Abstract
Southwestern (DNA-protein) screening of a murine L-cell cDNA library by using a probe for the microE3 site in the immunoglobulin heavy-chain enhancer yielded a clone, mTFE3, which is a member of the subset of basic helix-loop-helix (BHLH) proteins that also contain a leucine zipper (ZIP). Since the individual contribution of these domains is not well understood for proteins which contain them both, mutational analyses were performed to assess the functional roles of the HLH and ZIP regions for DNA binding and multimerization. The HLH region is stringently required for DNA binding but not for multimerization. The ZIP region is not stringently required for binding or multimerization, but stabilizes both multimer formation and DNA binding. A high degree of conservation at both the amino acid and nucleotide levels between the human transcription factor TFE3 and mTFE3 suggests that mTFE3 is the murine homolog of human TFE3. By using fluorescent in situ hybridization, mTFE3 was mapped to mouse chromosome X in band A2, which is just below the centromere. We show that in addition to the immunoglobulin heavy-chain microE3 site, mTFE3 binds to transcriptional elements important for lymphoid-specific, muscle-specific, and ubiquitously expressed genes. Binding of mTFE3 to DNA induces DNA bending.
Full text
PDF

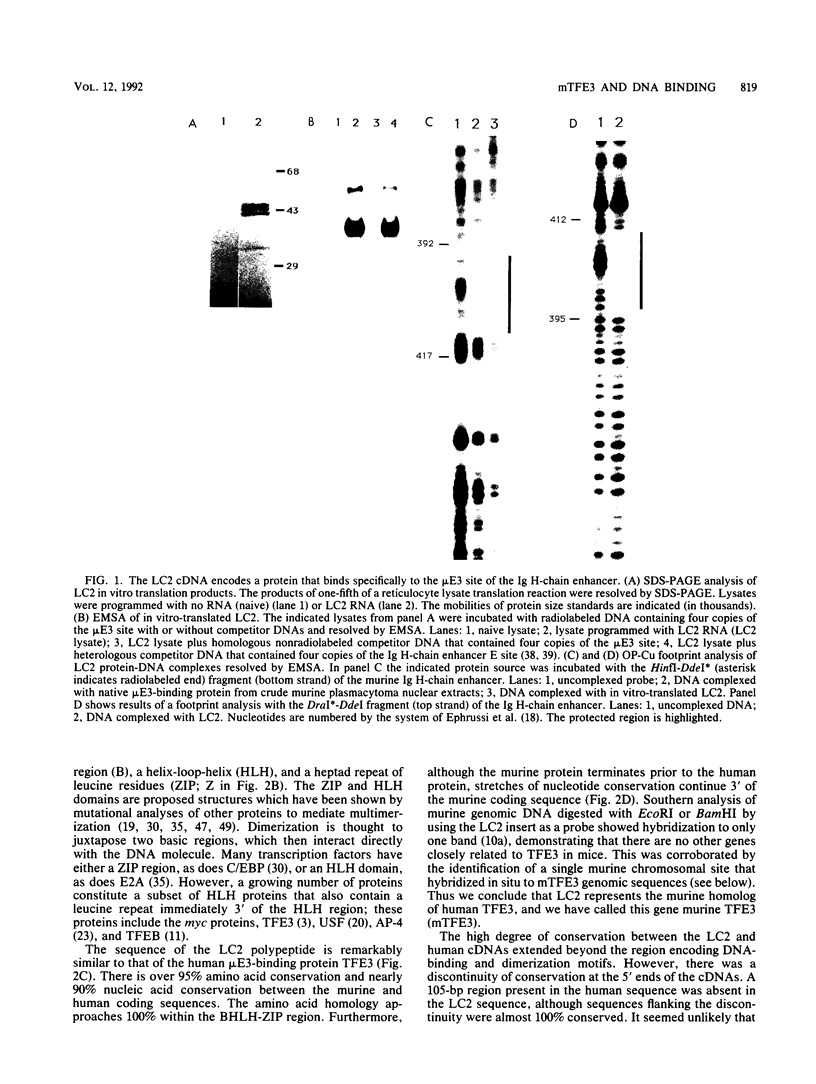


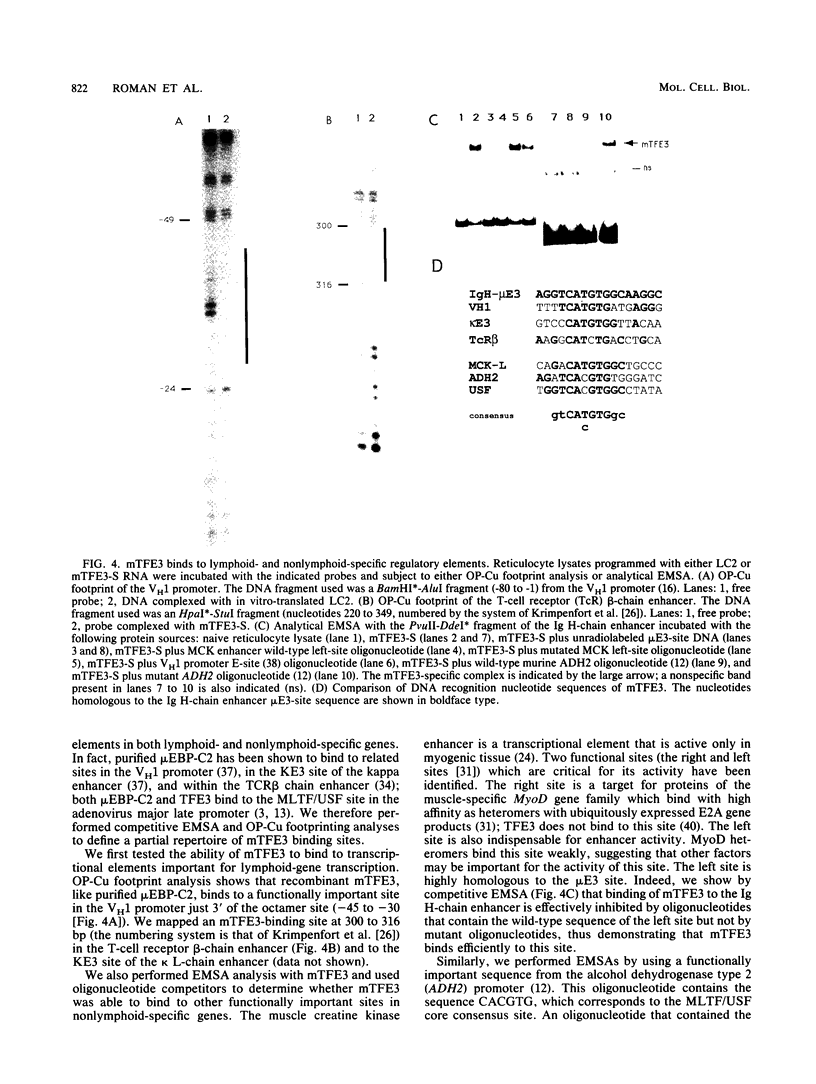





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckmann H., Kadesch T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991 Jun;5(6):1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Bickmore W. A., Sumner A. T. Mammalian chromosome banding--an expression of genome organization. Trends Genet. 1989 May;5(5):144–148. doi: 10.1016/0168-9525(89)90055-3. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Boyle A. L., Ballard S. G., Ward D. C. Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7757–7761. doi: 10.1073/pnas.87.19.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. L., Feltquite D. M., Dracopoli N. C., Housman D. E., Ward D. C. Rapid physical mapping of cloned DNA on banded mouse chromosomes by fluorescence in situ hybridization. Genomics. 1992 Jan;12(1):106–115. doi: 10.1016/0888-7543(92)90412-l. [DOI] [PubMed] [Google Scholar]
- Brigati D. J., Myerson D., Leary J. J., Spalholz B., Travis S. Z., Fong C. K., Hsiung G. D., Ward D. C. Detection of viral genomes in cultured cells and paraffin-embedded tissue sections using biotin-labeled hybridization probes. Virology. 1983 Apr 15;126(1):32–50. doi: 10.1016/0042-6822(83)90460-9. [DOI] [PubMed] [Google Scholar]
- Calame K. L. Immunoglobulin gene transcription: molecular mechanisms. Trends Genet. 1989 Dec;5(12):395–399. doi: 10.1016/0168-9525(89)90197-2. [DOI] [PubMed] [Google Scholar]
- Carr C. S., Sharp P. A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990 Aug;10(8):4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr L. G., Edenberg H. J. cis-acting sequences involved in protein binding and in vitro transcription of the human alcohol dehydrogenase gene ADH2. J Biol Chem. 1990 Jan 25;265(3):1658–1664. [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Disteche C. M., Tantravahi U., Gandy S., Eisenhard M., Adler D., Kunkel L. M. Isolation and characterization of two repetitive DNA fragments located near the centromere of the mouse X chromosome. Cytogenet Cell Genet. 1985;39(4):262–268. doi: 10.1159/000132155. [DOI] [PubMed] [Google Scholar]
- Eaton S., Calame K. Multiple DNA sequence elements are necessary for the function of an immunoglobulin heavy chain promoter. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7634–7638. doi: 10.1073/pnas.84.21.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman A. M., Blumenthal D. K., Krebs E. G. Protein serine/threonine kinases. Annu Rev Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Gentz R., Rauscher F. J., 3rd, Abate C., Curran T. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science. 1989 Mar 31;243(4899):1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- Gregor P. D., Sawadogo M., Roeder R. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990 Oct;4(10):1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- Henthorn P. S., Stewart C. C., Kadesch T., Puck J. M. The gene encoding human TFE3, a transcription factor that binds the immunoglobulin heavy-chain enhancer, maps to Xp11.22. Genomics. 1991 Oct;11(2):374–378. doi: 10.1016/0888-7543(91)90145-5. [DOI] [PubMed] [Google Scholar]
- Holmquist G. P. Evolution of chromosome bands: molecular ecology of noncoding DNA. J Mol Evol. 1989 Jun;28(6):469–486. doi: 10.1007/BF02602928. [DOI] [PubMed] [Google Scholar]
- Hu Y. F., Lüscher B., Admon A., Mermod N., Tjian R. Transcription factor AP-4 contains multiple dimerization domains that regulate dimer specificity. Genes Dev. 1990 Oct;4(10):1741–1752. doi: 10.1101/gad.4.10.1741. [DOI] [PubMed] [Google Scholar]
- Johnson J. E., Wold B. J., Hauschka S. D. Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol Cell Biol. 1989 Aug;9(8):3393–3399. doi: 10.1128/mcb.9.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., Su L. K., Kadesch T. Identification and characterization of two functional domains within the murine heavy-chain enhancer. Mol Cell Biol. 1988 Jan;8(1):145–152. doi: 10.1128/mcb.8.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P., de Jong R., Uematsu Y., Dembic Z., Ryser S., von Boehmer H., Steinmetz M., Berns A. Transcription of T cell receptor beta-chain genes is controlled by a downstream regulatory element. EMBO J. 1988 Mar;7(3):745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Lichter P., Tang C. J., Call K., Hermanson G., Evans G. A., Housman D., Ward D. C. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990 Jan 5;247(4938):64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- McDougall S., Peterson C. L., Calame K. A transcriptional enhancer 3' of C beta 2 in the T cell receptor beta locus. Science. 1988 Jul 8;241(4862):205–208. doi: 10.1126/science.2968651. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Ono S. Ancient linkage groups and frozen accidents. Nature. 1973 Aug 3;244(5414):259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Calame K. Proteins binding to site C2 (muE3) in the immunoglobulin heavy-chain enhancer exist in multiple oligomeric forms. Mol Cell Biol. 1989 Feb;9(2):776–786. doi: 10.1128/mcb.9.2.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Eaton S., Calame K. Purified mu EBP-E binds to immunoglobulin enhancers and promoters. Mol Cell Biol. 1988 Nov;8(11):4972–4980. doi: 10.1128/mcb.8.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Orth K., Calame K. L. Binding in vitro of multiple cellular proteins to immunoglobulin heavy-chain enhancer DNA. Mol Cell Biol. 1986 Dec;6(12):4168–4178. doi: 10.1128/mcb.6.12.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Roman C., Cohn L., Calame K. A dominant negative form of transcription activator mTFE3 created by differential splicing. Science. 1991 Oct 4;254(5028):94–97. doi: 10.1126/science.1840705. [DOI] [PubMed] [Google Scholar]
- Roman C., Platero J. S., Shuman J., Calame K. Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes Dev. 1990 Aug;4(8):1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Why bend DNA? Cell. 1990 Jan 26;60(2):177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- Turner R., Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989 Mar 31;243(4899):1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., LaMarco K. L., Johnson P. F., Landschulz W. H., McKnight S. L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988 Jul;2(7):801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- Voronova A., Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienberg J., Jauch A., Stanyon R., Cremer T. Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization. Genomics. 1990 Oct;8(2):347–350. doi: 10.1016/0888-7543(90)90292-3. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]









