Abstract
Micro-RNAs (miRNAs) are small non-coding RNA molecules, which can act as either oncogenes or tumor suppressors. Dysregulated expression of miRNA genes have been implicated in the development of many different cancers. We hypothesize that genetic variations in miRNA biogenesis genes may be associated with the prognosis of bladder cancer. We genotyped 76 single nucleotide polymorphisms (SNPs) in eight miRNA biogenesis genes in 421 patients with non-muscle-invasive bladder cancer (NMIBC). We analyzed the associations of SNPs with recurrence and progression in all patients as well as stratified by treatment: transurethral resection (TUR) alone or TUR plus intravesical bacillus Calmette–Guérin (BCG) instillation. Two SNPs were significantly associated with tumor recurrence in TUR only subgroup after adjustment for multiple comparisons (Q < 0.1). The most significant SNP was rs197412 in DDX20: the variant allele conferred a decreased risk of recurrence [hazard ratio (HR) = 0.58, 95% confidence interval (95% CI) = 0.40–0.82]. This SNP was validated in a separate group of 586 NMIBC patients and the pooled HR was 0.62 (95% CI = 0.48–0.81, P < 0.001). Two linked SNPs (rs2073778 and rs720012) in DGCR8 showed significant association with tumor progression (HR = 4.00, 95% CI = 1.53–10.46, P = 0.005). A strong gene-dosage effect was observed with higher risk for tumor recurrence and progression with increasing number of unfavorable genotypes. Haplotype and survival tree analyses further characterized the association of miRNA-related SNPs with tumor recurrence and progression. Taken together, our results indicate that genetic variants in miRNA biogenesis pathway may influence bladder cancer clinical outcome in NMIBC patients.
Introduction
Bladder cancer accounts for approximately 4.6% of new primary cancers and 2.6% of cancer deaths in the USA, with an estimated 73 510 new cases and 14 880 deaths in 2012 (1). Most bladder cancers (75–85%) are non-muscle invasive at diagnosis. Among these non-muscle-invasive bladder cancers (NMIBCs), 70% present as stage Ta cancer, 20% as T1 cancer and 10% as carcinoma in situ (2). Although multimodal treatments have been standard treatment for many years, the prognosis for NMIBC remains diverse, as seen in the 30–80% recurrence rate and up to 45% progression rate within 5 years (2). Several factors are known to be associated with risk of NMIBC recurrence and progression, such as the presence of multiple concomitant tumors, tumor size of at least 3cm, prior recurrence, the presence of carcinoma in situ, tumor stage of T1 and tumor grade of at least G2 (3). Recent research has shown that genetic variations and protein expression profiles can also serve as biomarkers for predicting bladder cancer prognosis (4). Tumor cell implantation immediately after transurethral resection (TUR, the standard treatment for NMIBC) is responsible for many early recurrences (5). To reduce the risk of recurrence, perioperative intravesical therapy is used to prevent tumor reimplantation (6). The major choice of intravesical therapy is immunotherapy with bacillus Calmette–Guérin (BCG), which results in a massive local immune response characterized by the induced expression of cytokines in the urine and bladder wall and by the accumulation of granulocytes and mononuclear cells (7). Complete response to TUR plus BCG occurs in 36–71% of patients with NMIBC (8). For this reason, BCG is considered the best adjuvant therapy after TUR treatment in preventing recurrence and delaying progression in NMIBC (9,10). Nevertheless, approximately one-third of BCG responders later display recurrence or progress to muscle-invasive disease (11). Moreover, intravesical BCG administration is commonly accompanied by untoward side effects that may compromise patient’s quality of life and success of treatment. Most patients experience urinary frequency and urgency, and in some cases, serious sequelae and rare deaths have occurred (11). Therefore, it is important to identify new biomarkers that accurately predict a patient’s response to BCG therapy and NMIBC post-treatment outcomes.
Micro-RNA (miRNA) expression profiles differ between bladder cancer tissues and normal urothelia (12). miRNAs are a group of short non-protein-coding RNAs that regulate gene expression and are composed of 20–25 nucleotides (13). The biogenesis of miRNAs is a complex, multistep process with several intermediates. It begins with the generation of primary miRNAs that are 500–3000 nucleotides long. The primary miRNAs are then cleaved by Drosha/DGCR8 to form precursor miRNAs (pre-miRNAs), which are 60–70 nucleotides long. The pre-miRNAs are then transported from the nucleus to the cytoplasm by the Exportin-5/Ran-GTP complex and are subsequently processed by Dicer, an RNase III endonuclease, into short double-stranded miRNA duplexes. Finally, one strand of the duplex is incorporated into the RNA-induced silencing complex and binds to the 3′ untranslated regions of target mRNAs to either degrade the transcript or inhibit translation of the mRNAs (14). A single miRNA can bind to as many as 200 mRNA targets and function as either a tumor suppressor or an oncogene depending on the characteristics of the target gene (15).
Single nucleotide polymorphisms (SNPs) in miRNA-coding genes, miRNA biogenesis genes or in the miRNA-binding sites of mRNAs can affect the biogenesis and function of miRNAs (16,17). Several studies have shown that miRNA-related SNPs are associated with cancer development and prognosis (18–20). We previously reported that SNPs in miRNA biogenesis genes as well as in genes encoding for the miRNAs might affect bladder cancer risk (21). In the current study, we selected 76 SNPs in the miRNA biogenesis pathway and evaluated their association with clinical outcome of patients with NMIBC. To our knowledge, this is the first study of the effects of miRNA-related genetic polymorphisms on the clinical outcomes of bladder cancer.
Patients and methods
Study population and epidemiologic data
The study population and enrollment procedures were described previously (21). Briefly, 421 patients of non-Hispanic whites with newly diagnosed and histopathologically confirmed NMIBC were recruited from The University of Texas MD Anderson Cancer Center and the Baylor College of Medicine through a daily review of computerized appointment schedules as part of an ongoing project since 1995. For validation, we also included another group of 586 NMIBC patients of non-Hispanic whites from prior studies recruited at MD Anderson with clinical information available. All patients were chemotherapy and radiotherapy naive. There were no age or gender restrictions on recruitment. To minimize the effect of population stratification, we restricted the current study to Caucasian patients. All participants provided written informed consent and were interviewed using a structured questionnaire that asked about demographic characteristics and standard risk factors for bladder cancer. Then, a 40ml blood sample was drawn into a coded, heparinized tube, which was delivered to the laboratory for molecular analysis. Clinical and follow-up data such as tumor stage, grade, treatment protocol and dates of recurrence and progression events were abstracted from a computerized medical record system by trained chart reviewers.
The study end points were tumor recurrence, which was defined as a newly found bladder tumor subsequent to a previous cystoscopy negative follow-up and tumor progression, which was defined as the transition from NMIBC to invasive or metastatic disease. All patients received TUR as the initial treatment. Those with carcinoma in situ, T1, G2, G3 or multiple Ta/G1 tumors also received intravesical BCG instillation. This study was approved by the Institutional Review Boards of MD Anderson Cancer Center and Baylor College of Medicine.
SNP selection and genotyping
Seventy-six tagging or potential functional SNPs (coding region, 5′ and 3′ untranslated regions) in eight genes of the miRNA biogenesis pathway were identified through mining of the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/), dbSNP (http://www.ncbi.nlm.nih.gov/snp/) and the UCSC Genome Bioinformatics (http://genome.ucsc.edu) databases (Supplementary Table 1, available at Carcinogenesis Online). All SNPs selected had a reported minor allele frequency of >0.05 in the Caucasian population.
DNA was extracted from peripheral blood samples using a QIAamp DNA extraction kit (Qiagen, Valencia, CA). SNP genotyping was done using the Illumina Infinium II iSelect custom SNP array platform (Illumina, San Diego, CA) according to the standard protocol (22). Genotyping data were analyzed and exported using BeadStudio software (Illumina). For the assay, 2% of the samples were randomly selected for replication. The overall concordance rate was 99%. The genotyping call rate for the selected SNPs in this study was 93%. For validation, the significant SNPs (Q < 0.1) were genotyped in the validation population using Taqman genotyping assays in 7900HT real-time PCR sequence detection system following the standard protocol (Life Technologies, Grand Island, NY). For the latter assay, 5% of the samples were randomly selected for replication. The concordance rate was 100% and the call rate was 96%.
Statistical analysis
Statistical analyses were performed using the Stata 10.0 (StataCorp LP, College Station, TX). Either Pearson’s χ2 or Fisher’s exact test was used to assess the differences in the distributions of categorical variables, such as sex, smoking status, tumor stage, tumor grade and treatment. Student’s t-test was used to examine differences in the distributions of continuous variables, such as age. The main effect of a SNP on time to recurrence or progression was estimated as a hazard ratio (HR) and a 95% confidence interval (95% CI) using the multivariate Cox proportional hazards model adjusted for age, sex, smoking status, tumor stage, tumor grade and treatment. The patients who were lost to follow-up or died were censored. The effect of each SNP was tested based on three genetic models of inheritance (dominant, recessive and additive). The model with the smallest P value was reported as the best-fitting model. All P values were two sided, with a value of P < 0.05 being considered statistically significant. To control for multiple testing, a Q value [a false discovery rate (FDR)-adjusted P value] was used to adjust the significance level for individual SNPs (23). We used a threshold of 0.1 (FDR < 10%) for significance to minimize missing true discoveries in this pilot study. Q values were obtained using the R package software (http://genomics.princeton.edu/storeylab/qvalue/).
We applied a bootstrap resampling method to internally validate the results by generating 100 bootstrapped samples for SNPs that remained significant after multiple comparisons. Each bootstrap sample was drawn from the original data set, and a P value was obtained for each SNP. The cumulative effects of SNPs that showed significant main effects with risk of recurrence or progression were assessed by summing the number of unfavorable genotypes (i.e. genotypes associated with increased risk) in each subject and dividing the subjects into three groups according to the tertile distribution of their risks. Using a multivariate Cox proportional hazards model, we calculated HRs and 95% CIs for all groups and then compared them to those of the low-risk reference group with the fewest unfavorable genotypes. Kaplan–Meier estimates were used to plot event-free survival curves and log-rank tests were used to compare the survival distributions between the risk groups.
Haplotype analyses were performed in selected genes with at least two significant SNPs. The adjusted HRs and 95% CIs for each haplotype were calculated using multivariate Cox proportional hazard models using the most abundant haplotype as the reference group.
Survival tree analysis was performed to investigate higher order gene–gene interactions and to identify subgroups of individuals at higher risk of recurrence. In this analysis, the log-rank statistics was used as node splitting criteria. Each terminal node represents a group of patients with different HR based on distinct genotype combinations. HRs and 95% CIs were calculated for each terminal node using multivariate Cox proportional hazard models after adjusting for appropriate variables.
Results
Patient characteristics
In the discovery population, no significant differences for recurrence rate were identified based on age (P = 0.69), sex (P = 0.11), smoking status (P = 0.69), tumor stage (P = 0.13) or tumor grade (P = 0.19). There were no significant differences in progression based on age (P = 0.05) or smoking status (P = 0.96). Men had higher rates of progression than women (P < 0.01) and patients with a higher stage or grade were also more likely to experience progression (P < 0.01). BCG treatment was significantly associated with reduced risks of both recurrence and progression (P < 0.01) (Supplementary Table 2, available at Carcinogenesis Online). In the validation population, age, sex and smoking status did not significantly influence recurrence rate (P > 0.05); however, tumor stage, grade and types of treatment significantly affected NMIBC recurrence (P < 0.01) (Supplementary Table 3, available at Carcinogenesis Online). Additionally, 33% of the NMIBC cases progressed to invasive disease (data not shown) compared with 19% of the cases in the discovery population (Supplementary Table 2, available at Carcinogenesis Online). The median follow-up time for the validation group was 32.3 months (95% CI = 0.20–318.4) compared with 55.2 months (95% CI = 0.56–200) (P < 0.001) for the discovery group.
Individual SNPs and NMIBC recurrence
Among the 76 SNPs examined, 8 were significantly associated with recurrence in TUR-only patients (Table I). The top two SNPs were rs197412 in DDX20 (HR = 0.58, 95% CI = 0.40–0.82, P = 0.002) and rs12186785 in RNASEN (HR = 2.15, 95% CI = 1.25–3.68, P = 0.005). After adjustment for multiple comparisons, these two SNPs remained statistically significant at FDR of 10% (Q < 0.1). For internal validation, we also performed bootstrap analysis. In 100 bootstrap samplings, rs197412 and rs12186785 reached significance (P < 0.05) 100 and 99 times, respectively, indicating that these findings were robust.
Table I.
Significant SNPs associated with recurrence and cumulative effect of unfavorable genotypes in NMIBC patients receiving TUR treatment alone
| SNP | Gene | Genotype | MOIa | Recurrence (yes/no) | HRb (95% CI) | P | Q | Bootstrap (P < 0.05) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| WW | WV | VV | ||||||||
| rs197412 | DDX20 | C>T | add | 35/9 | 49/27 | 7/9 | 0.58 (0.40–0.82) | 0.002 | 0.087 | 100 |
| rs12186785 | RNASEN | T>C | dom | 70/37 | 21/8 | 0/0 | 2.15 (1.25–3.68) | 0.005 | 0.095 | 99 |
| rs2257082 | XPO5 | C>T | add | 52/18 | 33/23 | 6/4 | 0.67 (0.47–0.95) | 0.026 | 0.196 | 60 |
| rs2740349 | GEMIN4 | A>G | dom | 62/35 | 29/9 | 0/1 | 1.70 (1.06–2.71) | 0.027 | 0.196 | 74 |
| rs1640299 | DGCR8 | G>T | add | 35/13 | 37/17 | 19/15 | 0.73 (0.54–0.98) | 0.033 | 0.196 | 41 |
| rs639174 | RNASEN | T>C | dom | 63/22 | 24/22 | 4/1 | 0.60 (0.38–0.97) | 0.035 | 0.196 | 40 |
| rs197383 | DDX20 | G>A | dom | 76/30 | 15/14 | 0/1 | 0.55 (0.30–0.99) | 0.046 | 0.196 | 1 |
| rs563002 | DDX20 | T>C | dom | 58/23 | 31/18 | 2/4 | 0.63 (0.40–0.99) | 0.047 | 0.196 | 13 |
| Group (number of unfavorable genotypesc) | Recurrence (yes/no) | HRb (95% CI) | P | MST (months) | Bootstrap (95% CI) | |||||
| Low-risk group (0–2) | 17/20 | Ref. | 42.3 | |||||||
| Medium-risk group (3–4) | 44/17 | 2.29 (1.27–4.15) | 0.006 | 7.3 | 1.14–4.82 | |||||
| High-risk group (5–7) | 33/8 | 4.05 (2.15–7.62) | 1.47×10–5 | 4.3 | 1.91–8.23 | |||||
| P for trend | 9.44×10–6 | |||||||||
Note: SNPs that continued to have a significant effect after correcting for multiple comparisons by Q value with a FDR of ≤10% are in boldface. WW, homozygous wild-type genotype; WV, heterozygous variant genotype; VV, homozygous variant genotype; HR, hazard ratio; CI, confidence interval.
aMOI: model of inheritance, the model with the smallest P value; add, additive; dom, dominant.
bAdjusted by age, sex, smoking status, cancer stage and cancer grade.
cUnfavorable genotypes: DDX20 rs197412 (ww); RNASEN rs12186785 (wv + vv); XPO5 rs2257082 (ww); GEMIN4 rs2740349 (wv + vv); DGCR8 rs1640299 (ww); RNASEN rs639174 (ww); DDX20 rs197383 (ww); DDX20 rs563002 (ww).
In 205 patients who received intravesical BCG therapy, three SNPs showed significant association with recurrence (Table II): rs3744741 in GEMIN4 (HR = 0.61, 95% CI = 0.38–0.99, P = 0.045), rs563002 in DDX20 (HR = 2.80, 95% CI = 1.30–6.04, P = 0.009) and rs720012 in DGCR8 (HR = 2.34, 95% CI = 1.00–5.46, P = 0.049). However, none of these SNPs retained significance after adjustment for multiple comparisons.
Table II.
SNPs associated with recurrence in NMIBC patients receiving BCG after TUR
| SNP | Gene | Genotype | MOIa | Recurrence (yes/no) | HRb (95% CI) | P | Q | Bootstrap (P < 0.05) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| WW | WV | VV | ||||||||
| rs563002 | DDX20 | T>C | rec | 70/49 | 32/32 | 8/1 | 2.80 (1.30–6.04) | 0.009 | 0.412 | 95 |
| rs3744741 | GEMIN4 | C>T | dom | 87/60 | 23/21 | 0/1 | 0.61 (0.38–0.99) | 0.045 | 0.466 | 41 |
| rs720012 | DGCR8 | G>A | rec | 78/60 | 24/21 | 6/0 | 2.34 (1.00–5.46) | 0.049 | 0.466 | 84 |
WW, homozygous wild-type genotype; WV, heterozygous variant genotype; VV, homozygous variant genotype; HR, hazard ratio; CI, confidence interval.
aMOI: model of inheritance, the model with the smallest P value; dom, dominant; rec, recessive.
bAdjusted by age, sex, smoking status, cancer stage and cancer grade.
Individual SNPs and NMIBC progression
Only 85 patients had a progression of their cancer. Because of the limited number of patients, we were unable to stratify patients based on treatment protocols. As seen in Table III, seven SNPs were significantly associated with tumor progression. The most significant SNP was rs2073778 in DGCR8, whose variant homozygous genotype was associated with a 4-fold increased risk of progression compared with wild-type genotypes (HR = 4.00, 95% CI = 1.53–10.46, P = 0.005). After adjustment for multiple comparisons, two SNPs remained significant (Q < 0.1; rs2073778 and rs720012, both in DGCR8). Out of 100 bootstrap samplings, rs2073778 and rs720012 reached significance (P < 0.05) in 70 and 78 iterations, respectively.
Table III.
Significant SNPs associated with progression and cumulative effect of unfavorable genotypes in all NMIBC patients
| SNP | Gene | Genotype | MOIa | Progression (yes/no) | HRb (95% CI) | P | Q | Bootstrap (P < 0.05) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| WW | WV | VV | ||||||||
| rs2073778 c | DGCR8 | C>T | rec | 51/243 | 19/75 | 5/8 | 4.00 (1.53–10.46) | 0.005 | 0.099 | 70 |
| rs720012 c | DGCR8 | G>A | rec | 51/243 | 19/73 | 5/8 | 3.97 (1.52–10.36) | 0.005 | 0.099 | 78 |
| rs1187652 | DICER1 | T>C | dom | 68/313 | 7/14 | 0/0 | 2.62 (1.15–5.98) | 0.022 | 0.220 | 25 |
| rs11160231 | DICER1 | C>A | add | 41/224 | 30/93 | 4/10 | 1.52 (1.15–2.23) | 0.035 | 0.220 | 30 |
| rs10773831 | RAN | C>G | dom | 48/152 | 18/148 | 9/27 | 0.60 (0.37–0.97) | 0.036 | 0.220 | 97 |
| rs12318549 | RAN | A>G | dom | 52/254 | 22/68 | 1/4 | 1.69 (1.02–2.80) | 0.042 | 0.220 | 12 |
| rs16901165 | RNASEN | G>A | dom | 61/284 | 13/42 | 1/1 | 1.86 (1.02–3.39) | 0.043 | 0.220 | 12 |
| Group (number of unfavorable genotyped) | Progression (yes/no) | HRb (95% CI) | P | MST (months) | Bootstrap 95% CI | |||||
| Low-risk group (0–1) | 42/227 | Ref. | — | |||||||
| Medium-risk group (2–3) | 32/103 | 2.04 (1.26–3.32) | 0.004 | — | 1.22–3.41 | |||||
| High-risk group (4–5) | 7/4 | 10.79 (4.40–26.46) | 2.05×10–7 | 7.1 | 2.23–33.01 | |||||
| P for trend | 3.45×10–6 | |||||||||
Note: SNPs that continued to have a significant effect after correcting for multiple comparisons by Q value with a FDR of ≤10% are in boldface. WW, homozygous wild-type genotype; WV, heterozygous variant genotype; VV, homozygous variant genotype; HR, hazard ratio; CI, confidence interval.
aMOI: model of inheritance, the model with the smallest P value; add, additive; dom, dominant; rec, recessive.
bAdjusted by age, sex, smoking status, cancer stage and cancer grade.
crs2073778 is in strong linkage with rs720012, with r 2 = 1.0.
dUnfavorable genotypes: DGCR8 rs2073778 (vv); DICER1 rs1187652 (wv + vv); DICER1 rs11160231 (wv + vv); RAN rs10773831 (ww); RAN rs12318549 (wv + vv); RNASEN rs16901165 (wv + vv).
Cumulative effects of unfavorable genotypes
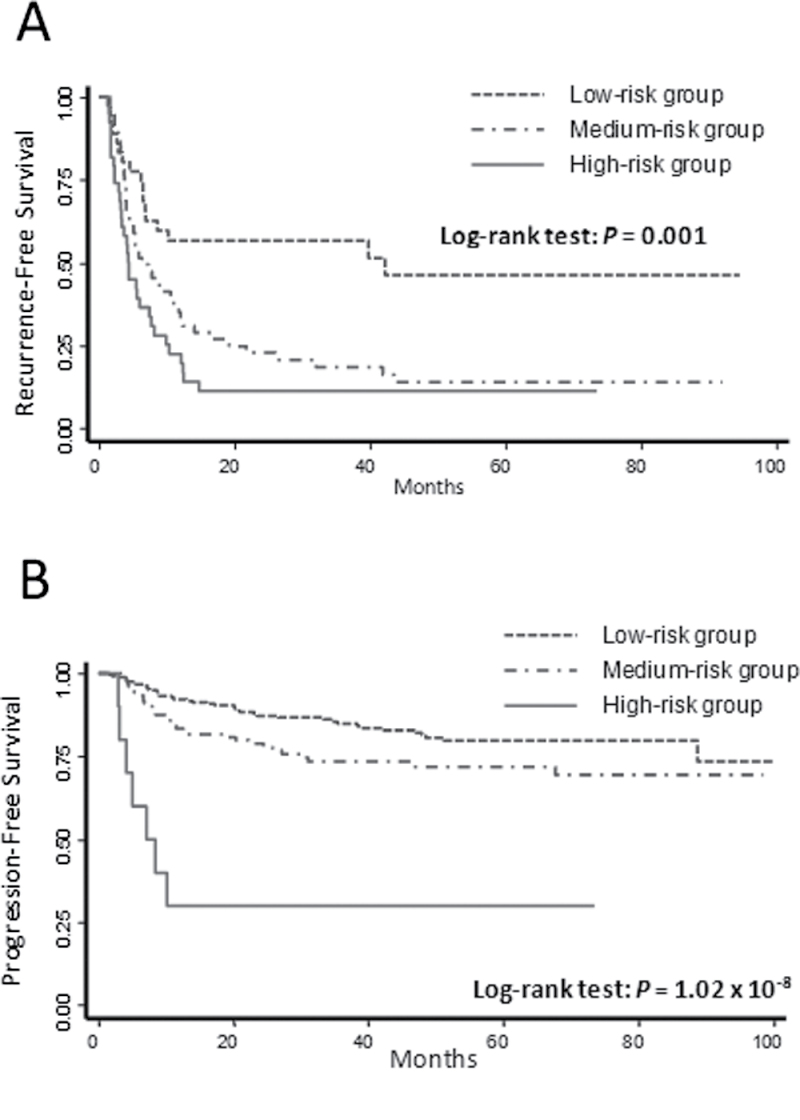
We assessed the cumulative effects of unfavorable genotypes identified from the significant SNPs in the main effect analysis on bladder cancer recurrence and progression. We observed that elevated risks of recurrence and progression were associated with increased numbers of these genotypes. Table I shows results for recurrence risk and recurrence-free survival in patients of the TUR treatment group. Compared with the low-risk group (patients with 0–2 unfavorable genotypes), the medium-risk group (patients with 3 or 4 unfavorable genotypes) and the high-risk group (patients with 5–7 unfavorable genotypes) had HRs for recurrence of 2.29 (95% CI = 1.27–4.15, P = 0.006) and 4.05 (95% CI = 2.15–7.62, P = 1.47×10–5), respectively (P trend = 9.44× 10–6). Kaplan–Meier estimate of recurrence-free survival (Figure 1A) indicated that the high-risk group and the medium-risk group had recurrence-free median survival times (MSTs) of 4.3 and 7.3 months, respectively, which were much shorter than that of the low-risk group (MST = 42.3 months) (P log-rank = 0.001).
Fig. 1.
Kaplan–Meier curves of (A) cumulative effect of unfavorable genotypes on recurrence of NMIBC in the TUR group and (B) cumulative effect of unfavorable genotypes on progression of NMIBC.
Table III shows the cumulative effect of unfavorable genotypes on cancer progression. When compared with the low-risk group (patients with 0 or 1 unfavorable genotypes), the medium-risk group (patients with 2 or 3 unfavorable genotypes) and the high-risk group (patients with 4 or 5 unfavorable genotypes) had increased progression risk of 2.04 (95% CI = 1.26–3.32, P = 0.004) and 10.79 (95% CI = 4.40–26.46, P = 2.05×10–7), respectively (P trend = 3.45×10–6). Kaplan–Meier curve of progression-free survival (Figure 1B) showed that the high-risk group had a progression-free MST of 7.1 months, which was shorter than that of the medium- or low-risk group (P log-rank = 1.02×10–8). The MSTs of the medium- and low-risk groups were not assessable because less than half of the patients encountered progression events prior to censor date.
Validation of the top SNPs with recurrence
To validate the top SNPs, we assessed the association of rs197412 in DDX20 and rs12186785 in RNASEN, two SNPs that reached significance at FDR of 10%, in a second group of 586 NIMBC patients for which recurrence information is available. In stratified analysis by TUR treatment, the DDX20 SNP but not the RNASEN variant showed similar effect and trend of association with borderline significance (HR = 0.71, 95% CI = 0.47–1.06, P = 0.09; pooled HR = 0.62, 95% CI = 0.48–0.81, P = 0.00077) (Table IV).
Table IV.
Replication result of top SNPs associated with recurrence in NMIBC patients receiving TUR treatment alone
| SNP | Gene | MOIa | Recurrence (yes/no) | Replication HRb (95% CI) | P | Pooled HRb (95% CI) | P | ||
|---|---|---|---|---|---|---|---|---|---|
| WW | WV | VV | |||||||
| rs197412 | DDX20 | add | 31/11 | 40/36 | 10/17 | 0.71 (0.47–1.05) | 0.092 | 0.62 (0.48–0.81) | 0.00077 |
| rs12186785 | RNASEN | dom | 67/51 | 13/11 | 0/0 | 1.2 (0.59–2.43) | 0.611 | — | — |
WW, homozygous wild-type genotype; WV, heterozygous variant genotype; VV, homozygous variant genotype; HR, hazard ratio; CI, confidence interval.
aMOI: model of inheritance, the model with the smallest P value; add, additive; dom, dominant.
bAdjusted by age, sex, smoking status, cancer stage, and cancer grade
Haplotype analyses
We conducted haplotype analyses of the significant SNPs in miRNA biogenesis genes associated with recurrence risk after TUR and with progression risk (Supplementary Table 4, available at Carcinogenesis Online). Two haplotypes in DDX20 and one haplotype in RNASEN showed altered risk in NMIBC recurrence. Specifically, VWV and VVW haplotypes in DDX20 involving rs197412, rs197383 and rs563002 (W, wild-type allele; V, variant-type allele) displayed reduction in recurrence risk [HR of 0.67 (95% CI = 0.45–1.00, P = 0.049) and 0.56 (95% CI = 0.32–0.98, P = 0.04), respectively] compared with most common haplotype (WWW). The VW haplotype in RNASEN (rs12186785 and rs639174) had a significantly increased risk (HR = 1.87; 95% CI = 1.87–3.03, P = 0.012) compared with the WW haplotype. In NMIBC progression, WV haplotype in RAN (rs10773831 and rs12318549) showed 2-fold increase in progression risk (HR = 2.00; 95% CI = 1.18–3.38, P = 0.01), whereas VW haplotype showed significantly reduced HR of 0.57 (95% CI = 0.33–0.98, P = 0.044) compared with WW haplotype.
Survival tree analysis
To explore potential high-order gene–gene interactions between the miRNA biogenesis genes, we conducted a survival tree analysis using the significant SNPs identified from the individual SNP analysis. In NMIBC recurrence study, no interactions were found either in TUR group or in BCG group. In progression study, the tree structure resulted in four terminal nodes with a range of low- to high-risk subgroups (data not shown). The initial split was rs10773831 in RAN, followed by rs11160231 in DICER1 and rs16901165 in RNASEN. Using low-risk node 1 as reference, node 2 had a HR of 1.38 (95% CI = 0.80–2.38, P = 0.246), node 3 had a HR of 1.83 (95% CI = 0.94–3.55, P = 0.073) and node 4 had a HR of 6.67 (95% CI = 2.75–16.2, P = 2.72×10–5) (P trend = 0.00049).
Discussion
In this study, we systematically evaluated the effects of 76 SNPs in eight miRNA biogenesis genes on bladder cancer recurrence and progression. We have shown that miRNA-related SNPs may impact bladder cancer recurrence and progression. In stratified analysis by treatment, the most significant SNP associated with risk of NMIBC recurrence was rs197412 in DDX20, a missense SNP that alters codon 636 from isoleucine to threonine near the C-terminus of the protein. It is not known if this SNP affects protein function although it is located in a protein region that contains several phosphorylation sites. Its variant allele is not predicted to generate a new threonine phosphoration site based on in silico analysis (NetPhos 2.0 Server prediction; http://www.cbs.dtu.dk/services/NetPhos). Compared with patients with the wild-type allele, those carrying the variant allele had reduced risk of recurrence for patients treated with TUR only in both the discovery and the validation groups; however, the association was not significant in the latter population. In contrast, patients with the homozygous variant genotype of rs563002 in DDX20 had a 2.8-fold increased risk of recurrence after intravesical BCG therapy although the association was not significant after adjustment for multiple comparisons. Intravesical immunotherapy with BCG results in a massive local immune response activating cell-mediated cytotoxic mechanisms that work to prevent cancer recurrence and progression (7). These cytotoxic mechanisms involve modulation of miRNAs that have significant impact on the function of antitumor T cells and immune-mediated recognition of cancer cells (24,25). Several miRNAs have been utilized to improve T-cell-based cancer immunotherapy (24). DDX20, also known as GEMIN3, encodes one of the DEAD box proteins, which are characterized by the conserved amino acid motif Asp-Glu-Ala-Asp (DEAD). These DEAD box proteins are involved in many cellular processes including the alteration of RNA secondary structure during translation initiation, nuclear and mitochondrial splicing and ribosome and spliceosome assembly. Some members of the DEAD box protein family are believed to be involved in embryogenesis, spermatogenesis and cellular growth and division (26). Since the DDX20 protein is involved as a member of the AGO protein family pivotal to miRNA processing (27), it is possible that genetic variations in miRNA biogenesis genes could change the immunologic response in bladder cancer. Interestingly, improvement in overall survival was shown for renal cell carcinoma patients carrying DDX20 variants (28). In addition, other variants in DDX20 have been shown to significantly increase risk of bladder (21) and esophageal cancers (29), but not renal cell carcinoma (20) or lung cancer (30). Taken together, genetic variants in DDX20 are potential predictive markers for NMIBC treatment and clinical outcome.
One SNP in RNASEN, rs12186785, also showed significant impact on bladder cancer recurrence although this variant was not significant in our validation, likely due to the low frequency of the variant allele and small sample size after stratification by treatment. RNASEN, also known as DROSHA, encodes an RNase III protein that is part of the microprocessor complex. In a study of genome-wide miRNA expression in salivary gland pleomorphic adenomas, increased DROSHA expression correlated with deregulated expression of miRNAs (31). Moreover, genetic variants of this gene have been associated with recurrence in renal cell carcinoma (28) and head and neck cancer (32). Since miRNA expression profiles vary in different cancer tissues, the effects of RNASEN genetic variation on disease prognosis could differ between different cancer types. Further studies to correlate the effect of RNASEN genetic variations with miRNA expression in various cancer tissues may reveal their influence on cancer prognosis.
Other SNPs that were associated with NMIBC progression in the discovery population include those in the DGCR8 gene. DGCR8 is a double-stranded RNA-binding protein that functions as the non-catalytic subunit of the microprocessor complex and facilitates RNA cleavage by the RNase III protein Drosha. As referenced earlier, both DGCR8 and DROSHA expressions were correlated with deregulated expression of miRNAs (30). In vitro knockdown of DROSHA, DGCR8 and DICER1 impaired miRNA processing and thereby promoted oncogenic transformation in mouse lung cancer cells and tumor development in vivo (33). Han et al. (34) reported that RNASEN and DGCR8 regulate each other post-transcriptionally and that DGCR8 stabilizes RNASEN via protein–protein interactions. Because of the direct effect of DGCR8 and RNASEN on miRNA biogenesis and the associations between miRNA expression and cancer development and progression, it follows that variations in either gene might affect bladder cancer recurrence and progression. Interestingly, the DGCR8 variant, rs720012, was found to be associated with NMIBC recurrence in TUR- and BCG-treated patients and NMIBC progression in the discovery population; however, the association with recurrence was not significant after adjustment for multiple comparisons and the correlation with progression was not replicated in the validation group. We should note that the two linked DGCR8 SNPs, rs720012 and rs2073778, were significantly associated with both recurrence and progression in the overall discovery group even after adjustment for multiple testing (Supplemental Table 5, available at Carcinogenesis Online). Nevertheless, these associations were not replicated in the overall validation set (P > 0.05). Therefore, the jury is still out on whether these SNPs are predictive markers of treatment or prognostic of clinical outcomes. Further validation in larger independent population is necessary to confirm these findings.
We also evaluated the cumulative effects of these miRNA biogenesis SNPs on the risk of bladder cancer recurrence and progression. We observed a dose-dependent correlation between an increased recurrence or progression risk and a higher number of unfavorable genotypes. In addition, haplotype and survival tree analyses of a combination of SNPs revealed the effects of significant genes and higher order gene–gene interactions. These results support the hypothesis that the development of cancer recurrence or progression after definitive treatment of NMIBC is a polygenic process.
There are some limitations in our study. For example, our relatively small sample size after stratification by treatment limited the power to identify and validate significant SNPs associated with recurrence and progression. Nevertheless, for large effect variants (HR > 1.5 or HR < 0.7), the study has sufficient power (>80%) to detect significant associations, even after correction for multiple testing. It should be noted that several of our identified top SNPs have large main effects. Another limitation is that there might be population heterogeneity between the discovery and validation populations since only in the latter group was recurrence influenced by tumor stage and grade as well as treatment although these covariates were adjusted during the logistic regression analysis. Moreover, the discrepancies between the two populations in terms of frequency of disease progression and follow-up times may suggest dissimilar proportion of high-risk patients. Due to the small number of patients with progressive disease, we did not perform stratified analysis and the results were not validated in the second population.
In conclusion, we conducted the first study to evaluate the association between genetic variants in miRNA biogenesis pathway genes and bladder cancer clinical outcomes. Using stratified analysis, we also determined the effect of miRNA-related polymorphisms in various treatment subgroups and the joint effects of these SNPs on NMIBC recurrence and progression. Some of the identified SNPs may be potential predictive markers for bladder cancer treatment. However, the underlying biological mechanisms through which these genetic variants affect clinical behavior of bladder cancers are currently unknown. Further larger independent studies and functional characterization are needed to confirm these variants as prognostic or predictive markers.
Supplementary material
Supplementary Tables 1–5 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health grants (U01 CA 127615, R01 CA 74880 and P50 CA 91846).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- BCG
bacillus Calmette–Guérin
- CI
confidence interval
- FDR
false discovery rate
- HR
hazard ratio
- miRNA
micro-RNA
- MST
median survival time
- NMIBC
non-muscle-invasive bladder cancer
- SNP
single nucleotide polymorphism
- TUR
transurethral resection.
References
- 1. Siegel R., et al. (2012). Cancer statistics, 2012. CA Cancer J. Clin., 62, 10–29 [DOI] [PubMed] [Google Scholar]
- 2. van Rhijn B.W., et al. (2009). Recurrence and progression of disease in non-muscle-invasive bladder cancer: from epidemiology to treatment strategy. Eur. Urol., 56, 430–442 [DOI] [PubMed] [Google Scholar]
- 3. Babjuk M., et al. (2008). EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur. Urol., 54, 303–314 [DOI] [PubMed] [Google Scholar]
- 4. Lintula S., et al. (2010). Developing biomarkers for improved diagnosis and treatment outcome monitoring of bladder cancer. Expert Opin. Biol. Ther., 10, 1169–1180 [DOI] [PubMed] [Google Scholar]
- 5. Heney N.M., et al. (1981). Prognostic factors in carcinoma of the ureter. J. Urol., 125, 632–636 [DOI] [PubMed] [Google Scholar]
- 6. Klän R., et al. (1991). Residual tumor discovered in routine second transurethral resection in patients with stage T1 transitional cell carcinoma of the bladder. J. Urol., 146, 316–318 [DOI] [PubMed] [Google Scholar]
- 7. Böhle A., et al. (2003). Immune mechanisms in bacillus Calmette-Guerin immunotherapy for superficial bladder cancer. J. Urol., 170, 964–969 [DOI] [PubMed] [Google Scholar]
- 8. Catalona W.J., et al. (1990). Bacillus Calmette-Guérin and superficial bladder cancer. Clinical experience and mechanism of action. Surg. Annu., 22, 363–378 [PubMed] [Google Scholar]
- 9. Davis J.W., et al. (2002). Superficial bladder carcinoma treated with bacillus Calmette-Guerin: progression-free and disease specific survival with minimum 10-year followup. J. Urol., 167, 494–500 [DOI] [PubMed] [Google Scholar]
- 10. Lamm D.L., et al. (1991). A randomized trial of intravesical doxorubicin and immunotherapy with bacille Calmette-Guérin for transitional-cell carcinoma of the bladder. N. Engl. J. Med., 325, 1205–1209 [DOI] [PubMed] [Google Scholar]
- 11. Oddens J.R., et al. (2004). One immediate postoperative instillation of chemotherapy in low risk Ta, T1 bladder cancer patients. Is it always safe? Eur. Urol., 46, 336–338 [DOI] [PubMed] [Google Scholar]
- 12. Wang G., et al. (2010). Up-regulation of microRNA in bladder tumor tissue is not common. Int. Urol. Nephrol., 42, 95–102 [DOI] [PubMed] [Google Scholar]
- 13. Grosshans H., et al. (2008). Molecular biology: the expanding world of small RNAs. Nature, 451, 414–416 [DOI] [PubMed] [Google Scholar]
- 14. Mishra P.J., et al. (2008). MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle, 7, 853–858 [DOI] [PubMed] [Google Scholar]
- 15. Esquela-Kerscher A., et al. (2006). Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer, 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 16. Cai Y., et al. (2009). A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics, 7, 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen K., et al. (2008). Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis, 29, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 18. Ye Y., et al. (2008). Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. (Phila.), 1, 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu Z., et al. (2008). Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest., 118, 2600–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horikawa Y., et al. (2008). Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res., 14, 7956–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang H., et al. (2008). Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res., 68, 2530–2537 [DOI] [PubMed] [Google Scholar]
- 22. Chen M., et al. (2010). Genetic variations in the sonic hedgehog pathway affect clinical outcomes in non-muscle-invasive bladder cancer. Cancer Prev. Res. (Phila.), 3, 1235–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Storey J.D. (2002). A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B, 64, 479–498 [Google Scholar]
- 24. Steemers F.J., et al. (2006). Whole-genome genotyping with the single-base extension assay. Nat. Methods, 3, 31–33 [DOI] [PubMed] [Google Scholar]
- 25. Okada H., et al. (2010). MicroRNAs in immune regulation—opportunities for cancer immunotherapy. Int. J. Biochem. Cell Biol., 42, 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charroux B., et al. (1999). Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol., 147, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mourelatos Z., et al. (2002). miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin J., et al. (2010). Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis, 10, 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ye Y., et al. (2008). Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. (Phila.), 1, 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim J.S., et al. (2010). Association of a common AGO1 variant with lung cancer risk: a two-stage case-control study. Mol. Carcinog., 49, 913–921 [DOI] [PubMed] [Google Scholar]
- 31. Zhang X., et al. (2009). Alterations in miRNA processing and expression in pleomorphic adenomas of the salivary gland. Int. J. Cancer, 124, 2855–2863 [DOI] [PubMed] [Google Scholar]
- 32. Zhang X., et al. (2010). MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis, 31, 2118–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar M.S., et al. (2007). Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet., 39, 673–677 [DOI] [PubMed] [Google Scholar]
- 34. Han J., et al. (2009). Posttranscriptional crossregulation between Drosha and DGCR8. Cell, 136, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.