Abstract
Background
Genome-wide association studies (GWAS) have had limited success when applied to complex diseases. Analyzing SNPs individually requires several large studies to integrate the often divergent results. In the presence of epistasis, multivariate approaches based on the linear model (including stepwise logistic regression) often have low sensitivity and generate an abundance of artifacts.
Methods
Recent advances in distributed and parallel processing spurred methodological advances in nonparametric statistics. U-statistics for structured multivariate data (μStat) are not confounded by unrealistic assumptions (e.g., linearity, independence).
Results
By incorporating knowledge about relationships between SNPs, μGWAS (GWAS based on μStat) can identify clusters of genes around biologically relevant pathways and pinpoint functionally relevant regions within these genes.
Conclusion
With this computational biostatistics approach increasing power and guarding against artifacts, personalized medicine and comparative effectiveness will advance while subgroup analyses of Phase III trials can now suggest risk factors for ad verse events and novel directions for drug development.
Keywords: common disease, epilepsy, epistasis, genome-wide association study, GWAS, Phase III trials, Ras pathway, U-statistics
Almost a decade after the completion of the Human Genome Project [1], the scientific and medical advances hoped for from genome-wide association studies (GWAS) have not yet been realized. After early successes with diseases where a single haplotype confers all or most risk [2], the same statistical approaches have often produced ambiguous results when applied to complex diseases [3,4]. Increasing the sample size (to tens of thousands of subjects as suggested [5]) is impractical for rare disease forms, and also greatly increases the duration and cost of data collection. Improving accrual by broadening the inclusion criteria increases variance and thus requires yet larger samples; a vicious cycle. Moreover, increasing sample size in a nonrandomized study may, somewhat paradoxically, increase the risk of false positives [6,7].
Several mutations within a gene may contribute to the risk of common diseases and several SNPs may have become associated with the same mutation over time. One risk factor’s contribution may depend on the presence of others and sets of mutations may confer more risk if they affect both chromosomes (compound heterozygosity). Hence, any statistical approach based on p-values derived one SNP at a time (ssGWAS) is ill-suited to identify the short-range epistasis involved [8] (following Fisher [9], the term ‘epistasis’ will be used for any deviation from independence, be it between neighboring SNPs, intragenic regions or genes). Analyzing diplotypes (sets of neighboring SNPs with unknown phase) comprehensively would be preferable [10], yet traditional multivariate methods [11] including linear/logistic regression (lr) assume independence and additivity/multiplicativity of risk factors to yield computationally simple algorithms. Making unrealistic assumptions, such as linearity, may easily lead to meaningful nonlinear relationships being overlooked (false negatives). More importantly, random errors, not subject to biological constraints, may occasionally fulfill these assumptions, so that the most ‘significant’ results are often false positives.
Association studies, in general, are exploratory ‘selection procedures’ [12] to generate rather than confirm hypotheses. Even though the same algorithms are used as in confirmatory tests, ‘p-values’ merely serve to sort candidates, so that a sufficiently large selection of candidate genes will include the most interesting genes with high power. Even minor differences in the composition of the study population can result in different subsets of genes being selected [13], and each could help with understanding a different aspect of the disease etiology when confirmed using mouse studies or clinical trials. Hence, the challenge in improving GWAS is to reduce artifacts caused by applying oversimplifying approaches to complex diseases (analyzing one SNP at a time, assuming independence and additivity of effects) while incorporating more knowledge to increase the sensitivity for detecting biologically relevant subsets of the genes involved.
With the advent of mainframe computers, more complex calculations (e.g., factor analysis) became feasible. More recently, personal computers triggered the development of resampling methods. Now we are, again, entering an era of advances in computational biostatistics, where massively parallel computing has spurred the methodological advances making wide-locus GWAS based on a nonparametric approach (μGWAS, based on u-statistics for structured multivariate data) feasible [14]. Below, we introduce two novel concepts.
First, several ‘tag’ sets of ‘genetically indistinguishable’ SNPs [15] are typically scattered across a linkage disequilibrium (LD) block, yet traditional methods cannot differentiate between ‘permuted’ diplotypes containing members of the same tag sets in different order. μGWAS draw on the spatial structure of SNPs within a diplotype and expected LD from HapMap [16] to improve the resolution of GWAS to intragenic regions. Second, we apply the concept of ‘information content of multivariate data’ (μIC) [14] at several stages of the ana lysis to guard against artifacts. With these methodological advances, disease-relevant genes and intragenic regions can now be suggested from a single study, often of only a few hundred narrowly defined cases, rather than from a variety of large studies, turning GWAS from a technique to identify isolated SNPs into a powerful tool to generate plausible and testable hypotheses about the etiology of complex diseases.
Methods
μ-scores for diplotypes
It is often reasonable to assume that risk conferred by a heterozygous SNP lies somewhere between baseline risk and a homozygous SNP (having two risk alleles) that is, between the risk of a recessive and dominant allele, respectively. U-statistics (including the Wilcoxon/Mann–Whitney U test [17]) treat SNPs as ordinal (wild-type = xx < xX < XX = homozygous), but do not require the degree of dominance to be known. Treating diplotypes as multivariate data then avoids the need for assumptions about independence and relative importance of the SNPs [18], yet the theory was never broadly developed owing to prohibitively high computational demand [19]. With GWAS, for instance, the number of ‘polarities’ (combinations of −1 = bad; 0 = irrelevant; +1 = good) increases exponentially with diplotype length, yet with massively parallel computing we were now able to include diplotypes up to length six.
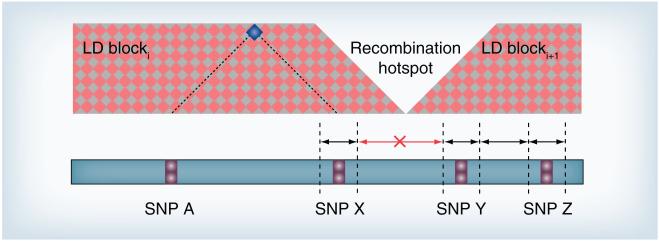
Traditionally, one would have more confidence in a ‘significant’ locus if neighboring loci also show association [20] and add recombination information to the data displayed. Here we integrate the concepts behind this intuitive visual inspection into the statistical approach itself. Recently, U-scores for multivariate data (μ-scores) have been extended to reflect structures among variables with applications including sports [21], policy-making [22] and medicine [14]. The proposed GWAS-specific structure is based on the notion that neighboring disease loci may have similar effects and that a disease locus may be in LD with both adjacent SNPs, unless the SNPs are separated by a recombination hotspot (boundary between LD blocks; Figure 1).
Figure 1. SNP-related chromosomal intervals.
Conceptual structure of chromosomal SNP-related intervals for disease loci in LD with three consecutive SNPs (SNPs X, Y and Z), but not with a more distant SNP (SNP A). SNPs X and Y are part of different LD blocks, separated by a recombination hotspot. Hence, the interval between these two SNPs is excluded. The location indicating LD between SNPs A and X is highlighted in blue. The inter-regional boundaries need not be known.
LD: Linkage disequilibrium.
μGWAS starts with computing matrices representing the partial order of each SNP, combining pairs of these matrices into matrices representing the intervals and, finally, combining SNP and interval matrices into a diplotype matrix from which the μ-scores are computed [14,22]. As diplotype profiles are built from intervals around and between neighboring SNPs, diplotypes where members Xi, Yi and Zi of the tag sets (X), (Y) and (Z) appear in different order (permuted diplotypes), such as (X1, Y1 and Z1) versus (Y2, X2 and Z2) can be distinguished. This novel approach to incorporate knowledge of neighborhood relationships between SNPs increases power over merely combining all SNPs within a diplotype in a single step [14], yet avoids the need for assumptions about dependencies and relative importance required when using linear combinations (weighted sums) of univariate scores. With GWAS based on lr (lrGWAS), one could work towards a similar goal by adding sequential interaction terms. Hence, we will compare μGWAS not only with ssGWAS for dominant, linear trend [23], and recessive effects, but also with stepwise logistic regression with and without sequential interaction terms.
Subjects
Childhood absence epilepsy (CAE) [24], formerly known as ‘petit mal’, is characterized by frequent, short episodes of ‘daydreaming’. Through trial and error of different combinations of valproic acid and various ion channel blockers, these absences can be controlled in approximately 75% of affected children [25]. For adult patients, etiracetam, an IL-1β inhibitor [26] was approved in November 1999, and a caspase 1 inhibitor (VRT-765) is under going a controlled Phase IIb study [101]. CAE does not follow a simple Mendelian pattern of inheritance, although recurrence of epilepsy in families is high. A high concurrence in monozygotic twins and the absence of known exogenic factors make CAE an ideal model for studying the genetics of complex diseases and approaches to unravel their genetic risk factors to better match patients to existing drugs and identify new drug targets for patients who do not respond to existing drugs.
The 185 CAE patients in this study were predominantly Caucasian (83%) and white Hispanic (10%) with the well-known female preponderance (115 female vs 70 male patients). Average age of onset for absence seizures was 5.7 years. Patients were required to be seizure free on antiepileptic medication. Controls were selected from a publicly available database [102]; see Supplementary Material at www.futuremedicine.com/doi/suppl/10.2217/pgs.13.28 for details.
Results
Identifying genes
As is typical for ssGWAS, especially with small samples, only two SNPs reached the customary s = −log10(p) > 7.5 level of significance with univariate tests (Figure 2, black foreground), one in a noncoding region (chromosome 1, lr only), the other in the pseudogene EE1A1P12 (chromosome 2).
Figure 2. Comparison of 185 childhood absence epilepsy cases versus matched controls.
Unadjusted −log10(p) by chromosomal location; top: μ-statistics for structured multivariate data (μGWAS); bottom: linear/logistic regression (lr; without interaction terms). Univariate results, shown in black, are consistently similar across the approaches, as expected. For μGWAS, dots vary in size by diplotype length and are color coded, with red indicating results with low μ-scores for reliability (high significance, low μIC). Lr results are overlayed with the Cochran–Armitage (squares) and Mantel–Haenszel (×/+) test results. Genes known to be directly related to the NOD/intellectual disability–axonal guidance signaling/ataxin pathway are shown in bold. Genes indicated in the center header row (pink) of each chromosome have support in both μGWAS and lrGWAS; genes ranking higher in μGWAS or linear/logistic regression GWAS appear in the first row (blue) or third row (red), respectively. Darker colors indicate more significant results. Other implicated genes are shown against the dark background of univariate results.
?: No gene in the region implicated; …: Several genes within the same linkage disequilibrium block.
Since ssGWAS was inconclusive and sequential interaction terms created an abundance of likely false positives with lrGWAS (Supplementary Figure 1), even with regularization (AIC [27]), the following discussion focuses on μGWAS versus traditional lrGWAS. In the spirit of conducting a selection procedure [12,28], rather than confirmatory tests, p-values were used solely for the purpose of ranking the loci and at any given level, lrGWAS had more ‘significant’ results in general, including many likely false positives. Hence, methods were compared using similar arbitrary numbers of top regions (first comparison used only the top 6, second comparison used ~20 and third comparison used ~40; see Supplementary Table 1), the latter cutoffs adjusted for display purposes (Figure 2) to match commonly used s-values (μ: 7.5/7.0; lr: 8.0/7.5).
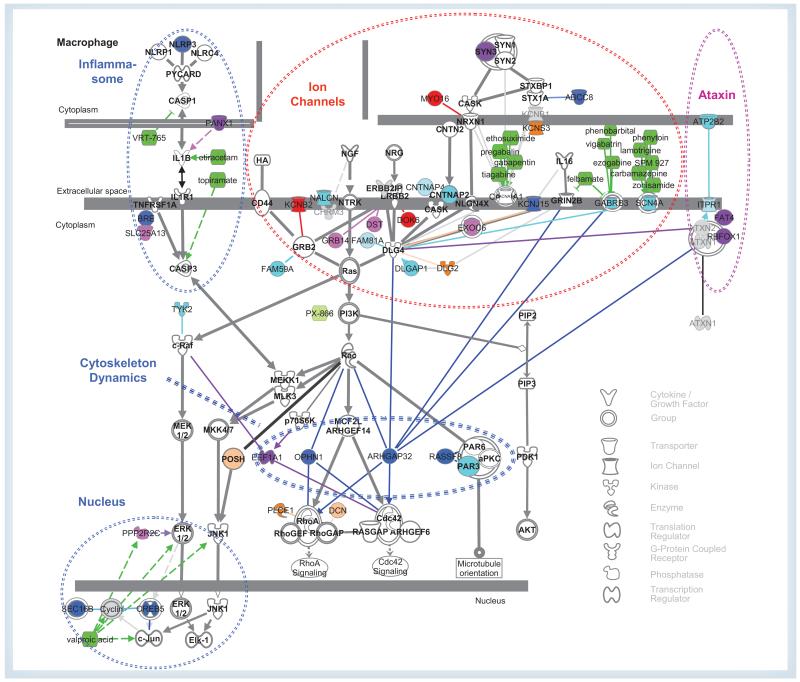
Only one of the top six genes in lrGWAS (RBFOX1) ranks higher than rμ = 73rd in μGWAS (5th), while the other four among the top six regions in μGWAS are also among the top 22 in lrGWAS (the above elongtion factor pseudo gene EEF1A1P12; synapsin III, SYN3; FAT4; and CREB5; Supplementary Table 1). Of the top 17 μGWAS regions (s > 7.5), 14 (82%) are known to be in genes directly related to the NOD/ID–axonal guidance signaling/ataxin pathway (Figure 3), including PANX1, SEC16B, the Rho GTPase activating proteins OPHN1/ARHGAP41 and RICS/ARHGAP32, ABCC8, the potassium channel KCNJ5, BRE, NLRP3 and RASSF8, compared with only eight genes (36%) of the top 22 (s > 8), including KCNB2, DOK6 and MYO16, or 16 (40%) of the top 40 lrGWAS (s > 7.5) regions.
Figure 3. Published direct relationships between the minimal subset of the NOD/intellectual disability–axonal guidance signaling/ataxin pathway directly related to significant genes by μGWAS (23 of 40, s > 6.5) and lrGWAS (17 of 40, s > 7.0), respectively.
The members of the pathway are shown and labeled in bold. Methods are indicated in colors (blue: μGWAS; red: lrGWAS; pink: both). The most significant genes (μGWAS: >7.5, lrGWAS: >8.0) are shown in darker shades. (See for details). Supplementary Table 1 Dotted circles relate to functional clusters mentioned in the text. Drugs are indicated in green.
μGWAS: μ-statistics for structured multivariate data; lr: Linear/logistic regression.
Channelopathies
Epilepsy is commonly seen as a channelopathy, and lrGWAS identify both postsynaptic (KCNB2, rlr = 3rd; DOK6, rlr = 10th) and presynaptic (MYO16, 13th) membrane processes. μGWAS adds KCNJ15 (rμ = 14th), confirms CNTNAP2 [29] and CNTNAP4 (27th, and 48th, respectively), and hints at two targets for approved antiepileptic drugs, the ion channels SCN4A and GABRB3 (43rd and 57th, respectively) [30]. Both methods implicate SYN3 (rμ/rlr = 3rd/22nd), a presynaptic vesicle-associated protein [31]. μGWAS adds OPHN1 and ABCC8 (8th and 12th, respectively).
Inflammasome
Two approved antiepileptic drugs, topiramate and levetiracetam, and the investigational drug VRT-765 target the NOD-like receptor signaling pathway [32]. While both approaches suggest genetic variations in PANX1 (rμ/rlr = 13th/16th), μGWAS adds the TNFRSF1A modulator BRE (15th) as involved and NLRP3 as a risk factor (16th). Hence, VRT-765 might be particularly effective for patients with a ‘gain-of-function’ mutation in NLRP3.
Cytoskeleton dynamics
RHOA was upregulated in patients with intractable epilepsy [33], yet the mechanism involved is unknown. Two genes known to regulate RHOA, OPHN1 (also known as ARHGAP41) and ARHGAP32 are among the top ten genes with μGWAS, but rank only 99th and 58th, respectively, in lrGWAS. The risk of epilepsy is increased in children with intellectual disability (ID), where ARHGAP32 has been implicated. Binding between ARHGAP32 and ATXN1 has been implicated in inherited ataxias [34]. OPHN1 is known to affect X-linked ID [35] and thus might explain the preponderance of CAE among girls. μGWAS adds a pair of binding partners downstream of RAC1 to the picture, RASSF8 (17th) and PARD3 (26th). Finally, the ‘pseudogene’ EEF1A1P12, being among the top ten regions in both approaches, hints at an involvement of EEF1A1, which regulates CDC42. Hence, μGWAS uniquely provides a testable hypothesis about the mechanism by which RHOA is upregulated in some forms of epilepsy.
Ataxin
Ataxias and epilepsy share genetic risk factors [36,37], including OPHN1 [38,39], and both methods implicate two genes binding ataxins, RBFOX1 (rμ = rlr = 5th) and FAT4 (rμ/rlr = 6th/17th). μGWAS also hints at the calcium transporter ATPB2 (39th) and the calcium channel ITPR1 (42nd) as potential drug targets.
Nucleosome
The effectiveness of valproic acid in treating epilepsies hints at a role of nucleosome assembly in epilepsies and, in fact, μGWAS implicates mutations in CREB5 and SEC16B (4th and 10th, respectively).
Detecting epistasis & selection
Among the genes involved in cytoskeleton dynamics, ARHGAP32, with known direct interactions with many of the key players, ranked 11th in μGWAS, but only 58th in lrGWAS. Moreover, it had two separate ‘peaks’ in μGWAS, one in the promoter region.
Epistasis between neighboring SNPs
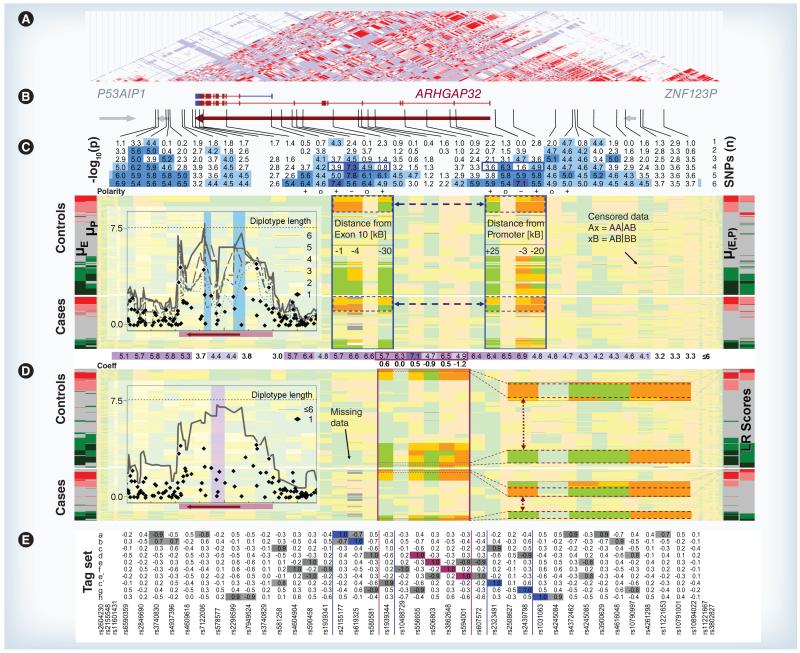
The most significant SNPs in ARHGAP32 by ssGWAS (s = 4.3–4.7) are all members of tag set a (Figure 4e). The two μGWAS peaks, separated by a clear trough (Figure 4C), pinpoint two loci where the effects of different haplotypes converge, centered within 4 kb of exon 10 and the promoter region (exon 0), respectively. Both regions contain a set c SNP as a distant member (≈20 kb), indicating a common ‘background’ risk factor, and two members of region specific tag sets (exon 10: a/b, exon 0: g/h). Both regions belong to a recently identified alternative splice variant, which is expressed during neural development and involved in axon and dendrite extension [40,41]. lrGWAS results are also elevated, yet without discriminating intragenic regions (Figure 4D, insert).
Figure 4. Microarray genotyping results for the linkage disequilibrium block containing ARHGAP32.
(A) Linkage disequilibrium (LD) map; (B) coding regions; (C) μ-statistics for structured multivariate data (μGWAS) test results by diplotype length followed by the polarities of the SNPs contributing and the SNP pattern (orange: homozygous; yellow: heterozygous; green: wild-type) for controls and cases sorted by μ(E, P) = μ((E1, E2, E4), (P1, P3, P4)) (near right stub). Diplotypes ranked high (red) and low (green) by μ-scores for each region (left stub: μE = μ(E1, E2, E4), μP = μ(P1, P3, P4)) are highlighted as more saturated. Horizontal arrows indicate consistently paired diplotypes. The insert shows the ‘Manhattan plot’ of the −log10(p) values. (D) lrGWAS results followed by the lr coefficients (coeff.) of the SNPs involved. SNP pattern are sorted by lr scores (far right stub). The enlarged profiles with extreme lr scores differ in one the five SNP only (vertical arrow). The insert shows the values based on univariate and stepwise lr. (E) LD between each of the ten SNPs included in the two μGWAS regions (blue) and the lrGWAS region (purple) and the members of the same tag set (gray). Tag set c is represented in both μGWAS diplotypes and the lrGWAS diplotype, which contains two members of tag set e.
Coeff: Coefficient; E: Exon; (E,P): Exon 10 and promoter region; lr: Linear/logistic regression; P: Promoter.
Intragenic epistasis/selection
No case or control subject had more than four risk alleles among the three relevant SNPs in either region, although homozygous variants for each SNP are present. Hence, the unobserved combinations must have been selected against, for example, because of a more severe phenotype.
Intergenic epistasis
As approximately one-third of all subjects with the ARHGAP32 genetic risk factor lack the phenotype, other genetic cofactors are yet to be identified. Figure 3 suggests the possibility of epistasis in trans between regulatory and functional factors; that is, between the plasma membrane (NGF/NRG–RAS) and the cytoplasm (RAC–RHOA/CDC42).
Validation
In this analysis, we have reduced the potential for false-positive results by taking advantage of the novel internal validation features made possible with μGWAS. During data preparation, we used a data quality μ-score based on a comprehensive assessment of missing data, Hardy–Weinberg equilibrium, short-range LD, and expected LD from HapMap information. During ana lysis, we have drawn on polarity conflict and lower than expected μIC (Supplementary Figure 2). Finally, we utilized μIC to indicate highly significant results with low μIC. Notably, none of the pathway related genes flagged as potentially unreliable are related to the genes downstream of RAC1 (Figure 2).
Larger genes are both more likely to carry mutations and to have false positives. Still, although several of the genes identified are among the largest 5% (>200 kb) in the human genome, only two of the top 11 unique genes in μGWAS (CREB5 and BRE) and three of the top 13 unique genes in lrGWAS (DYSF, DOK6 and TMCO7) are ‘direct hits’ within the coding region (Supplementary Table 1). ARHGAP32 and OPHN1 were implicated by ‘hits’ in the stop or promoter regions, respectively, and thus are not at an increased risk for being false positives owing to their size.
The results on ARHGAP32 (Figure 4) are supported by further evidence. First, each of the six SNPs included in the two diplotypes is in high LD with several other SNPs (Figure 4e), for which the probe sequences differ and thus are not subject to the same calling errors. Second, only the two pairs of diplotypes having the highest association with disease risk by μ-scores were in high LD between the intragenic regions (Figure 4C, horizontal dashed arrows), while lower risk diplotypes were unrelated. Not only is it highly unlikely for each of these results to occur by chance alone, it is virtually impossible that they could occur together, and in both independent populations. While this cannot rule out a false-positive result due to association with factors beyond the etiology of epilepsy, these findings validate the ability of μGWAS to detect intragenic regions of biologically relevant epistatic patterns. Finally, the diplotype with the highest overall (exon 10 and promoter region [E,P]) score μ(E,P) is clearly over-represented among cases, with a prevalence of 14.1% (26 out of 185) and 6.5% (23 out of 354) in cases and controls, respectively, compared with 3.8% (7 out of 185) and 6.2% (22 out of 354) for the diplotypes with the lowest μ-scores, confirming that μ-scores are, in fact, reflecting disease risk.
As one would expect, μ and lr scores (Figure 4, right border) are correlated. The subjects with the pair of diplotypes having the highest μ(E,P)-scores (Figure 4D) also share a diplotype with a high lr-score (Figure 4e), but the subjects scoring even higher in lr-scores comprise four different diplotypes. Interestingly, the largest of these groups differs only in the first SNP from a diplotype with low lr- and μ-scores (vertical arrows in Figure 4D), consistent with the sensitivity of linear model results to outliers. As the partial ordering underlying μ-scores, which directly reflects an underlying functional model, results in more genetic uniformity among subjects with extreme scores, these more homogeneous subpopulations could then be selected for identification of functional variations through sequencing.
Conclusion
With GWAS of complex diseases, only a few solitary SNPs typically stand out from the noise, especially in small studies, and this study is no exception. Different compositions of rare disease variants across studies almost inevitably result in different SNPs being ‘significant’, so that validation in independent ssGWAS requires many large studies until a testable hypothesis emerges. μGWAS, by contrast, related approved and experimental drugs to functional clusters of genes along a known pathway in a study of 185 well-characterized cases only.
ssGWAS can efficiently screen for loci, where a single haplotype confers all or most of the risk (EEF1A1P12). lrGWAS has advantages when the effects of SNPs are at least approximately independent and additive (as they might be in some transporters and ion channels). With more complex processes, however, like the interactions of ARHGAP32 with its various binding and activation partners, not constraining results by making overly simplistic assumptions leads to biologically relevant hypotheses about functionally related genes clustered around biologically relevant pathways.
Pathway-based approaches [42] and gene set enrichment analyses [43] combine results of univariate statistics using assumptions regarding the relative importance of genes and prior declarations of relatedness among genes instead of observed interactions. How ever, this ana lysis suggests that few, if any, pathway genes themselves may carry mutations in common diseases, unless they are members of redundant complexes (NLRP3, SYN3 and PARD3; Figure 3), in which case multiple genes may need to be knocked out to produce a phenotype [44].
Wide-locus GWAS aim at accounting for compound heterozygosity, different haplotypes carrying the same mutation and epistasis between nearby disease loci. Hence, functional regions can be identified more easily, even when the contribution of individual SNPs would be difficult – if not impossible – to detect. Many traditional statistical methods, however, have deficiencies for relevant types of epistasis. ARHGAP32, which ranked 10th among μGWAS genes and was validated through the distinct epistatic pattern among the highest-risk allelotypes confirmed in sequencing (Figure 4), did not even appear among the top 50 lrGWAS regions.
μGWAS requires neither Hardy–Weinberg equilibrium nor independence or additivity/multiplicativity of genetic effects, thereby improving sensitivity for nonlinear effects (including evolutionary selection; Figure 4, horizontal dashed arrows and Supplementary Table 1). Adding sequential interactions and recombination hotspots improves resolution, rather than creating artifacts. Together with OPHN1 (also unique to μGWAS at rank 8), this study provides a plausible hypothesis why expression of RHOA is upregulated in some forms of epilepsy [33].
Increased expression of RHOA was recently associated with some epilepsies [33]. Both OPHN1 and ARHGAP32 interact with both RHOA and PI3K (Figure 3), a drug target currently being investigated in cancer [45] and inflammatory diseases [46]. Wortmannin, an inhibitor of PI3K, attenuates the effects of seizures in rats [47] and PX-866 (a oral drug derivative of wortmannin in a Phase II prostate cancer trial [103]), targets PI3K. If our results are confirmed and hold for patients with other epilepsies as well, this might lead to novel therapeutic approaches to treat patients whose seizures do not respond to drugs targeting ion channels, the inflammasome or the nucleosome. As this study included only patients whose seizures were controlled by valproic acid and/or ion channel blockers, these genes may play an even larger role in other populations.
A particular advantage of μGWAS is the ability to guide the interpretation of data patterns in terms of biological function. Sorting diplotypes by the overall risk they confer (Figure 4C), rather than by linear weight scores lacking direct biological interpretation (Figure 4D) provided compelling evidence for intragenic epistasis (Figure 4C), facilitated validation (Figure 4e), and generated testable hypotheses regarding the function of underlying mutations. By utilizing the order of neighboring SNPs and HapMap information about their expected LD, μGWAS can often identify functional intragenetic regions, whereas the resolution of lrGWAS, irrespective of sample size, is typically limited to an LD block as a whole. For instance, this ana lysis suggests that the combinations of diplotypes with the highest μ-score, in either of the ARHGAP32 regions, have been selected for because they partially compensate for each other. Epistasis might also explain why knocking out the entire ARHGAP32 gene produced no obvious phenotype in mice [48].
In summary, our results show that genetic risk factors for complex diseases cannot be adequately addressed with ssGWAS alone and that the computationally simple lrGWAS approach may be insensitive to complex forms of epistasis. Reducing artifacts by avoiding models motivated by computational convenience, rather than biological plausibility, reduces the need for independent studies to guard against false-positive results from model misspecifications. For comparative effectiveness research and personalized diagnostics to live up to their expectations, cases and controls need to be closely matched to the population or patient involved. Adequately controlling for genetic and environmental confounders when selecting appropriate cases and controls is essential to tease out predictive factors. This goal is much easier to achieve with only a few hundred subjects, rather than several thousands to be matched. Finally, subset analyses of Phase III trials and published epidemiological studies could rapidly reveal novel insights for drug development.
Future perspective
The Ras pathway is known to be involved in both cancers and many developmental disorders [49], so the findings here suggest that identifying genetic risk factors modulating this pathway may help in better using information from sequencing patients when targeting pharmacological interventions not only in cancers, but also in other neurodevelopmental diseases other than CAE, including ID and autism spectrum disorders [31].
With more appropriate statistical methods and more powerful computational tools becoming available, the focus in screening for genetic risk factors of complex diseases can now shift from individual SNPs scattered across the genome to clusters of genes around biologically meaningful pathways. With further advances in computational resources, μGWAS can be extended from epistasis across recombination hotspots (Figure 1) to epistasis between intragenic regions (Figure 4), and between genes (Figure 3).
As μGWAS can provide therapeutically relevant information from substantially smaller sample sizes, decisions in personalized medicine and comparative effectiveness research can be based on samples fine-tuned to the particular patient or population, respectively.
As a few hundred subjects experiencing adverse events or lack of a treatment effect and matched controls from the same population suffice to determine genetic risk factors, data from previous or upcoming Phase III trials can now be effectively mined to determine subpopulations at risk of adverse events and identify directions for development of drugs with a broader target population.
Supplementary Material
Executive summary.
Background
-
■
The requirement for (tens of) thousands of subjects with univariate statistical approaches limits the usefulness of genome-wide association studies (GWAS) for comparative effectiveness research, personalized diagnostics/treatment and subgroup analyses of Phase III trials.
-
■
Several mutations within an intragenic or promoter region may contribute to the risk of common diseases.
-
■
GWAS using multivariate statistical approaches based on unrealistic assumptions (e.g., independence and additivity) implicit to linear/logistic regression (lrGWAS) has low power to detect meaningful relationships and carries a risk of false positives.
-
■
The advent of massively parallel computing has spurred the development of statistical methods that require fewer unrealistic assumptions, including GWAS based on U-statistics for structured multivariate data (μGWAS).
Methods
-
■
Extending μ-statistics to reflect linkage disequilibrium (LD) structures in the data increases the power and avoids artifacts.
-
■
A well-characterized sample of 185 children with childhood absence epilepsy was analyzed as an example.
Results
-
■
With single-SNP GWAS, only two SNPs reached the customary level of significance.
-
■
Of the top 17 regions in μGWAS, 14 (82%) were in genes related to a known disease-related signaling pathway, compared to only eight (36%) of the top 22 regions in linear/logistic regression GWAS.
-
■
μGWAS was able to detect intragenic regions (i.e., exon and promoter) and LD structures, suggesting evolutionary selection.
Conclusion
-
■
Avoiding overly simplistic assumptions leads to biologically relevant hypotheses about functionally related genes clustered around biologically relevant pathways.
-
■
The pathway identified by μGWAS contains targets of approved antiepileptic drugs and a gene being investigated as a cancer drug target.
-
■
Reducing artifacts by avoiding biologically implausible assumptions guards against false-positive results from model misspecifications.
-
■
By reducing the GWAS sample sizes to a few hundred subjects only, μGWAS enables personalized medicine, comparative effectiveness research, and subset analyses of epidemiological studies/Phase III trials.
Acknowledgements
The authors would like to acknowledge S Wrigley, D Politis and R Cauley for collecting the samples, K Olden, LE Thorpe, M Dornbaum and F Steen of the City University of New York School of Public Health for providing access to computational resources for the grid operation, D Kaplun, B Zhou and E Schiffmiller for help with data inspection and ana lysis, WH Greer and GS Zhang for implementing GPU cloud instances, E Horn for editorial assistance, and the reviewers and editors for many helpful suggestions.
KM Wittkowski, V Sonakya and T Song are in part funded by grant #2 UL1 RR024143 from the US National Center for Research Resources (NCRR) Clinical and Translational Science Award (CTSA) and #8 UL1 TR000043 from the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS). M Durner and V Sonakya are in part funded by grant #2 R01NS037466 from the US National Institute of Neurological Disorders and Stroke (NINDS). MP Seybold was in part funded by the German National Academic Foundation and the German Academic Exchange Agency.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422(6934):835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. ■ Describes the first successful genome-wide association study (GWAS), which identified a variation in a single SNP, rs1061170, as causing age-related macular degeneration, yet it did not yield a treatment.
- 3.Sullivan P. Don’t give up on GWAS. Mol. Psychiatry. 2012;17(1):2–3. doi: 10.1038/mp.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein C, Lohmann K, Ziegler A. The promise and limitations of genome-wide association studies. JAMA. 2012;308(18):1867–1868. [Google Scholar]
- 5.Cichon S, Craddock N, Daly M, et al. Psychiatric GWAS Consortium Coordinating Committee. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am. J. Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meehl PE. Theoretical risks and tabular asterisks: Sir Karl, Sir Ronald, and the slow progress of soft psychology. J. Consult. Clin. Psychol. 1978;46:806–834. [Google Scholar]
- 7.Waller NG. The fallacy of the null hypothesis in soft psychology. Appl. Prev. Psychol. 2004;11:83–86. [Google Scholar]
- 8.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat. Rev. Genet. 2003;4(9):701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 9.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- 10.Goldstein DB. Common Genetic Variation and Human Traits. N. Engl. J. Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 11.Ballard DH, Cho J, Zhao HY. Comparisons of multi-marker association methods to detect association between a candidate region and disease. Genet. Epidemiol. 2009;34(3):201–212. doi: 10.1002/gepi.20448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bechhofer RE. A single-sample multiple decision procedure for ranking means of normal populations with known variances. Ann. Math. Statist. 1954;25:16–39. ■■ Introduces the concept of ‘selection procedures’ as opposed to confirmatory tests.
- 13.Rosenthal R. Cumulating evidence. In: Keren G, Lewis C, editors. A Handbook For Data Analysis in the Behavioral Sciences: Methodological Issues. Erlbaum; NJ, USA: 1993. pp. 519–559. [Google Scholar]
- 14.Morales JF, Song T, Auerbach AD, Wittkowski KM. Phenotyping genetic diseases using an extension of μ-scores for multivariate data. Stat. Appl. Genet. Mol. 2008;7(1):19. doi: 10.2202/1544-6115.1372. ■■ Introduces the mathematical underpinning of extending u-scores by including knowledge about a hierarchical factor structure, of which the SNP-related chromosomal intervals are a special case.
- 15.Lawrence R, Evans DM, Morris AP, et al. Genetically indistinguishable SNPs and their influence on inferring the location of disease-associated variants. Genome Res. 2005;15(11):1503–1510. doi: 10.1101/gr.4217605. ■■ Discusses the problems tag sets of genetically indistingushable SNPs may cause for identifying intragenic regions.
- 16.A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Statist. 1947;18(1):50–60. [Google Scholar]
- 18.Hoeffding W. A class of statistics with asymptotically normal distribution. Ann. Math. Statist. 1948;19:293–325. ■■ Derives the asymptotic distribution of linear rank tests based on u-scores.
- 19.Li H. U-statistics in genetic association studies. Hum. Genet. 2012;131(9):1395–1401. doi: 10.1007/s00439-012-1178-y. ■ Discusses the potential of U-statistics for GWAS, in general, including their limitations, for example, when ‘irrelevant SNPs’ within a diplotype are not considered.
- 20.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299(11):1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 21.Wittkowski KM, Song T, Anderson K, Daniels JE. U-scores for multivariate data in sports. J. Quant. Anal. Sports. 2008;4(3):7. doi: 10.2202/1559-0410.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diana M, Song T, Wittkowski KM. Studying travel-related individual assessments and desires by combining hierarchically structured ordinal variables. Transportation. 2009;36(2):187–206. doi: 10.1007/s11116-009-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilton JF. The appropriateness of the Wilcoxon test in ordinal data. Stat. Med. 1996;15(6):631–645. doi: 10.1002/(SICI)1097-0258(19960330)15:6<631::AID-SIM206>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Loiseau P, Panayiotopoulos CP, Hirsch E. Childhood absence epilepsy and related syndromes. In: Roger J, Bureau M, Dravet C, Genton P, Tassinari CA, Wolf P, editors. Epilepsy Syndromes in Infancy, Childhood and Adolescence. John Libbey; Montrouge, France: 2002. pp. 285–303. ■ Provides an extensive discussion of childhood absence epilepsy.
- 25.Glauser TA, Cnaan A, Shinnar S, et al. Ethosuximide, valproic acid, and lamotrigine in childhood absence epilepsy. N. Engl. J. Med. 2010;362(9):790–799. doi: 10.1056/NEJMoa0902014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JE, Choi HC, Song HK, et al. Levetiracetam inhibits interleukin-1 beta inflammatory responses in the hippocampus and piriform cortex of epileptic rats. Neurosci. Lett. 2010;471(2):94–99. doi: 10.1016/j.neulet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Akaike H. A new look at statistical-model identification. IEEE Trans. Autom. Control. 1974;AC19(6):716–723. [Google Scholar]
- 28.Lehmann EL. Some model I problems of selection. Annals of Mathematical Statistics. 1961;32:990–1012. [Google Scholar]
- 29.Friedman JI, Vrijenhoek T, Markx S, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol. Psychiatry. 2008;13(3):261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 30.Crunelli V, Leresche N. Childhood absence epilepsy: genes, channels, neurons and networks. Nat. Rev. Neurosci. 2002;3(5):371–382. doi: 10.1038/nrn811. [DOI] [PubMed] [Google Scholar]
- 31.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu. Rev. Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. ■ Provides a comprehensive review of the intellectual disability pathway.
- 32.Oprica M, Eriksson C, Schultzberg M. Inflammatory mechanisms associated with brain damage induced by kainic acid with special reference to the interleukin-1 system. J. Cell Mol. Med. 2003;7(2):127–140. doi: 10.1111/j.1582-4934.2003.tb00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Wang L-Y, Li J-M, et al. Altered expression of the small guanosine triphosphatase RhoA in human temporal lobe epilepsy. J. Mol. Neurosci. 2010;42(1):53–58. doi: 10.1007/s12031-010-9330-4. [DOI] [PubMed] [Google Scholar]
- 34.Lim J, Hao T, Shaw C, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 35.Billuart P, Bienvenu T, Ronce N, et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392(6679):923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 36.Gribaa M, Salih M, Anheim M, et al. A new form of childhood onset, autosomal recessive spinocerebellar ataxia and epilepsy is localized at 16q21-q23. Brain. 2007;130(7):1921–1928. doi: 10.1093/brain/awm078. [DOI] [PubMed] [Google Scholar]
- 37.Imbrici P, Jaffe SL, Eunson LH, et al. Dysfunction of the brain calcium channel Ca(V)2.1 in absence epilepsy and episodic ataxia. Brain. 2004;127:2682–2692. doi: 10.1093/brain/awh301. [DOI] [PubMed] [Google Scholar]
- 38.Tentler D, Gustavsson P, Leisti J, et al. Deletion including the oligophrenin-1 gene associated with enlarged cerebral ventricles, cerebellar hypoplasia, seizures and ataxia. Eur. J. Hum. Genet. 1999;7(5):541–548. doi: 10.1038/sj.ejhg.5200320. [DOI] [PubMed] [Google Scholar]
- 39.Bergmann C, Zerres K, Senderek J, et al. Oligophrenin 1 (OPHN1) gene mutation causes syndromic X-linked mental retardation with epilepsy, rostral ventricular enlargement and cerebellar hypoplasia. Brain. 2003;126(Pt 7):1537–1544. doi: 10.1093/brain/awg173. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi T, Okabe T, Nasu-Nishimura Y, et al. PX-RICS, a novel splicing variant of RICS, is a main isoform expressed during neural development. Genes Cells. 2007;12(8):929–939. doi: 10.1111/j.1365-2443.2007.01101.x. ■ First report on the extended splice variant of ARHGAP32/RICS.
- 41.Nakamura T, Hayashi T, Mimori-Kiyosue Y, et al. The PX-RICS-14-13-3 zeta/theta complex couples N-cadherin-beta-catenin with dynein-dynactin to mediate its export from the endoplasmic reticulum. J. Biol. Chem. 2010;285(21):16145–16154. doi: 10.1074/jbc.M109.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, Zhang H, Kugathasan S, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn disease. Am. J. Hum. Genet. 2009;84(3):399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha V, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ketzef M, Kahn J, Weissberg I, Becker AJ, Friedman A, Gitler D. Compensatory network alterations upon onset of epilepsy in synapsin triple knock-out mice. Neuroscience. 2011;189:108–122. doi: 10.1016/j.neuroscience.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 45.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 2010;28(6):1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris SJ, Foster JG, Ward SG. PI3K isoforms as drug targets in inflammatory diseases: lessons from pharmacological and genetic strategies. Curr. Opin. Investig. Drugs. 2009;10(11):1151–1162. [PubMed] [Google Scholar]
- 47.Xue Y, Xie N, Cao L, Zhao X, Jiang H, Chi Z. Diazoxide preconditioning against seizure-induced oxidative injury is via the PI3K/Akt pathway in epileptic rat. Neurosci. Lett. 2011;495(2):130–134. doi: 10.1016/j.neulet.2011.03.054. [DOI] [PubMed] [Google Scholar]
- 48.Nasu-Nishimura Y, Hayashi T, Ohishi T, et al. Role of the Rho GTPase-activating protein RICS in neurite outgrowth. Genes Cells. 2006;11(6):607–614. doi: 10.1111/j.1365-2443.2006.00966.x. [DOI] [PubMed] [Google Scholar]
- 49.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
Websites
- 101.A Study to Evaluate the Efficacy and Safety of VX-765 in Subjects With Treatment-Resistant Partial Epilepsy. http://clinicaltrials.gov/ct2/show/NCT01501383.
- 102.Illumina Science / Illumina iControlDB. www.illumina.com/science/icontroldb.ilmn.
- 103.A Phase II Study of PX-866 in Patients With Recurrent or Metastatic Castration Resistant Prostate Cancer. http://clinicaltrials.gov/show/NCT01331083.
Information resources
- Wittkowski KM. Friedman-type statistics and consistent multiple comparisons for unbalanced designs. J. Am. Statist. Assoc. 1988;83(404):1163–1170. extension: 87(417), 258 (1992) [Google Scholar]
- Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for deter-mining continental origin and admixture proportions in common populations in America. Hum. Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Wittkowski KM, Song T. Nonparametric methods for molecular biology. Methods Mol. Biol. 2010;620:105–153. doi: 10.1007/978-1-60761-580-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajek J, Sidak Z. Theory of Rank Tests. Academic Press; New York, NY, USA: 1967. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.