Abstract
The Y-family DNA polymerase IV or PolIV (Escherichia coli) is the founding member of the DinB family and is known to play an important role in stress-induced mutagenesis. We have determined four crystal structures of this enzyme in its pre-catalytic state in complex with substrate DNA presenting the four possible template nucleotides that are paired with the corresponding incoming nucleotide triphosphates. In all four structures, the Ser42 residue in the active site forms interactions with the base moieties of the incipient Watson–Crick base pair. This residue is located close to the centre of the nascent base pair towards the minor groove. In vitro and in vivo assays show that the fidelity of the PolIV enzyme increases drastically when this Ser residue was mutated to Ala. In addition, the structure of PolIV with the mismatch A:C in the active site shows that the Ser42 residue plays an important role in stabilizing dCTP in a conformation compatible with catalysis. Overall, the structural, biochemical and functional data presented here show that the Ser42 residue is present at a strategic location to stabilize mismatches in the PolIV active site, and thus facilitate the appearance of transition and transversion mutations.
INTRODUCTION
Many prokaryotic organisms respond to environmental stress through activation of the SOS response, which involves upregulation of a number of genes. One major aspect of this response involves enhancement in the frequency at which mutations appear in the genome (1). As evolution operates through the selection of mutations that confer fitness for a specific environment, it is believed that this strategy will ultimately aid in relieving selection pressure imposed by an adverse environment. This phenomenon is sometimes termed as adaptive mutagenesis (2,3).
Error-prone DNA polymerases (dPols) belonging to the Y-family have been implicated in stress-induced mutagenesis (4–10). Escherichia coli possesses two members belonging to the Y-family of DNA polymerases (dPols)—PolIV (gene:dinB) and PolV or UmuCD2’ (genes: umuC and umuD) (4,11,12). The expression of both these enzymes is upregulated during the SOS response. Mutant strains of E. coli lacking these polymerases exhibit substantial reduction in fitness during starvation conditions as compared with wild-type strains (13). In addition to stress-induced mutagenesis, it has been shown that the PolIV is responsible for enhancing the frequency of mutations during the stationary phase. The expression of this enzyme is upregulated during the SOS response (LexA-dependent) and stationary phase (RpoS dependent); therefore, this enzyme is involved in both stress-induced and spontaneous mutagenesis (1,7,14–17). During the stationary phase, PolIV has been shown to be responsible for as much as 85% of adaptive point mutations (14). Additionally, upregulation of PolIV is sufficient for stress-induced mutagenesis in E. coli and in conjunction with other molecules can lead to the transition of a subpopulation of cells to a hypermutable state (18). Also, transcription of the dinB gene can be induced independent of the SOS response because of β-lactam antibiotics like ceftazidime even in RpoS-deficient strains (19).
A number of groups have shown that PolIV can promote base substitutions and frameshift mutations (20–23). As predicted from its observed role in adaptive mutagenesis, PolIV and homologous molecules do play a role in the appearance of resistance towards antibiotics in bacteria (24–27). It is expected that the ability of PolIV to create mutations has to be regulated to exist within a certain range, as too many mutations can be deleterious and too few will not provide enough templates for natural selection. The structural attributes of the PolIV active site that allow calibration of mutagenesis within the correct limits are not known.
We present four crystal structures of PolIV in complex with DNA substrates that present dA, dT, dG and dC at the templating position paired with dTTP, dATP, dCTP and dGTP, respectively. An unusual feature of PolIV evident from the structures is the presence of a serine residue (Ser42) in the active site that forms interactions with the base moieties of all four incipient base pairs. When this Ser residue was mutated to Ala, there was a substantial enhancement in the fidelity of PolIV. In addition, the structure of a ternary complex of PolIV with the A:C mismatch in the active site showed that Ser42 forms direct and water-mediated interactions with the template and incoming nucleotides, respectively. Overall, structural, biochemical and functional studies presented here show that the Ser42 is present at a strategic location to stabilize mismatches in the PolIV active site; thus, this residue plays a critical role in the mutator activity of DNA polymerase IV.
MATERIALS AND METHODS
Cloning, purification and crystallization
The dinB gene was amplified from genomic DNA of Escherichia coli and cloned into pGEX_6P1 vector and expressed using the C41DE3 strain. The GST–PolIV fusion polypeptide with an N-terminal GST-tag was purified by affinity chromatography using GST–sepharose resin (GE Healthcare). The GST-tag was removed by PreScission protease, and the PolIV protein was further purified by gel filtration using a Superdex 200 column. For phasing, selenomethionine-labelled PolIV was prepared using B834 cells, and the labelled protein was purified and crystallized using identical protocols as the native protein. Purified oligonucleotides were purchased from Sigma Genosys and annealed to provide duplex DNA substrates. In addition, oligonucleotide with dA at the templating position and a dideoxy nucleotide at the 3′-end were purchased from Keck Centre (Yale University), purified using ion exchange chromatography, desalted, lyophilized, solubilized and annealed. The ternary complexes were reconstituted by mixing PolIV (0.3 mM) with non-terminated DNA at a 1:1.2 molar ratio followed by addition of 5 mM dNMPnPP (Jena Biosciences) and 5 mM MgCl2. In the case of chain terminated DNA, dCTP (GE Amersham) was added to a final concentration of 5 mM. After screening and optimization, the best crystals were obtained in 0.1 M acetate (pH 4.8) and 5–12% (w/v) MPD.
Structure determination and crystallographic refinement
X-ray diffraction data were collected at the BM14 beamline of ESRF (PolIVdG:dCMPnPP, SeMet–PolIVdG:dCTP and PolIVdT:dAMPnPP) and the PXIII beamline of SLS (PolIVdC:dGMPnPP, PolIVdA:dTMPnPP and PolIVdA:dCTP). The structure of the SeMet–PolIVdG:dCTP complex was determined by multiwavelength anomalous dispersion method using the SHELX suite of programs (28). The initial model was built manually using O, and the available structures of MsPolIV (4DEZ) and that of the Polymerase associated domain (PAD) region of PolIV (1UNN) were used to guide the model building (29–31). There are two complexes in the asymmetric unit based on the chain ID of the protein, template strand, primer strand, incoming nucleotide and ion, the two complexes were named ABCDE and FGHIJ. The other structures were determined by molecular replacement using the structures of SeMet–PolIVdG:dCTP as a search model (with template and incoming nucleotide removed). All the structures were refined using CNS until convergence after which TLS refinement was carried out using Refmac in CCP4 (32,33). The presence and conformation of the incoming nucleotide in all the complexes was confirmed through calculation of simulated annealed omit maps. Majority of the residues are in the favourable regions of the Ramachandran plot with only 1% residues in the disallowed regions. The data and refinement statistics are provided in Tables 1 and 2. In all the structures, the electron density map of the FGHIJ complex was better than that of the ABCDE complex, and this complex was selected for analysis.
Table 1.
Data collection and refinement statistics (matched base pairs)
| PolIVdG:dCMPnPP | PolIVdC:dGMPnPP | PolIVdA:dTMPnPP | PolIVdT:dAMPnPP | |
|---|---|---|---|---|
| Data Collection | ||||
| Wavelength (Å) | 1.0 | 1.0 | 1.0 | 1.0 |
| Space group | P21 | P21 | P21 | P21 |
| Cell (Å) dimensions | 86.14, 56.95, 110.81 | 86.76, 57.04, 110.7 | 85.75, 57.25, 110.3 | 86.69, 56.88, 110.93 |
| (°) | 90.00, 93.53, 90.00 | 90.00, 94.76, 90.00 | 90.00, 90.44, 90.00 | 90.00, 93.99, 90.00 |
| Resolution (Å) | 2.67 (2.81–2.67)a | 2.33 (2.46–2.33)a | 2.38 (2.38–2.51)a | 2.48 (2.61–2.48)a |
| Rsym or Rmerge | 7.4 (52.9) | 6.1 (51.4) | 5.4 (51.8) | 5.9 (52.2) |
| I/δI | 10.7 (2.6) | 10.0 (2.2) | 12.7(2.3) | 11.3(2.3) |
| Completeness (%) | 99.7 (100) | 99.4 (99.5) | 99.9 (100) | 100 (99.9) |
| Redundancy | 3.5 (3.5) | 3.7 (3.4) | 3.6 (3.6) | 3.8 (3.8) |
| Refinement | ||||
| Resolution (Å) | 45.30–2.67 | 47.66–2.33 | 47.6–2.38 | 43.28–2.48 |
| No. of reflections | 29 306 | 44 007 | 41 474 | 36 659 |
| Rwork/Rfree | 22.5/27.5 | 22.1/27.4 | 21.2/26.1 | 22.6/27.3 |
| No. of atoms | ||||
| Protein | 5376 | 5376 | 5376 | 5376 |
| DNA | 1356 | 1356 | 1356 | 1356 |
| dNMPnPP | 58 | 62 | 60 | 56 |
| Ion | 4 | 4 | 4 | 4 |
| Water | 93 | 123 | 301 | 106 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.011 | 0.008 | 0.007 | 0.011 |
| Bond angles (°) | 1.424 | 1.108 | 1.136 | 1.727 |
aHighest resolution shell is shown in parentheses.
Table 2.
Data collection and refinement statistics (mismatched base pair)
| PolIVdA:dCTP | |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.0 |
| Space group | P21 |
| Cell (Å) dimensions | 86.8, 56.98, 111.34 |
| (°) | 90.00, 94.34, 90.00 |
| Resolution (Å) | 2.32 (2.45–2.32)a |
| Rsym or Rmerge | 10.2 (53.0) |
| I/δI | 8.4 (2.7) |
| Completeness (%) | 99.2 (99.2) |
| Redundancy | 3.4 (3.5) |
| Refinement | |
| Resolution (Å) | 35.2–2.32 |
| No. of reflections | 85474 |
| Rwork/Rfree | 23.4/27.8 |
| No. of atoms | |
| Protein | 5376 |
| DNA | 1296 |
| dNMPnPP | 56 |
| Ion | 2 |
| Water | 324 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.906 |
aHighest resolution shell is shown in parentheses.
Primer extension assays for wild-type and mutant
The Ser42Ala mutant was generated using the Quickchange kit (Stratagene), and the presence of the mutation was confirmed by sequencing. The mutant enzyme was purified using a protocol similar to that for wild-type (wt) PolIV. The presence of the mutation was also confirmed using mass spectrometry. Primer extension assays to generate fidelity profiles for wild-type and mutant enzymes were carried out as described elsewhere (34). Four template oligonucleotides (TA, TT, TG and TC) were purchased from Sigma Genosys. TA, TT, TG and TC were designed to present four different nucleotides A, T, G and C, respectively, at the templating position in the active site. DNA substrates TAP*, TTP*, TGP* and TCP* prepared by annealing the fluorescently labelled primer P* (5′-6-FAM-CGTACTCGTAGGCAT-3′) with each of the four 50mer templates TA (TCCTACCGTGCCTACCTGAACAGCTGGTCTCGCTAATGCCTACGAGTACG), TT (TCCTACCGTGCCTACCTGAACAGCTGGTCACACATATGCCTACGAGTACG), TG (TCCTACCGTGCCTACCTGAACAGCTGGTCACATAGATGCCTACGAGTACG) and TC (TCCTACCGTGCCTACCTGAACAGCTGGTCATAGTCATGCCTACGAGTACG), respectively. For primer extension assay, the reaction mixture (20 μl) consisted of 5 μM of dNTPs, 20 nM of DNA substrate, 0.1 mM ammonium sulphate, 2.5 mM MgCl2, 4 μl of 5× assay buffer (125 mM Tris–Cl, pH 8.0, and 5 mM DTT) and 10 nM of PolIV (wt or mutant). After incubation for 2 h at 37°C, the reaction was terminated by adding 10 μl of stop solution (80% formamide, 1 mg/ml of xylene cyanol, 1 mg/ml bromophenol blue and 20 mM ethylenediaminetetraacetic acid) followed by 2 min of incubation at 95°C. This mixture was immediately transferred to ice for 10 min. In all, 15 μl of this sample was loaded onto a 20% polyacrylamide gel containing 8 M urea, and 1× TBE was used to resolve the reaction products. The gel was pre-run for at least 60 min before loading the samples. Resolved products on the gel were observed by excitation at 488 nm, and the bands were visualized and recorded using Biorad Pharos FX Plus Molecular Imager. The intensity of the observed bands was quantified using Quantity one, 1D analysis software. The level of incorporation was calculated using the following equation: Percentage incorporation = 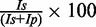 where Is = intensity of band that has shifted upwards because of incorporation, and Ip = intensity of the non-shifted primer band in the same lane. All assays were repeated thrice, and the results were reproducible. The standard error in the measured values of the intensities of the bands was <15%.
where Is = intensity of band that has shifted upwards because of incorporation, and Ip = intensity of the non-shifted primer band in the same lane. All assays were repeated thrice, and the results were reproducible. The standard error in the measured values of the intensities of the bands was <15%.
In vivo assay to assess mutator activity of wild-type and mutant PolIV
Wild-type and mutated dinB genes were cloned into a modified pET22b(+) vector to generate the plasmids pDJNNdb and pDJNNdbSER42ALA. The PolIV deleted strain dinB749 (del)::kan was obtained from the Coli Genetic Stock Centre (Yale university). Freshly prepared competent cells of this strain were transformed separately with the plasmids pDJNNdb, pDJNNdbSER42ALA and empty plasmid. A single colony for each of these samples was used to inoculate starter cultures at 37°C. Twenty-five millilitres of sterile LB media was prepared for each sample and inoculated with 100 µl of log-phase cells from each starter culture. These cultures were allowed to grow overnight at 37°C after which the cells were pelleted down by centrifugation. The cells were then resuspended in 500 µl of LB media, and 100 µl each of this suspension was plated on five different plates containing LB-agar media with 100 μg/ml of rifampicin. In addition, dilutions were also plated on LB agar without rifampicin to determine viability of the cells. The plates were incubated overnight at 37°C, and the number of colonies obtained were counted and compared. This assay was repeated three times to ensure reproducibility of the observations.
Data deposition
Atomic coordinates and structure factors have been deposited in the PDB with accession codes 4IRC (PolIVdG:dCMPnPP), 4IR9 (PolIVdC:dGMPnPP), 4IR1 (PolIVdA:dTMPnPP), 4IRD (PolIVdT:dAMPnPP) and 4IRK (PolIVdA:dCTP).
RESULTS
Structure of ternary complexes of PolIV
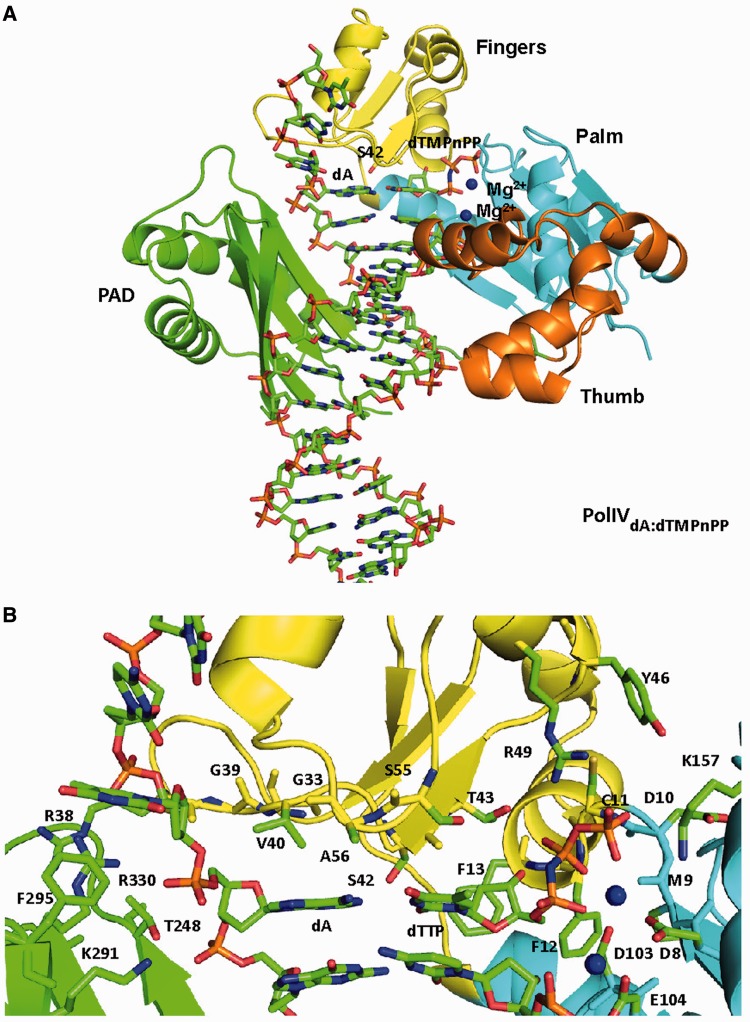
We have determined the structure of four functional complexes of PolIV corresponding to the four possible Watson–Crick pairs. To our knowledge, these are the first structures of a prokaryotic Y-family DNA polymerase captured in the functional state. The four complexes PolIVdG:dCMPnPP, PolIVdC:dGMPnPP, PolIVdA:dTMPnPP and PolIVdT:dAMPnPP crystallized in the space group P21 with roughly similar cell constants of a = 86.7 Å, b = 56.9 Å, c = 111.3 Å, α = γ = 90° and β = 94.3° (Table 1). X-ray diffraction data could be collected to a maximal resolution of 2.67, 2.33, 2.38 and 2.48 Å for the PolIVdG:dCMPnPP, PolIVdC:dGMPnPP, PolIVdA:dTMPnPP and PolIVdT:dAMPnPP complexes, respectively. In all the structures, the asymmetric unit is composed of two complexes. The final refined structure for each complex includes PolIV residues 1–342, nucleotides 1–18 for the template strand, nucleotides 1–14 to 17 for the primer strand, incoming dNTP, two Mg2+ ions and water molecules (Figure 1A). The structures show the presence of the four domains characteristic of Y-family dPols, namely, the palm (residues 1–10, 74–165), fingers (11–73), thumb (166–230) and the PAD or little finger (241–341) (Figure 1A). The palm domain houses the catalytic residues—Asp8, Asp103 and Glu104.
Figure 1.
Structure of ternary complex (PolIVdA:dTMPnPP). (A) The structure of PolIV is displayed in ribbon representation with the palm domain coloured cyan, fingers domain coloured yellow, thumb domain coloured orange, PAD domain coloured green and linker between Thumb and PAD domain coloured wheat. DNA is displayed in stick representation, and atoms are coloured according to element. The template and incoming nucleotide—dA and dTMPnPP—are coloured according to element. The Mg2+ ions are shown in the form of blue spheres. (B) Active site of PolIV. A close up of the active site of PolIVdA:dTMPnPP is shown with the residues that form interactions with the template dA (G33, R38, G39, V40, S42, A56, T248, K291, F295 and R330) and incoming dTMPnPP (D8, M9, D10, C11, F12, F13, S42, T43, R49, S55, D103 and K157) displayed in stick representation. DNA residues and incoming nucleotide are also displayed in stick representation, and all the residues are coloured according to constituent elements. The Mg2+ ions are shown in the form of a blue spheres.
In all the complexes, one Mg2+ ion is coordinated in octahedral geometry by the oxygen atoms of β- (2.3 Å) and γ-(2.3 Å) phosphates of the dNTP molecule and the carboxylates of Asp8 (2.2 Å) and Asp103 (2.3 Å) in a plane. The coordination sphere is completed by the oxygen atom of the α-phosphate (2.2 Å) and the main chain carbonyl of Met9 (2.3 Å) at the apical positions. This Mg2+ is the equivalent of metal ‘B’ of replicative polymerases and should be involved in the two-metal ion mechanism of DNA synthesis (35). The 3′ oxygen of the primer is present close to the α-phosphate of the incoming dNTP and is aligned almost linearly with the bond between the α-phosphate and O3A, suggesting that the enzyme is primed for catalysis and has been captured in the pre-catalytic state. The second Mg2+ ion equivalent of the metal ‘A’ is located in a position similar to that observed in the functional complexes of human DNA Polη and Polβ (36,37).
Active site residues that interact with the incipient base pair
In all four structures, the incipient base pair is present in a cavity and is lined by the residues of three domains—palm, fingers and PAD (Figure 1B). Arg330 of the PAD forms interactions with the phosphodiester backbone of the template nucleotide. The template nucleotide interacts with residues of the fingers domain (Gly33, Arg38, Gly39, Val40, Ser42 and Ala56) and the PAD region (Thr248, Lys291, Phe295 and Arg330). The triad of Arg, Tyr and Lys that is present in all Y-family dPols and serves to stabilize the triphosphate moiety of the incoming nucleotide is also present in PolIV (Tyr46, Arg49 and Lys157) (38). However, unlike other Y-family dPols, the Tyr residue (Tyr46) from this invariant triad is not involved in making contacts with the incoming nucleotide. In addition to Arg49 and Lys157, the residues Met9, Cys11, Phe12 and Thr43 also form polar interactions with the triphosphate moiety of the incoming nucleotide. Overall, the incoming nucleotide is present in the close proximity of residues of the palm (Asp8, Met9, Asp10 and Asp103) and fingers (Cys11, Phe12, Phe13, Ser42, Thr43, Arg49 and Ser55) domains (Figure 1B).
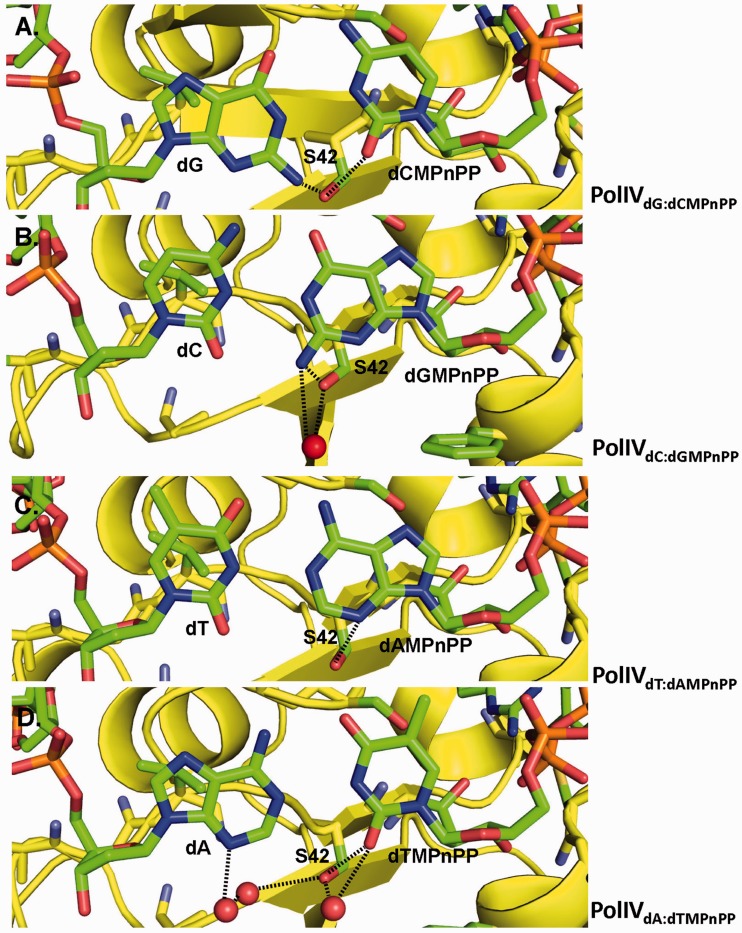
Ser42 forms interactions with the base moieties of the nascent base pair
The side chains of majority of the residues that line the active site form van der Waal interactions with the atoms of the base moieties of the incipient base pair. However, unlike the other residues, Ser42 forms direct and water-mediated polar interactions with the atoms of the bases in the nascent base pair in all four complexes (Figure 2). This residue is located close to the midline of the Watson–Crick base pair towards the minor groove and is, therefore, positioned correctly to interact with the base atoms. In the case of PolIVdG:dCMPnPP complex, the –OH group of the side chain of this residue forms a hydrogen bond with the N2 atom (2.7 Å) of the template guanine and the O2 atom (3.3 Å) of the incoming nucleotide dCMPnPP (Figure 2A). In the PolIVdC:dGMPnPP complex, this residue forms both direct (3.0 Å) and water-mediated hydrogen bonds with the N2 atom of dGMPnPP (Figure 2B). For the PolIVdA:dTMPnPP complex, Ser42 forms direct (2.9 Å) and water-mediated interactions with the O2 atom of dTMPnPP (Figure 2C). In addition, Ser42 interacts with the N3 atom of dA through two bridging water molecules. Finally, in the case of the PolIVdT:dAMPnPP complex, Ser42 forms an interaction with the N3 atom (3.2 Å) of dAMPnPP (Figure 2D). Overall, it is clear that the Ser42 residue interacts with the bases of the nascent base pair and could influence fidelity of replication.
Figure 2.
Ser42 interacts with bases of the nascent base pair. The figure shows the interactions between Ser42 and (A) template dG and incoming dCMPnPP in the case of PolIVdG:dCMPnPP, (B) incoming dGMPnPP in the case of PolIVdC:dGMPnPP, (C) template dA and incoming dTMPnPP in the case of PolIVdA:dTMPnPP and (D) incoming dAMPnPP in the case of PolIVdT:dAMPnPP. The amino acid and nucleotide residues are shown in stick representations and coloured according to element, and water molecules are shown in the form of red spheres. The direct and water-mediated hydrogen bonds formed between Ser42 and the bases are highlighted by dotted lines.
Sequence analysis of orthologues using the KEGG database shows that a Ser residue at position 42 is conserved in a majority of close prokaryotic homologues of PolIV (Supplementary Figure S1). Also, PolIV structure was superimposed with available structures of other Y-family dPols to ascertain the identity of the residue present at a position equivalent to that of Ser42. A serine residue is present at the equivalent position in the eukaryotic dPol kappa (human; S137). In the only available structure of Polκ with undamaged DNA, (PolκdA:dTTP) S137 is located at a distance of 3.5 Å (Supplementary Figure S2) from the O2 atom of dTTP (39). Serine is not found in an equivalent position in other eukaryotic Y-family dPols, such as ι (human; V64) (40), η (human, L48; yeast L60) (36,41) and Rev1 (human, A509; yeast A401) (42,43) and known archaeal dPols Dpo4 (Sulfolobus solfataricus, A44) (44) and Dbh (Sulfolobus acidocaldarius, A44) (45). Thus, a polar residue is present at this location only in case of orthologues of PolIV and Polκ. Additionally, available structures of ternary complexes of dPols belonging to the A, B, C, X and Y families do not show the presence of an active site residue like Ser42 that forms direct and water-mediated polar interactions with the base moieties in all four nascent Watson–Crick base pairs.
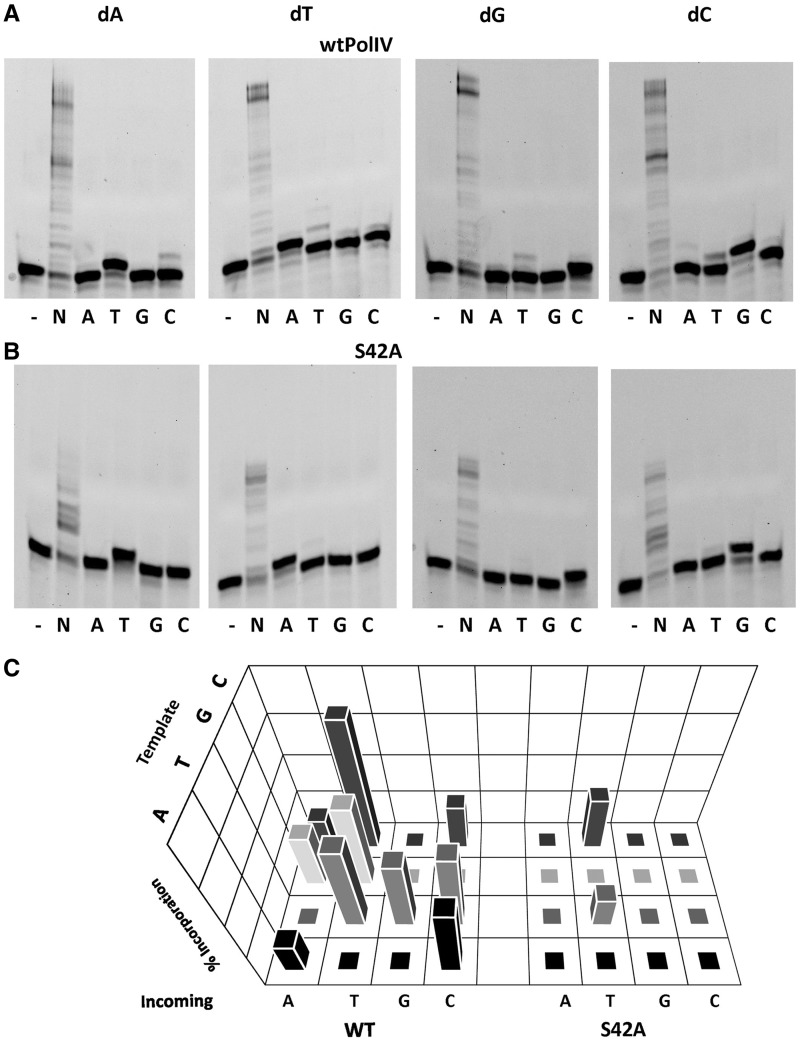
The Ser42Ala mutant exhibits higher fidelity than wild-type enzyme
To further probe the role of Ser42 in influencing fidelity, we mutated this residue to Ala. Primer extension assays were carried out to assess the ability of the mutant protein to incorporate the correct and incorrect nucleotide opposite all four possible template nucleotides in comparison with native PolIV. In the presence of the correct nucleotide, both wt and mutant enzyme showed maximal extension of the primers. For the reaction conditions used, ∼80% of the primer was extended on addition of the correct nucleotide in case of all the templates (Figure 3A). However, the wild-type (wt) enzyme exhibited significant misincorporation for a number of different combinations of template and incoming nucleotide (Figure 3A). For template nucleotide dA, the wtPolIV exhibited significant incorporation of dCTP. Also, all three incorrect nucleotides dTTP, dGTP and dCTP could be incorporated opposite template dT. Additionally, dATP and dTTP were added opposite template dG. Finally, when the template nucleotide was dC, there was significant misincorporation in the presence of all three incorrect nucleotides (Figure 3A) with maximal extension in case of dTTP (∼20%). In comparison, the Ser42Ala mutant exhibited substantially higher fidelity than the native enzyme with no misincorporation opposite the two template nucleotides dA and dG (Figure 3B). Marginal incorporation of dTTP opposite dT was observed and misincorporation opposite dC was considerably reduced and observed only in case of dTTP (∼7%). Also, in case of wtPolIV, significant incorporation of dTTP, dATP and dCTP was observed opposite nucleotides 5′ of the correct template nucleotide dA, dT and dG, respectively (Figure 3A). However, in case of the Ser42Ala, these additions were not observed (Figure 3B). Overall, the primer extension assays show that the fidelity of the Ser42Ala mutant is substantially higher than the wtPolIV (Figure 3C).
Figure 3.
Fidelity profile of wt PolIV and the mutant Ser42Ala. The results for primer extension assay wherein wtPolIV (A) or the Ser42Ala mutant (B) was incubated with DNA duplexes (TAP*, TTP*, TGP* and TCP*) and different dNTPs are displayed. (B) 3D plot exhibiting the misincorporation profile of wtPolIV and the mutant. Incorporation was quantitated in the form of percentage of primer extended. The level of incorporation of the incorrect nucleotide is plotted on the z-axis for different template nucleotides (dA, dT, dG and dC; y-axis), for all four incoming nucleotides (dATP, dTTP, dGTP and dCTP; x-axis) for the two proteins (wtPolIV and Ser42Ala; x-axis).
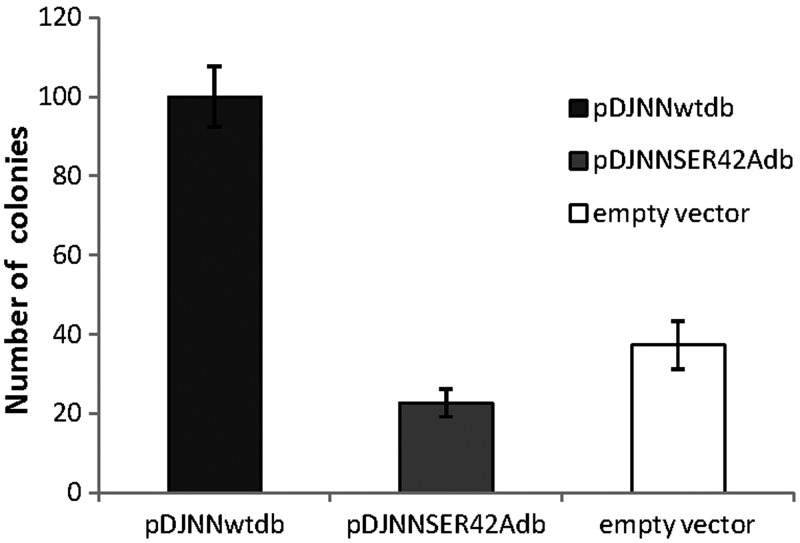
We carried out an in vivo assay to estimate the mutator activity of Ser42Ala mutant in comparison with wtPolIV. The assay probed for the appearance of rifampicin-resistant colonies when PolIV was expressed ectopically from a plasmid in a strain wherein the dinB gene had been deleted. The bactericidal activity of rifampicin has been attributed to its ability to bind close to the active site of the β-subunit of RNA polymerase (Rpo) and prevent elongation of the nascent RNA transcripts (46). It has been seen earlier that substitution mutations at key locations in the rpoB gene render resistance to E. coli against this antibiotic. Consequently, the designed assay is an appropriate tool to evaluate the ability of wtPolIV and the Ser42Ala mutant to generate substitution mutations. The results clearly show that a plasmid bearing the wtPolIV gene gave rise to nearly five-fold the number of colonies observed for a plasmid with the Ser42Ala mutant or for empty vector (Figure 4). Also, in this assay, the number of colonies obtained with the plasmid bearing the mutated dinB gene and empty vector were nearly equal. These observations show that the Ser42 residue plays a critical role in the ability of PolIV to create substitution mutations.
Figure 4.
Mutator activity of wtPolIV and Ser42Ala mutant. The bar graph displays the number of colonies obtained on plates containing rifampicin (100 µg/ml) after overnight incubation (37°C) of a dinB deleted strain [dinB749(del)::kan] transformed with (i) pDJNNwtdb, (ii) pDJNNSER42ALAdb and (iii) empty vector.
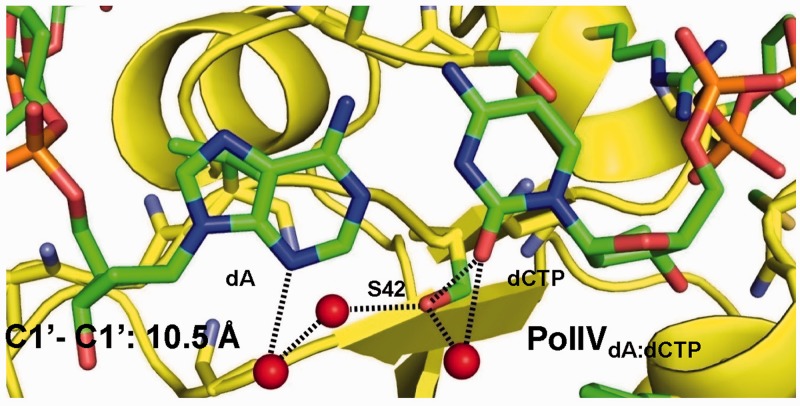
Structure of PolIVdA:dCTP
The PolIVdA:dCTP complex was crystallized in the same conditions as the other complexes, and X-ray diffraction data could be collected to a maximal resolution on 2.32 Å. The final structure refined to a final Rfree and Rcryst of 27.8 and 23.4%, respectively (Table 2). The real space correlation coefficient for the incoming dCTP was found to be 1.0 using PHENIX. The average B-factor for incoming dCTP was 46 Å2. The presence and conformation of the dCTP was confirmed through calculation of a simulated annealed omit map (Supplementary Figures S3A). Superimposition of the PolIVdA:dCTP with that of the PolIVdG:dCMPnPP and PolIVdA:dTMPnPP yields rmsds of 0.35 and 0.43 Å, respectively (341 Cα atoms). Additionally, the residues lining the active site do not exhibit any change in conformation (Supplementary Figure S3B). Hence, PolIV can stabilize the dA:dCTP base pair with minimal distortion in the enzyme structure. This is unlike what has been reported for Polβ and Dpo4. In the case of Polβ, stabilization of the mismatches A:C and T:C can only occur if the N-subdomain attains a conformation that is not compatible with productive catalysis (47). Additionally, in the case of Dpo4dT:dGTP complex, the bases of the template and incoming nucleotides are not coplanar and the two nucleotides refine to high B-factors (>100 Å2) (48).
In the PolIVdA:dCTP complex, the Ser42 residue forms direct interactions with the O2 atom of dCTP nucleotide (3.0 Å). In addition, this residue also forms water-mediated hydrogen bonds with the incoming and template nucleotide (Figure 5). The enzyme stabilizes the mismatch in a configuration similar to that of the Watson–Crick base pair with a C1′–C1′ distance of 10.5 Å (Figure 5). In the observed configuration, two hydrogen bond acceptors (N1 from dA and N3 from dCTP:2.7 Å) and two hydrogen bond donors (N6 from dA and N4 from dCTP:2.8 Å) are placed close to each other. Unless the N3 of dCTP is protonated, there does not seem to be any hydrogen-bonding possible between the adenine and cytosine bases. The interaction of the Ser42 residue with the template dA and the incoming dCTP, therefore, ensures that the incoming nucleotide is stabilized in a conformation compatible with productive catalysis. This observation is consistent with the inference that the Ser42 residue plays a critical role in the ability of PolIV to create substitution mutations.
Figure 5.
A:C base pair in the Pol IV active site. The Ser42 and other residues lining the active site are coloured according to element and shown in stick representation. Water molecules are shown in the form of red spheres. The direct and water-mediated interactions formed between Ser42 and the O2 atom of incoming dCTP and N3 atom of dA, respectively, are shown in the form of dotted lines.
DISCUSSION
A number of studies over the years suggest that replicative dPols ensure high fidelity of DNA synthesis primarily through geometric selection of the 3D shape of Watson–Crick base pairs (49–51). To ensure this, the active site residues will have to generate a 3D mould into which the Watson–Crick base would fit perfectly in an orientation that would facilitate catalysis. Any perturbation in the active site should, therefore, lead to a loss of fidelity. In line with this, mutational analysis of different replicative dPols, such as RB69, Polδ and Polγ, has shown that active site residues generally play a role in ensuring fidelity. Substitution of key active site residues with Ala or Gly results in significant decrease in the fidelity of the enzyme (52–56). In case of extensive studies carried out on RB69, some of the designed mutations did not impair the ability of the polymerase to select the correct nucleotide but allowed stabilization of mismatches in the active site to reduce fidelity. Conversely, in case of E. coli PolIV, substitution of Ser42 with Ala enhances fidelity and reduces the ability of the enzyme to promote mismatches. The structures presented in this study suggest that the Ser42 residue interacts with incorrect incoming nucleotides and stabilizes them in the PolIV active site to ensure their addition to the growing primer strand. Hence, the Ser42 residue plays a critical role in the ability of PolIV to create transition and transversion mutations. Overall, it seems that placement of specific side chains at key locations in the active site of DNA polymerases can have distinct effects on the fidelity of these enzymes.
Although, Ser42 forms interactions with the base atoms of the four possible incoming nucleotides, wtPolIV exhibits variation in its ability to promote different mismatches. It is seen that PolIV is unable to stabilize purine:purine mismatches with dGTP as the incoming nucleotide in its active site. Additionally, only low level of incorporation is seen for purine:purine mismatches with dATP as the incoming nucleotide. The C1′–C1′ distance in the case of canonical Watson–Crick base pairs is 10.6 Å. An analysis of non–Watson–Crick base pairs by Leontis et al. (57) shows that the C1′–C1′ distance and profile of hydrogen-bonding between bases can exhibit variation for different mispairs. For purine–purine base pairs, this distance is ∼12.5 Å, and this increase in the horizontal extent of the base pairs could give rise to steric clashes with the enzyme residues. These steric clashes would adversely influence stabilization of purine–purine mismatches in the PolIV active site. Overall, it is possible that the observed variability in incorporation of different mismatches could be due to the variation in the destabilizing effects of steric interactions that differentially offset the positive contributions arising from interaction of the incoming nucleotide with the Ser42 residue and variable number of hydrogen bonds formed between the mispaired bases.
It should be noted here that the error profiles reported here show marked differences from that observed in earlier studies. Kobayashi et al. (22) assessed fidelity of replication of the lacZα gene, whereas Wagner and Nohmi (21) did the same for the cII gene from λ-phage). Although both studies showed that frameshift mutations dominate the mutation spectrum, the contribution of substitution mutations was significantly higher in case of replication of the cII gene. Additionally, it was seen that although substitution mutations in case of the LacZα gene were dominated by T–G transversion (19 of 66 substitutions) and T–C transition (16 of 66 substitutions), these mutations were not observed in case of replication of the cII gene. Thus, the frequency of different substitution mutations exhibited substantial differences in the two studies. The propensity of PolIV to promote different mismatches reported in the present study is distinct from those observed earlier, suggesting that sequence context in substrate DNA influences the ability of PolIV to stabilize different mismatches. Thus, the mutation spectrum generated by PolIV could be complex, and it is possible that this attribute might ensure improved sampling of the mutation space to aid adaptive mutagenesis.
PolIV is part of the machinery responsible for transient enhancement of mutation rates in subpopulations of a culture of E. coli (14,18,58,59). The frequency of mutation during adaptive mutagenesis has to be low enough to prevent cell death of the entire pool because of mutational load and high enough to generate enough number of distinct genomic templates for natural selection. As only 5–25% of primer is extended for the different mismatches, the presence of a Ser residue at the correct location in the PolIV active site could allow calibration of mutagenesis within these required limits. The observations made during the course of this study suggest that Ser42 reduces fidelity to a level that will ensure enough genomic variability for natural selection without compromising genetic viability. The strategically placed Ser42 residue could, therefore, play an important role in the appearance of cells that possess sets of mutations that might allow survival in a stressful environment, and thus relieve selection pressure.
ACCESSION NUMBERS
4IRC, 4IR9, 4IR1, 4IRD and 4IRK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
NCBS-TIFR (Department of Atomic Energy); Council of Scientific and Industrial Research (CSIR: Sanction Letter No. 37/(1539)/12/EMR-II); Ramanujan fellowship from the Department of Science and Technology (Government of India) (to D.T.N.); Senior and Junior research fellowship from CSIR (Government of India) (to A.S. and J.K., respectively); Junior research fellowship from the Indian Council of Medical Research (ICMR) (to N.N.). Data collection at the BM14 beamline of ESRF (Grenoble, France) was funded by the BM14 project—a collaboration between Department of Biotechnology, EMBL and ESRF. Funding for open access charge: Extramural grant from the council of Scientific and Industrial Research (CSIR).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
D.T.N. thanks the X-ray diffraction facility located in the Molecular Biophysics Unit of the Indian Institute of Science [funded by Departments of Biotechnology (DBT) and Science and Technology (DST), Government of India] for facilitating screening and data collection. D.T.N. acknowledges the help rendered by Dr Hassan Belrhali (ESRF) and Dr Babu Manjashetty during data collection at the BM-14 beamline of ESRF and by Meitian Wang and Sandro Waltersperger at the PXIII beamline of SLS. Figures were generated using PyMOL (Schroedinger Inc.). The authors would like to thank Prof. Jayant B. Udgaonkar for critically reading the manuscript.
REFERENCES
- 1.Saint-Ruf C, Matic I. Environmental tuning of mutation rates. Environ. Microbiol. 2006;8:193–199. doi: 10.1111/j.1462-2920.2005.00968.x. [DOI] [PubMed] [Google Scholar]
- 2.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster PL. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Tippin B, Pham P, Goodman MF. Error-prone replication for better or worse. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Pham P, Bertram JG, O’Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 7.Tompkins JD, Nelson JL, Hazel JC, Leugers SL, Stumpf JD, Foster PL. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J. Bacteriol. 2003;185:3469–3472. doi: 10.1128/JB.185.11.3469-3472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl Acad. Sci. USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brotcorne-Lannoye A, Maenhaut-Michel G. Role of RecA protein in untargeted UV mutagenesis of bacteriophage lambda: evidence for the requirement for the dinB gene. Proc. Natl Acad. Sci. USA. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv. Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 12.Walsh JM, Hawver LA, Beuning PJ. Escherichia coli Y family DNA polymerases. Front Biosci. 2012;17:3164–3182. doi: 10.2741/3904. [DOI] [PubMed] [Google Scholar]
- 13.Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl Acad. Sci. USA. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 15.Strauss BS, Roberts R, Francis L, Pouryazdanparast P. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 2000;182:6742–6750. doi: 10.1128/jb.182.23.6742-6750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 17.Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, Rosenberg SM. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Capilla T, Baquero MR, Gomez-Gomez JM, Ionel A, Martin S, Blazquez J. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J. Bacteriol. 2005;187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuban W, Jonczyk P, Gawel D, Malanowska K, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J. Bacteriol. 2004;186:4802–4807. doi: 10.1128/JB.186.14.4802-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi S, Valentine MR, Pham P, O’Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- 23.Jacob KD, Eckert KA. Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat. Res. 2007;619:93–103. doi: 10.1016/j.mrfmmm.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrosino JF, Galhardo RS, Morales LD, Rosenberg SM. Stress-induced beta-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J. Bacteriol. 2009;191:5881–5889. doi: 10.1128/JB.00732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpinets T, Greenwood D, Pogribny I, Samatova N. Bacterial stationary-state mutagenesis and mammalian tumorigenesis as stress-induced cellular adaptations and the role of epigenetics. Curr. Genomics. 2006;7:481–496. doi: 10.2174/138920206779315764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirz RT, Romesberg FE. Induction and inhibition of ciprofloxacin resistance-conferring mutations in hypermutator bacteria. Antimicrob. Agents Chemother. 2006;50:220–225. doi: 10.1128/AAC.50.1.220-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Sharma A, Subramanian V, Nair DT. The PAD region in the mycobacterial DinB homologue MsPol IV exhibits positional heterogeneity. Acta Crystallogr. D Biol. Crystallogr. 2012;D68:960–967. doi: 10.1107/S0907444912017623. [DOI] [PubMed] [Google Scholar]
- 31.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Sharma A, Nair DT. MsDpo4-a DinB homolog from mycobacterium smegmatis-Is an error-prone DNA polymerase that can promote G:T and T:G mismatches. J. Nucleic Acids. 2012;2012:285481. doi: 10.1155/2012/285481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 36.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RE, Trincao J, Aggarwal AK, Prakash S, Prakash L. Deoxynucleotide triphosphate binding mode conserved in Y family DNA polymerases. Mol. Cell Biol. 2003;23:3008–3012. doi: 10.1128/MCB.23.8.3008-3012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 41.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 43.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J. Mol. Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 45.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol. Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Feklistov A, Mekler V, Jiang Q, Westblade LF, Irschik H, Jansen R, Mustaev A, Darst SA, Ebright RH. Rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNA polymerase active center. Proc. Natl Acad. Sci. USA. 2008;105:14820–14825. doi: 10.1073/pnas.0802822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuriyan J, Konforti B, Wemmer D. Molecules of Life: Physical and Chemical Principles. 2012. 1st edn. Garland Science, New York, USA. [Google Scholar]
- 51.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Beckman J, Wang J, Konigsberg W. RB69 DNA polymerase mutants with expanded nascent base-pair-binding pockets are highly efficient but have reduced base selectivity. Biochemistry. 2009;48:6940–6950. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Rhee C, Bebenek A, Drake JW, Wang J, Konigsberg W. The L561A substitution in the nascent base-pair binding pocket of RB69 DNA polymerase reduces base discrimination. Biochemistry. 2006;45:2211–2220. doi: 10.1021/bi052099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 55.Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Copeland WC, Ponamarev MV, Nguyen D, Kunkel TA, Longley MJ. Mutations in DNA polymerase gamma cause error prone DNA synthesis in human mitochondrial disorders. Acta Biochim. Pol. 2003;50:155–167. [PubMed] [Google Scholar]
- 57.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenzie GJ, Harris RS, Lee PL, Rosenberg SM. The SOS response regulates adaptive mutation. Proc. Natl Acad. Sci. USA. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez C, Hadany L, Ponder RG, Price M, Hastings PJ, Rosenberg SM. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS Genet. 2008;4:e1000208. doi: 10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.