence indicates that splicing can have a major impact on chromatin organization and transcriptional output. Early indications of this “reverse-coupling” were that the efficient expression of transgene constructs required the presence of an intron (Brinster et al., 1988). Such effects were later shown to arise in part as a consequence of enhanced transcription (Furger et al., 2002). Subsequent studies have demonstrated several mechanisms by which the splicing of nascent transcripts can impact chromatin organization and transcription. For example, H3K4me3 and H3K9ac, both of which are associated with active genes and widely assumed to peak in proximity to promoters together with increased Pol II occupancy, are in fact concentrated over first exon-intron boundaries (Bieberstein et al., 2012) (Figure 3A). In genes with long first exons, these marks are reduced at promoters, whereas in genes with short first exons, the marks are increased at promoters, as are transcription levels. Confirming a role for first intron splicing in establishing promoter proximal architecture, intron deletion reduced H3K4me3 levels and transcriptional output (Bieberstein et al., 2012). Taken together with previous observations of associations between U1 and Pol II (Damgaard et al., 2008), and between U2 snRNP and H3K4me3 (Sims et al., 2007), a picture emerges in which first intron splicing serves to establish or perhaps reinforce promoter proximal marks, that in turn recruit general transcription factors and Pol II to enhance initiation.
The enrichment of H3K36me3 at exons, which is established by the methyltransferase SETD2 as it travels with elongating Pol II, also arises in part as a consequence of splicing (Figure 3A). Global inhibition of splicing (via depletion of specific spliceosome components and/or exposure to the inhibitor spliceostatin) decreased H3K36me3 levels at particular exons, but also broadly altered its distribution within gene bodies (De Almeida et al., 2011; Kim et al., 2011). To what degree these effects are direct remains unclear, as global inhibition of splicing would also be expected to perturb transcription, for example, by affecting the expression and/or deposition of transcription and chromatin factors (Bieberstein et al., 2012). Nonetheless, a direct role also seems likely. For example, reciprocal H3K79me2 and H3K36me3 histone marks at first intronic 3' splice site-first internal exon boundaries, but not at the corresponding boundaries of pseudo-exons (Huff et al., 2010) (ENCODE Project Consortium, 2012), suggests more direct roles of splicing-dependent transitions in chromatin-modifications (Figure 3A). Moreover, mass spectrometry data further suggests that SETD2 may associate with exon definition complexes (Schneider et al., 2010).
Splicing also impacts Pol II pausing and elongation. An association between snRNPs and the Pol II elongation factor TAT-SF1 can stimulate elongation in vitro, and this activity was further enhanced by the presence of splicing signals in RNA (Fong and Zhou, 2001). Since TAT-SF1 interacts with the positive elongation factor P-TEFb, which phosphorylates the S2 residues of the CTD to increase Pol II processivity, it was proposed that the assembly of splicing complexes on nascent RNA may facilitate Pol II elongation across a gene (Fong and Zhou, 2001).
Additional studies have reported roles for splicing factors in elongation. Since this topic has been reviewed elsewhere (Pandit et al., 2008), only a few examples will be highlighted here. Of particular interest are SR and SR-like proteins, which have long-established roles in splicing. The S. cerevisiae SR-like protein Npl3, for example, regulates the splicing of a subset of introns (Chen et al., 2010; Kress et al., 2008), but it also facilitates elongation by acting as an anti-termination factor (Dermody et al., 2008). Specific mutations in Npl3 lead to defects in the transcription elongation and termination of ~30% of genes (Dermody et al., 2008). Npl3 binds the S2 phosphorylated CTD (Lei et al., 2001), bringing it into close proximity to nascent RNA. Phosphorylation of Npl3 was found to negatively regulate its binding to the CTD and RNA, suggesting that unphosphorylated Npl3 specifically promotes elongation in association with Pol II (Dermody et al., 2008).
Depletion of the SR family protein SRSF2/SC35 increases Pol II pausing, most likely as a consequence of defective recruitment of P-TEFb and reduced S2 CTD phosphorylation (Lin et al., 2008) (Figure 3B). It is interesting to consider that Npl3, SRSF2, and possibly other RNA-binding proteins, may also facilitate elongation in part by preventing the formation of DNA-RNA hybrids (or R-loops) formed by nascent RNA during transcription (Pandit et al., 2008). Finally, it is also conceivable that SR proteins bound to nascent RNA indirectly promote CTD phosphorylation and/or histone modifications that facilitate transcription. In this regard, it was recently shown that Npl3 associates in an RNA-independent manner with Bre1, a ubiquitin ligase with specificity for H2B (Moehle et al., 2012) that facilitates transcription elongation in vitro (Pavri et al., 2006).
The studies summarized above emphasize important roles for nascent RNA splicing and the factors that control splicing in establishing chromatin architecture and in controlling transcription. It is interesting to consider, therefore, that a major determinant of gene-specific chromatin architecture emanates from information provided by cis-acting elements comprising the splicing code. The previously described case of the Hu family of hnRNP proteins is illustrative of a mechanism through which proteins bound to nascent RNA can “reach back” to alter proximal chromatin and affect Pol II elongation (Zhou et al., 2011) (Figure 3C). Notably, this mode of regulation also mediates highly “local” changes in chromatin structure that in turn regulate the AS regulation of nearby exons. A more systematic investigation of the roles of splicing components in establishing region-specific chromatin modifications and functions will be important for understanding the crosstalk between chromatin and splicing.
Integration of splicing with 3′end processing, turnover and transport
Coupling and coordination of splicing with 3′end formation
Numerous studies have demonstrated communication between factors involved in the splicing of 3′-terminal introns and factors involved in 3′-end cleavage and polyadenylation (CPA), and this topic has been reviewed in detail elsewhere (Di Giammartino et al., 2011; Proudfoot, 2011). Similar to the formation of exon-definition complexes, it has been proposed that U2AF binding to the 3′splice site of a terminal exon forms interactions with Cleavage Factor I and the CTD of poly(A) polymerase to mutually stimulate terminal intron splicing and CPA (Millevoi et al., 2002; Millevoi et al., 2006) (Figure 4A). SR proteins have also been implicated in terminal exon cross-talk (Dettwiler et al., 2004; McCracken et al., 2002). In certain cases, competition between binding of CPA factors and splicing factors can result in physiologically important changes in AS and transcript levels (Evsyukova et al., 2013) (see below).
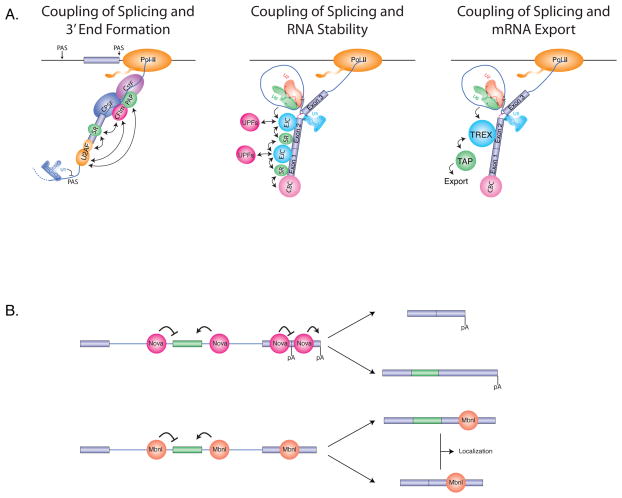
Figure 4. Splicing impacts the regulation of multiple downstream steps in gene regulation.
(A) Coupling connections between splicing and 3′ end formation, RNA stability, and mRNA export. Splicing and 3'-end formation are coupled by interactions between exon-bound SR proteins and the cleavage and polyadenylation factor CFIm, and between U2AF and both CFIm and PAP. Cryptic upstream polyadenlyation sites (PAS) are suppressed by U1 snRNP (left). Splicing is impacts RNA stability by interactions between SR proteins and the EJC, which in turn interacts with the UPF proteins involved in nonsense mediated decay (NMD) (middle). Splicing influences mRNA export through the splicing-dependent recruitment of the TREX complex, which in turn interacts with the RNA export factor TAP.
(B). Multi-tasking roles of RBPs in splicing and alternative polyadenylation, RNA export and RNA transport. (Top) The Nova RNA binding proteins have been shown to not only regulate alternative splicing, but also alternative polyadenylation (pA). Both of these processes are modulated in a position-dependent manner with some binding locations promoting splicing and polyadenylation, and other locations repressing these processes. The result of this regulation is the generation of mRNAs with different exons and 3' UTR sequences (Bottom) Similarly, Mbnl RNA binding proteins impact alternative splicing in a position-dependent manner, and to bind to 3' UTRs, where they function to control subcellular mRNA localization.
In addition to their roles in the control of large networks of alternative exons, splicing regulators such as Nova and hnRNP H1 function in the regulation of alternative polyadenylation (APA) through direct binding to recognition sites clustered around the CPA signals (Katz et al., 2010; Licatalosi et al., 2008) (Figure 4B). While these “moon-lighting” roles in APA regulation appear to be largely independent of the splicing of proximal exons/introns, regulation of AS and APA by the same RBPs presumably is important for globally coordinating these processes in a cell type or condition-dependent manner. For example, transcript profiling studies have shown that APA is widespread, affecting at least 50% of transcripts from human genes (Tian et al., 2005), and that it plays an important role in controlling the presence of miRNA and RNA binding protein target sites in UTR sequences, and therefore mRNA expression levels (Mayr and Bartel, 2009; Sandberg et al., 2008). Control of APA and AS by an overlapping set of RBP regulators may therefore constitute an effective mechanism for functionally coordinating these steps in RNA processing.
In an analogous manner, U1 snRNP also has dual roles in splicing and CPA. U1 snRNP is more abundant than other spliceosomal snRNPs, and this observation hinted that it may have additional functions in the nucleus. Indeed, recent studies have shown that, through binding to cryptic 5′ splice sites within pre-mRNAs, U1 snRNP can inhibit premature 3′-end formation at potential CPA sites that are distributed along pre-mRNAs (Berg et al., 2012) (Figure 4A). In situations where U1 snRNP becomes limiting, for example, during bursts of pre-mRNA transcription upon activation of neurons or immune cells, where the ratio of cryptic and bona-fide 5′ splice sites may be in excess of available U1 snRNP, premature CPA sites are activated leading to transcript shortening (Berg et al., 2012). Furthermore, reduced U1 snRNP to pre-mRNA ratios resulted in changes in terminal exon usage, consistent with the mutual stimulation between the splicing and CPA machineries in terminal exon definition. The discovery of a role for U1 snRNP in suppressing CPA has provided further insight into the mechanism by which certain mutations in 3′ UTRs cause disease. For example, a mutation in the 3′ UTR of the p14/ROBLD3 receptor gene that is causally linked to immunodeficiency, creates a 5′splice site that does not activate splicing, but suppresses CPA, leading to reduced p14/ROBLD3 expression (Langemeier et al., 2012).
Splicing modulates RNA stability and transport
The nonsense-mediated decay (NMD) pathway acts to prevent spurious expression of incompletely processed or mutant transcript (Rebbapragada and Lykke-Andersen, 2009). Although the NMD pathway appears to be present in some form in all eukaryotes, there are nonetheless species-specific differences, particularly in the way premature termination codons (PTCs) are recognized and in the nature of the degradation pathways involved. In mammalian cells, PTC recognition relies to a large extent on deposition of the exon junction complex (EJC) 20–24 nt upstream of exon-exon junctions. The EJC encompasses a stable tetrameric core consisting of eIF4AIII, MAGOH, MLN51, and Y14 proteins, which is deposited on mRNA during splicing (Tange et al., 2005). This core associates with a host of SR and SR-related proteins to form megadalton size complexes that presumably function in mRNP compaction as well as in facilitating coupling of splicing with downstream steps in gene expression (Singh et al., 2012) (Figures 1 and 4B). During the pioneer round of translation, EJCs are displaced by the ribosome (Isken et al., 2008). However, when the ribosome encounters a PTC more than 50–55 nt upstream of a terminal exon-exon junction, EJC components associate with upstream frame shift (UPF) proteins (Figure 4A) that trigger release of the ribosome through interaction with release factors eRFs. These and other interactions ultimately lead to mRNA decay through pathways that involve 5′-end decapping, deadenylation and exoribonucleotic enzymes (Schoenberg and Maquat, 2012).
Alternative splicing coupled to NMD controls the levels of specific subsets of genes. It has been estimated that approximately 10–20% of AS events that have the potential to introduce PTCs lead to substantial changes in overall total steady state transcript levels (Pan et al., 2006). In many cases, these AS-coupled NMD events serve to auto- and cross-regulate expression levels of regulatory and core factors involved in splicing and other aspects of RNA metabolism (Cuccurese et al., 2005; Lareau et al., 2007b; Mitrovich and Anderson, 2000; Ni et al., 2007; Plocik and Guthrie, 2012; Saltzman et al., 2008), but important roles in the regulation of other classes of proteins have also been reported (Barash et al., 2010; Lareau et al., 2007a).
It is important for a cell to prevent incompletely or aberrantly processed transcripts from being translated, as such transcripts may express truncated proteins with aberrant or dominant negative functions that have harmful consequences. One safeguarding mechanism is to prevent release of such transcripts from the nucleus. The TREX (transcription/export) complex is a conserved multi-protein complex that links transcription elongation with nuclear mRNA export (Katahira et al., 2009). While S. cerevisiae TREX is recruited to intronless transcripts (Strasser et al., 2002), its mammalian counterpart is incorporated into maturing mRNPs by the splicing machinery (Masuda et al., 2005), and further requires binding of the 5' cap by the TREX component Aly (Cheng et al., 2006). TREX then mediates association with the TAP nuclear export receptor to facilitate mRNA export through the nuclear pore complex (Stutz et al., 2000; Zhou et al., 2000) (Figure 4A). Natural intronless genes can circumvent the necessity for splicing to recruit TREX through sequence elements that directly mediate TREX- and TAP-dependent export (Lei et al., 2011). However, transcripts from some intron-containing yeast genes, for example the gene encoding the nuclear export factor SUS1, require introns for efficient nuclear mRNA export (Cuenca-Bono et al., 2011) (see below).
Regulated intron retention has been harnessed to play important regulatory roles in the control of transcript levels. For example, coordinated regulation of a set of alternative retained introns controls the expression of the neuron-specific genes Stx1b, Vamp2, Sv2a, and Kif5a. The splicing regulator Ptbp1, which is expressed widely in non-neural cells, represses splicing of these introns, such that the unspliced transcripts are retained in the nucleus where they are degraded by the exosome (Yap et al., 2012). Inhibition of Ptbp1 expression by miR-124 in neural cells results in splicing of these introns, allowing export and translation of the resulting mature mRNAs. With the wealth of available transcriptome profiling data, it can be expected that many additional examples of regulated intron removal linked to functions such as mRNA turnover and transport will soon emerge.
While the EJC appears to be seldom required for NMD in Drosophila, it is important for the localization of developmentally important transcripts. Localization of oskar mRNA to the posterior pole of the oocyte requires the deposition of the EJC core components together with an exon-exon junction-spanning localization element formed by splicing of the first intron (Ghosh et al., 2012). Changes in alternative splicing, particularly in UTR regions, have been observed to differentially regulate mRNA localization in mammalian cells (La Via et al., 2012; Terenzi and Ladd, 2010), and likely represent a more widely used mode of regulation than currently appreciated. Similar to previously mentioned examples in which specific RBPs have roles in both AS and APA, specific RBPs that function in AS regulation can also function in mRNA localization. Transcriptome profiling of cells and tissues deficient of MBNL1 and MBNL2, coupled with analysis of the in vivo target sites of these proteins, has revealed that they regulate large networks of alternative exons involved in differentiation and development (Charizanis et al., 2012; Wang et al., 2012) (Figure 4B). A transcriptomic and proteomic analysis of subcellular compartments further uncovered a widespread role for MBNL proteins in the regulation of transcript localization, translation, and protein secretion (Wang et al., 2012). These studies underscore the importance of integrative analyses that capture information from multiple aspects of mRNA processing and expression when analyzing the functions of individual RBPs. In particular, it is becoming increasingly evident that most if not all RBPs in the cell multitask, and the extent to which the multiple regulatory functions of RBPs arise through physical (i.e. direct) coupling between processes, as opposed to independently operating functions, will be important to determine.
Dynamic nuclear organization in splicing control
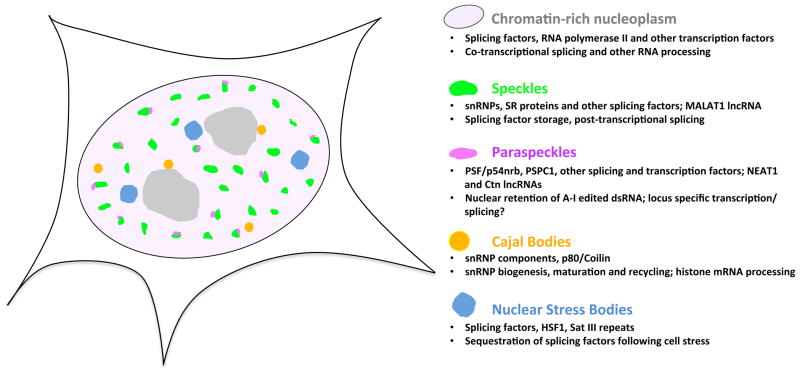
The majority of the mechanisms described thus far in this review invoke the formation and disruption of protein-protein and protein-RNA interactions in splicing control. However, of critical importance to any one of these mechanisms in vivo, is the local availability of active splicing components relative to the requirements for these factors presented by cognate cis-acting elements in nascent RNA. Regulation of the availability of splicing components provides a potentially powerful means by which constitutive and AS events may be controlled. The highly compartmentalized nature of the cell nucleus, which contains several different types of non-membranous substructures, or “bodies”, that concentrate RNA processing factors, provides such a regulatory architecture. Among the domains that concentrate splicing and other RNA processing factors are interchromatin granule clusters or “speckles”, paraspeckles, Cajal Bodies (CBs) and nuclear stress bodies (Figure 5) (Biamonti and Vourc'h, 2010; Machyna et al., 2013; Nakagawa and Hirose, 2012; Spector and Lamond, 2011).
Figure 5. Organization of the splicing components in the cell nucleus.
Major nuclear domains enriched in splicing and other factors in the mammalian cell nucleus are depicted, with known and putative roles indicated. Gray areas indicate nucleoli.
Mammalian cell nuclei typically contain 20–50 speckle structures that concentrate snRNP and non-snRNP splicing factors, including numerous SR family and SR-like proteins (Spector and Lamond, 2011). Experiments employing transcriptional inhibitors and inducible gene loci revealed that splicing factors can shuttle between speckles and nearby sites of nascent RNA transcription, and additional studies have shown that this shuttling behavior can be controlled by specific kinases and phosphatases that alter the post-translational modification status of SR proteins and other splicing factors. These and other observations led to the proposal that speckles primarily represent storage sites for splicing factors (Spector and Lamond, 2011). However, more recent studies using antibodies that specifically recognize the phosphorylated U2 snRNP protein SF3b155 (P-SF3b155), which is found only in catalytically-activated or active spliceosomes, paint a more complex picture (Girard et al., 2012). Immunolocalization using an anti-P-SF3b155 antibody showed spliceosomes localized to regions of decompacted chromatin at the periphery of – or within – nuclear speckles (Girard et al., 2012). Inhibition of transcription and splicing after SF3b155 phosphorylation further revealed that post-transcriptional splicing occurs in nuclear speckles. These results are consistent with results from earlier studies employing simultaneous fluorescence in situ hybridization detection of unspliced and spliced transcripts, which suggested that the introns of specific transcripts are spliced within speckles (Lawrence et al., 1993).
Paraspeckles are structures that form at the periphery of speckle domains and have been observed widely across mammalian cells and tissues (Fox and Lamond, 2010; Nakagawa and Hirose, 2012). They have been implicated in the regulation of gene expression by mediating the nuclear retention of adenosine-to-inosine (A-to-I) edited transcripts (Fox and Lamond, 2010). However, the recent discovery that these structures concentrate on the order of 40 multi-functional RNA binding proteins suggests yet undiscovered roles in other aspects of RNA processing (Naganuma et al., 2012).
Mammalian nuclei typically contain several Cajal Bodies, and these domains are thought to represent primary sites of spliceosomal and non-spliceosomal snRNP biogenesis, maturation and recycling (Machyna et al., 2013). The formation and size of CBs relates to the transcriptional and metabolic activity of cells, and these structures are prominent in rapidly proliferating cells. Since the in vivo concentration of basal spliceosomal components, including snRNPs, can impact specific subsets of AS events (Park et al., 2004), in particular those that are predicted to regulate levels of RNA processing factors (Saltzman et al., 2011), it is interesting to consider that processes that control the formation and activity of CBs could indirectly control AS of multiple genes to globally coordinate levels of RNA processing factors according to the metabolic requirements of the cell. Analogous to this proposed role for CBs, nuclear stress bodies are structures that form specifically in response to a variety of stress conditions including heat shock, oxidative stress or exposure to toxic materials (Biamonti and Vourc'h, 2010). These structures are thought to mediate global changes in gene expression, in part by sequestering splicing factors (Biamonti and Vourc'h, 2010).
An important facet of understanding the role of nuclear domains in the control of splicing and other steps in gene regulation is to determine how they are formed. Much in the way nucleoli form around tandem repeats of rRNA genes, formation of nuclear domains with connections to the splicing process may be nucleated by – or depend on for integrity – specific DNA or RNA sequences, including long (intergenic) non-coding RNAs (lnc/lincRNAs). CBs have been detected at U1 and U2 snRNA gene loci (Smith et al., 1995), although they may assemble via the association of multiple different protein and nucleic acid components (Machyna et al., 2013), and stress body formation is dependent on transcriptionally active, pericentric tandem repeats of satellite III sequences bound by heat shock transcription factor 1 (HSF1) (Biamonti and Vourc'h, 2010).
Speckle domains concentrate MALAT1, a nuclear lncRNA that appears to participate in controlling the phosphorylation state of SR proteins (Tripathi et al., 2010). Depletion of human MALAT1 was also reported to alter the nuclear distribution of SRSF1 and to lead to changes in SRSF1-dependent AS events (Tripathi et al., 2010), although a more recent study did not observe such effects (Zhang et al., 2012). Moreover, recent studies employing Malat1 knockout mice did not reveal an essential role for this lncRNA under normal laboratory conditions (Eissmann et al., 2012; Nakagawa et al., 2012), whereas another study reported that it is important for metastasis-associated properties of lung cancer cells (Gutschner et al., 2012). NEAT1, another lncRNA, is an integral structural component of paraspeckles (Clemson et al., 2009; Naganuma et al., 2012). A change in the alternative 3’-end processing of NEAT1 lncRNA by hnRNP K affects the formation of these domains (Naganuma et al., 2012). Very recently, a class of sno-lncRNAs transcribed from a genomic region linked to Prader–Willi syndrome was shown to sequester the RbFox2 splicing regulator, and to modulate AS (Yin et al., 2012). As additional ncRNAs are identified and characterized, it can be expected that many other examples of ncRNA-based control of splicing factor availability and functional activity will be discovered.
In addition to the aforementioned roles for DNA and RNA, it has recently emerged that the prevalence of low complexity or disordered protein regions in splicing and other RNA processing factors may play an important role in the formation and regulation of the activity of nuclear domains. Homotypic and heterotypic interactions involving these domains and RNA have been shown to form hydrogel-like structures, and it is intriguing to consider that such structures act as malleable interfaces or “matrices” with which to dynamically control (i.e. by differential phosphorylation or other post-translational modifications) the accessibility, assembly, and activity, of splicing and other highly integrated regulatory complexes in the cell nucleus (Han et al., 2012; Kato et al., 2012).
Conclusions and future perspectives
During the past several years remarkable strides have been made in our understanding of how splicing is dynamically integrated with other layers of gene regulation, and within the context of sub-nuclear structure and organization. Advancements in high-throughput technologies and computational approaches, together with focused biochemical, molecular and cell biological methods, have powered the discovery and characterization of the global principles by which splicing forms a nexus of extensive cross-talk between gene expression processes. This cross-talk temporally coordinates and enhances, and in some cases represses, the kinetics of physically-coupled steps in RNA metabolism, but it also serves to co-ordinately regulate different steps in the transcription, processing, export, stability and translation of mRNA.
Of key importance in future studies will be to determine the specific conditions and mechanisms by which chromatin- and transcription-associated components control splicing outcomes, and vice versa. Current models often propose networks of physical interactions between these processes. However, it is unclear to what extent regulatory mechanisms may rely on increased local concentrations of factors (i.e. through associations with chromatin and or other nuclear domains) that provide kinetic advantages, which in turn promote “coupled” effects. Regardless of the specific mechanisms by which cross-talk impacts splicing and coupled processes, it is exciting to consider that entirely new functional connections await discovery. For example, the role of splicing in the deposition of specific chromatin marks such as H3K36me3 could impact additional chromatin mark-regulated functions, such as DNA replication, repair and methylation (Wagner and Carpenter, 2012). The plethora of poorly characterized histone lysine methylation “readers” such as the tudor, chromodomain, PWWP and other “Royal family” domain-containing proteins are candidates for mediating possible new splicing-dependent regulation involving chromatin marks and their binding to reader proteins (Yap and Zhou, 2010).
Another important area of future investigation is to establish the extent to which nucleic acid binding proteins multi-task to coordinate different aspects of biology. While this review focuses on a few examples of multi-tasking RBPs, it is telling that almost every recent study employing in vivo mapping of binding sites of splicing regulators or other RBPs has uncovered previously unknown, additional functions of these proteins. Moreover, other in vivo cross-linking studies using polyadenylated RNA as bait to comprehensively identify RBPs, point to a much more extensive multitasking world in which transcription factors and proteins associated with other diverse cellular functions, including metabolism, may have unsuspected functions in association with RNA (Baltz et al., 2012; Castello et al., 2012). In this regard, it should be noted that among the largest group of uncharacterized nucleic acid binding factors are C2H2 and other zinc-finger domain proteins, defined examples of which can regulate gene expression through binding RNA.
Increasing examples of pivotal roles for switch-like AS events is providing a perspective in which a relatively small number of regulated exons can act to re-wire entire programs of gene regulation by modifying core domains of proteins that dictate the activities of regulators of chromatin, transcription and other steps in gene regulation (Irimia and Blencowe, 2012). Numerous other AS events remodel protein interaction and signaling networks that are important for establishing cell type-specific functions (Babu et al., 2011; Ellis et al., 2012; Weatheritt and Gibson, 2012). Such AS events are often found in disordered domains of proteins that are subject to phosphorylation and other types of post-translational modifications. Interestingly, these domains are often found in splicing factors and other nuclear gene expression regulators, with the RS-repeat domains of SR proteins and the CTD of Pol II representing notable examples. A very important area of future investigation will be to understand how these and other protein domains contribute to the assembly and disassembly of higher-order nuclear structures that function to organize and possibly catalyze splicing and other nuclear reactions (Han et al., 2012; Kato et al., 2012). Also central to this understanding will be to discover and characterize ncRNAs that participate in the dynamic integration of splicing with other nuclear processes.
Acknowledgments
We thank members of the Graveley and Blencowe laboratories for helpful discussions. B.R.G. acknowledges support from NIH grants R01 GM067842, R01 GM095296, U54 HG007005, and U54 HG006994. B.J.B. acknowledges funding from the Canadian Institutes of Health Research, Canadian Cancer Society, Natural Sciences and Engineering Research Council of Canada (NSERC), and the Ontario Research Fund. U.B. was supported by EMBO and HFSP Fellowships, S.G. was supported by an NSERC Studentship, and A.P. was supported by an NRSA Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012;1:543–556. doi: 10.1016/j.celrep.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Research. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Honig A, Berget SM, O'Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Current opinion in structural biology. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2:62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. Muscleblind-like 2-Mediated Alternative Splicing in the Developing Brain and Dysregulation in Myotonic Dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Milliman EJ, Goulet I, Cote J, Jackson CA, Vollbracht JA, Yu MC. Protein arginine methylation facilitates cotranscriptional recruitment of pre-mRNA splicing factors. Molecular and cellular biology. 2010;30:5245–5256. doi: 10.1128/MCB.00359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Chern TM, Paul N, van Nimwegen E, Zavolan M. Computational analysis of full-length cDNAs reveals frequent coupling between transcriptional and splicing programs. DNA research : an international journal for rapid publication of reports on genes and genomes. 2008;15:63–72. doi: 10.1093/dnares/dsm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel Ja, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara Sa, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P, East P, Dirac-Svejstrup AB, Hartmann H, Heron M, Maslen S, Chariot A, Söding J, Skehel M, Svejstrup JQ. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484:386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic acids research. 2005;33:5965–5977. doi: 10.1093/nar/gki905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca-Bono B, García-Molinero V, Pascual-García P, Dopazo H, Llopis A, Vilardell J, Rodríguez-Navarro S. SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic acids research. 2011;39:8599–8611. doi: 10.1093/nar/gkr496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5' splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. The RNA polymerase C-terminal domain: a new role in spliceosome assembly. Transcription. 2011;2:221–225. doi: 10.4161/trns.2.5.17272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Carmo-Fonseca M. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends in biochemical sciences. 2012;37:248–253. doi: 10.1016/j.tibs.2012.02.002. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. The Journal Of Biological Chemistry. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Dhami P, Saffrey P, Bruce AW, Dillon SC, Chiang K, Bonhoure N, Koch CM, Bye J, James K, Foad NS, et al. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PloS one. 2010;5:e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, et al. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsyukova I, Bradrick SS, Gregory SG, Garcia-Blanco MA. RNA. New York, NY: 2013. Cleavage and polyadenylation specificity factor 1 (CPSF1) regulates alternative splicing of interleukin 7 receptor (IL7R) exon 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gáspár I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol. 2012;19:441–449. doi: 10.1038/nsmb.2257. [DOI] [PubMed] [Google Scholar]
- Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, Urlaub H, Hartmuth K, Luhrmann R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nature communications. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko AS, Arun G, Stentrup M, Gross M, et al. The non-coding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2012 doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Tina W, Kato M, Xie S, Wu Leeju C, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends in biochemical sciences. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Research. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Current Opinion in Cell Biology. 2012 doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nature reviews Genetics. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. The EMBO journal. 2009;28:556–567. doi: 10.1038/emboj.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han Tina W, Xie S, Shi K, Du X, Wu Leeju C, Mirzaei H, Goldsmith Elizabeth J, Longgood J, Pei J, et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA. 2012;18:2174–2186. doi: 10.1261/rna.034090.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CHA, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genetics. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Via L, Bonini D, Russo I, Orlandi C, Barlati S, Barbon A. Modulation of dendritic AMPA receptor mRNA trafficking by RNA splicing and editing. Nucleic acids research. 2012 doi: 10.1093/nar/gks1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5'ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Langemeier, Schrom EM, Rabner A, Radtke M, Zychlinski D, Saborowski A, Bohn G, Mandel-Gutfreund Y, Bodem J, Klein C, et al. A complex immunodeficiency is based on U1 snRNP-mediated poly(A) site suppression. The EMBO journal. 2012;31:4035–4044. doi: 10.1038/emboj.2012.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007a;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007b;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome research. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J, Carter K, Xing X. Probing functional organization within the nucleus: is genome structure integrated with RNA metabolism? Cold Spring Harbor Symp Quan Biol. 1993;58:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Dias AP, Reed R. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17985–17990. doi: 10.1073/pnas.1113076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nature reviews Genetics. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science (New York, NY) 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA. 2013;4:17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature reviews Genetics. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997b;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997a;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3'-end cleavage. Mol Cell Biol. 2002;22:148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3'-end processing. EMBO Rep. 2002;3:869–874. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3' end processing machineries. The EMBO journal. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Ryan CJ, Krogan NJ, Kress TL, Guthrie C. The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing. PLoS Genet. 2012;8:e1003101. doi: 10.1371/journal.pgen.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell. 2010;37:223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. The EMBO journal. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Hirose T. Paraspeckle nuclear bodies--useful uselessness? Cellular and molecular life sciences : CMLS. 2012;69:3027–3036. doi: 10.1007/s00018-012-0973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic acids research. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Current Opinion in Cell Biology. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004:0407004101. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Plocik AM, Guthrie C. Diverse forms of RPS9 splicing are part of an evolving autoregulatory circuit. PLoS genetics. 2012:8. doi: 10.1371/journal.pgen.1002620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Current Opinion in Cell Biology. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Blencowe BJ. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3'-end cleavage. Rna. 2004;10:581–589. doi: 10.1261/rna.5207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Molecular and cellular biology. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011;25:373–384. doi: 10.1101/gad.2004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Luhrmann R. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol Cell. 2010;38:223–235. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nature reviews Genetics. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Llères D, Risso GJ, Pawellek A, Ule J, Lamond AI, Kornblihtt AR. Perturbation of chromatin structure globally affects localization and recruitment of splicing factors. PloS one. 2012;7:e48084. doi: 10.1371/journal.pone.0048084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Millhouse S, Chen CF, Lewis Ba, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The Cellular EJC Interactome Reveals Higher-Order mRNP Structure and an EJC-SR Protein Nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A, Hon GC, Jin F, Henry RE, Espinosa JnM, Ren B. CBX3 regulates efficient RNA processing genome-wide. Genome research. 2012;22:1426–1436. doi: 10.1101/gr.124818.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. Biochimica et Biophysica Acta. 2012 doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased Chromatin Signatures around Polyadenylation Sites and Exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Séraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA (New York, NY) 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F, Ladd AN. Conserved developmental alternative splicing of muscleblind-like (MBNL) transcripts regulates MBNL localization and activity. RNA Biol. 2010;7:43–55. doi: 10.4161/rna.7.1.10401. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucl Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Research. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstorukov MY, Goldman Ja, Gilbert C, Ogryzko V, Kingston RE, Park PJ. Histone variant H2A.Bbd is associated with active transcription and mRNA processing in human cells. Mol Cell. 2012;47:596–607. doi: 10.1016/j.molcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya Pa, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Waldholm J, Wang Z, Brodin D, Tyagi A, Yu S, Theopold U, Farrants AKÖ, Visa N. SWI/SNF regulates the alternative processing of a specific subset of pre-mRNAs in Drosophila melanogaster. BMC molecular biology. 2011;12 doi: 10.1186/1471-2199-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NAL, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatheritt RJ, Gibson TJ. Linear motifs: lost in (pre)translation. Trends in biochemical sciences. 2012;37:333–341. doi: 10.1016/j.tibs.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Aligianni S, Codlin S, Watt S, Bähler J. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome biology. 2011;12:R82. doi: 10.1186/gb-2011-12-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends in Genetics: TIG. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long Noncoding RNAs with snoRNA Ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Zeng C, Berget SM. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Molecular and cellular biology. 2000;20:8290–8301. doi: 10.1128/mcb.20.21.8290-8301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative Modeling Defines the Nova Splicing-Regulatory Network and Its Combinatorial Controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci USA. 2011;108:E627–635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- Zraly CB, Dingwall AK. The chromatin remodeling and mRNA splicing functions of the Brahma (SWI/SNF) complex are mediated by the SNR1/SNF5 regulatory subunit. Nucleic acids research. 2012;40:5975–5987. doi: 10.1093/nar/gks288. [DOI] [PMC free article] [PubMed] [Google Scholar]