Abstract
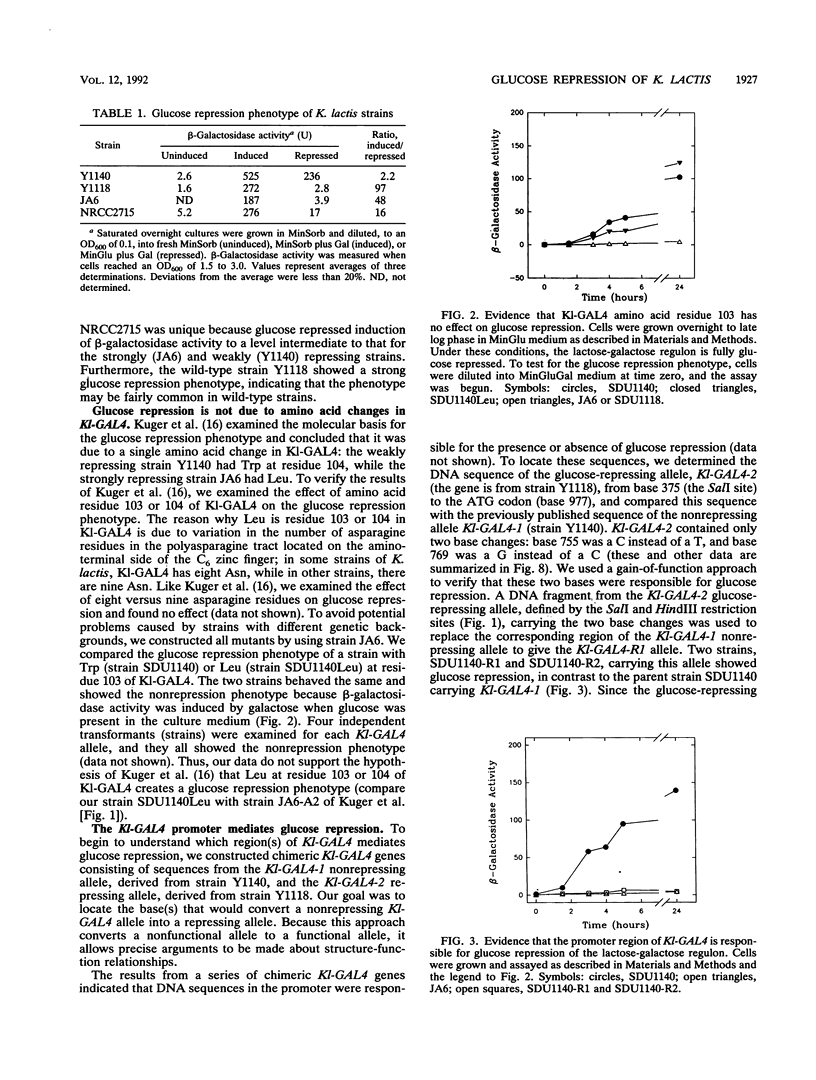
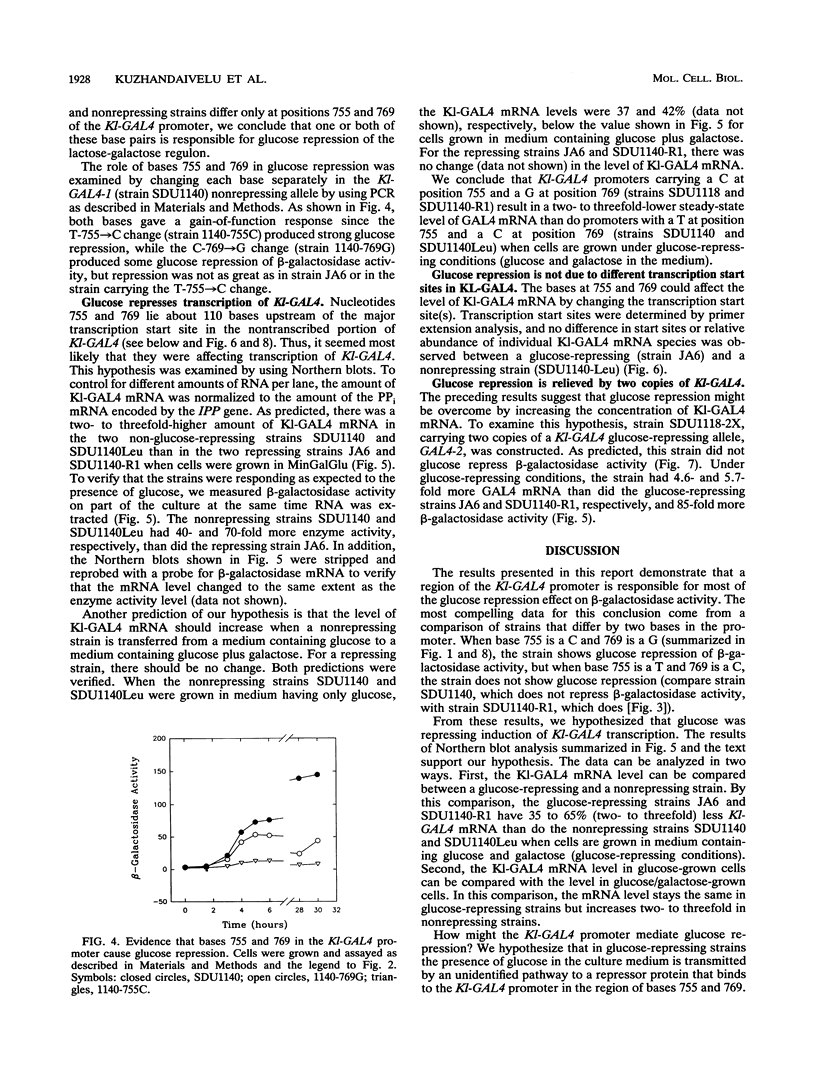
Induction of the lactose-galactose regulon is strongly repressed by glucose in some but not all strains of Kluyveromyces lactis. We show here that in strongly repressed strains, two to three times less Kl-GAL4 mRNA is synthesized and that expression of structural genes in the regulon such as LAC4, the structural gene for beta-galactosidase, is down regulated 40-fold or more. Comparative analysis of strains having a strong or weak repression phenotype revealed a two-base difference in the promoter of the Kl-GAL4 (also called LAC9) positive regulatory gene. This two-base difference is responsible for the strong versus the weak repression phenotype. The two base changes are symmetrically located in a DNA sequence having partial twofold rotational symmetry (14 of 21 bases). We hypothesize that this region functions as a sensitive regulatory switch, an upstream repressor sequence (URS). According to our model, the presence of glucose in the culture medium signals, by an unidentified pathway, a repressor protein to bind the URS. Binding reduces transcription of the Kl-GAL4 gene so that the concentration of the Kl-GAL4 protein falls below the level needed for induction of LAC4 and other genes in the regulon. For strains showing weak glucose repression, we hypothesize that the two base changes in the URS reduce repressor binding so that the regulon is not repressed. Our results illustrate an important principle of genetic regulation: a small (2- to 3-fold) change in the concentration of a regulatory protein can produce a large (40-fold or greater) change in expression of structural genes. This mechanism of signal amplification could play a role in many biological phenomena that require regulated transcription.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Breunig K. D. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet. 1989 Apr;216(2-3):422–427. doi: 10.1007/BF00334386. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987 Dec;7(12):4400–4406. doi: 10.1128/mcb.7.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990 May 24;345(6273):361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Deshler J. O., Larson G. P., Rossi J. J. Kluyveromyces lactis maintains Saccharomyces cerevisiae intron-encoded splicing signals. Mol Cell Biol. 1989 May;9(5):2208–2213. doi: 10.1128/mcb.9.5.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Physiological studies of beta-galactosidase induction in Kluyveromyces lactis. J Bacteriol. 1980 Jun;142(3):777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Riley M. I. The lactose-galactose regulon of Kluyveromyces lactis. Biotechnology. 1989;13:19–40. [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci U S A. 1988 Jan;85(2):382–386. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Herlitze S., Koenen M. A general and rapid mutagenesis method using polymerase chain reaction. Gene. 1990 Jul 2;91(1):143–147. doi: 10.1016/0378-1119(90)90177-s. [DOI] [PubMed] [Google Scholar]
- Kuger P., Gödecke A., Breunig K. D. A mutation in the Zn-finger of the GAL4 homolog LAC9 results in glucose repression of its target genes. Nucleic Acids Res. 1990 Feb 25;18(4):745–751. doi: 10.1093/nar/18.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo J. M., Bhairi S. M., Dickson R. C. Identification of upstream activator sequences that regulate induction of the beta-galactosidase gene in Kluyveromyces lactis. Mol Cell Biol. 1987 Dec;7(12):4369–4376. doi: 10.1128/mcb.7.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAGASANIK B. Catabolite repression. Cold Spring Harb Symp Quant Biol. 1961;26:249–256. doi: 10.1101/sqb.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. GAL4 transcription factor is not a "zinc finger" but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Halvorsen Y. D., Dickson R. C., Coleman J. E. The transcription factor LAC9 from Kluyveromyces lactis-like GAL4 from Saccharomyces cerevisiae forms a Zn(II)2Cys6 binuclear cluster. J Biol Chem. 1990 Dec 15;265(35):21427–21429. [PubMed] [Google Scholar]
- Riley M. I., Hopper J. E., Johnston S. A., Dickson R. C. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol. 1987 Feb;7(2):780–786. doi: 10.1128/mcb.7.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzi M., Breunig K. D., Ficca A. G., Hollenberg C. P. Positive regulation of the beta-galactosidase gene from Kluyveromyces lactis is mediated by an upstream activation site that shows homology to the GAL upstream activation site of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):991–997. doi: 10.1128/mcb.7.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Mutations affecting synthesis of beta-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980 Aug;95(4):877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekrishna K., Webster T. D., Dickson R. C. Transformation of Kluyveromyces lactis with the kanamycin (G418) resistance gene of Tn903. Gene. 1984 Apr;28(1):73–81. doi: 10.1016/0378-1119(84)90089-1. [DOI] [PubMed] [Google Scholar]
- Stark M. J., Milner J. S. Cloning and analysis of the Kluyveromyces lactis TRP1 gene: a chromosomal locus flanked by genes encoding inorganic pyrophosphatase and histone H3. Yeast. 1989 Jan-Feb;5(1):35–50. doi: 10.1002/yea.320050106. [DOI] [PubMed] [Google Scholar]
- Witte M. M., Dickson R. C. Cysteine residues in the zinc finger and amino acids adjacent to the finger are necessary for DNA binding by the LAC9 regulatory protein of Kluyveromyces lactis. Mol Cell Biol. 1988 Sep;8(9):3726–3733. doi: 10.1128/mcb.8.9.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte M. M., Dickson R. C. The C6 zinc finger and adjacent amino acids determine DNA-binding specificity and affinity in the yeast activator proteins LAC9 and PPR1. Mol Cell Biol. 1990 Oct;10(10):5128–5137. doi: 10.1128/mcb.10.10.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]