Abstract
A number of proteins form covalent bonds with DNA as obligatory transient intermediates in normal nuclear transactions. Drugs that trap these complexes have proven to be potent therapeutics in both cancer and infectious disease. Nonetheless, current assays for DNA–protein adducts are cumbersome, limiting both mechanistic studies and translational applications. We have developed a rapid and sensitive assay that enables quantitative immunodetection of protein–DNA adducts. This new ‘RADAR’ (rapid approach to DNA adduct recovery) assay accelerates processing time 4-fold, increases sample throughput 20-fold and requires 50-fold less starting material than the current standard. It can be used to detect topoisomerase 1-DNA adducts in as little as 60 ng of DNA, corresponding to 10 000 human cells. We apply the RADAR assay to demonstrate that expression of SLFN11 does not increase camptothecin sensitivity by promoting accumulation of topoisomerase 1-DNA adducts. The RADAR assay will be useful for analysis of the mechanisms of formation and resolution of DNA–protein adducts in living cells, and identification and characterization of reactions in which covalent DNA adducts are transient intermediates. The assay also has potential application to drug discovery and individualized medicine.
INTRODUCTION
DNA–protein covalent complexes (DPCCs) form as transient intermediates in a variety of DNA transactions. In human cells, more than 20 different proteins are currently known to form DPCCs, including topoisomerases (Top) 1, 2 and 3α (1); DNA repair factors with AP lyase activity, like PARP-1 (2) and Ku (3); DNA glycosylases that repair oxidative and chemical DNA damage, including 8-oxoguanine DNA glycosylase, thymine DNA glycosylase, and endonuclease three like (NTH) and endonuclease eight-like (NEIL) family members (4,5); O6-methylguanine-DNA methyltransferase and O6-alkylguanine alkyltransferase, which repair alkylated DNA lesions (6); tyrosyl–DNA phosphodiesterase 1 (7); DNA polymerases, including Pol β (8) and Y family polymerases ι, η and κ (9); and DNA methyltransferases (DNMT) 1, 3A and 3B (10). It is likely that other proteins form transient covalent intermediates with DNA but have not yet been shown to do. Many of the proteins that can form DNA adducts are involved in DNA repair; therefore the levels of adducts are likely to increase in response to general DNA damage. However, little is known about this because it has been difficult to assay DPCC.
Difficulty in assaying DPCCs has also limited experimental analysis of mechanisms of adduct repair. Some very potent drugs function by stabilizing normally transient DPCCs to produce persistent protein–DNA adducts. These adducts block DNA replication and RNA transcription and create local DNA damage, resulting in cytotoxicity. Among drugs that trap DPCCs are the quinolone antibiotics that trap DNA gyrase to combat bacterial infection; chemotherapeutics including camptothecin (CPT), which targets Top1; etoposide and doxorubicin, which target Top2; and 5-aza-deoxycytidine (5-aza-dC) and 5-aza-C, which target DNMTs; as well as non-specific crosslinkers such as cisplatin and melphalan (11). The potency of drugs known to trap DPCCs suggests that cells have limited capacity to repair DNA adducts, and that it will be useful to develop drugs against new DPCC targets.
One commonly used assay for DPCCs is the immunocomplex of enzyme (ICE) assay, which relies on physical separation of DPCC from the bulk cellular protein by cesium chloride gradient ultracentrifugation (12). The ICE assay is unsuitable for many applications because ultracentrifugation requires large amounts of starting material (typically 2 to 10 × 106 cells per sample) and is tedious and low throughput (13). In addition, many laboratories no longer have easy access to an ultracentrifuge. The TARDIS (trapped in agarose DNA immunostaining) assay detects DPCC in as few as 100–150 cells immobilized in agarose, using antibody specific for the protein adduct (14). However, throughput is limited because the assay requires considerable sample handling, including determining fluorescent intensity of sufficient numbers of individual cells to produce statistically significant data. Another method uses chaotropic salts to isolate genomic DNA carrying covalently bound proteins, and it eliminates free proteins very effectively (15). However, that method was designed for bulk DPCC identification by mass spectroscopy, and it requires large amount of starting material and extensive handling and was not validated for immunodetection. Very recently, total cross-linked protein has been quantitated by fluorescein isothiocyanate-labelling followed by fluorimetric detection or by western blotting (16), but that approach does not identify or distinguish among specific bound proteins, and it requires ultracentrifugation.
We set out to develop a robust assay for DPCC detection that would be convenient for mechanistic studies. Such an assay must be rapid, sensitive and must use conventional reagents and equipment. Here we report development of a new assay, the RADAR (rapid approach to DNA adduct recovery) assay (Figure 1A). The RADAR assay uses a combination of chaotropic salts and detergents that effectively separates DPCC from free protein without cesium gradient centrifugation. Bound protein is quantified by immunodetection. The RADAR assay accelerates processing time 4-fold, increases sample throughput 20-fold and requires 50-fold less starting material than current standard assays. The RADAR assay opens the way to detailed studies of the mechanism and kinetics of DPCC formation and repair. It permits mechanism-based assays for drug activity, independent of cell killing or other effects. It also has potential for application to discovery of new drugs for treatment of cancer or infectious disease, for rational optimization of existing drugs and for validating drug activity on patient cells to guide treatment.
Figure 1.
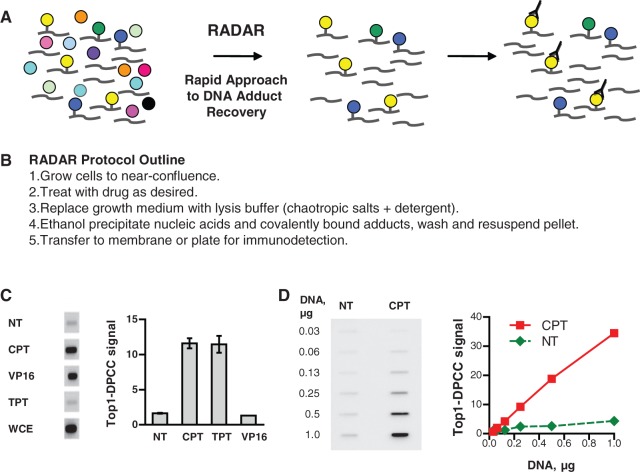
DPCC isolation and specific detection. (A) Diagram of DPCC recovery and detection. Left, nuclei contain DNA and proteins, some of which are covalently bound. Middle, DPCC are isolated along with free DNA. Right, specific DPCC are detected with antibody. (B) Outline of the protocol of the RADAR assay. (C) Specific detection of Top1–DPCC. Left, blot of DPCC isolated from HCT116 cells treated for 30 min with 10 µM Top1 inhibitors CPT or TPT; or Top2 inhibitor, VP16; and probed with anti-Top1 antibodies. NT, untreated; WCE, whole-cell extract. Right, quantification of Top1–DPCC signal in samples following each treatment. (D) Left, detection of Top1–DPCC in serial dilutions containing indicated amounts of DNA from untreated (NT) or CPT-treated HCT116 cells. Right, quantification of Top1–DPCC signal at each DNA dilution.
MATERIALS AND METHODS
Cell culture, drug treatment, viability assays and siRNA knockdown
Human cell lines were obtained from American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium or RPMI medium with 10% fetal calf serum. Cells were grown to 40–80% confluence before treatment with CPT (Sigma or Enzo Life Sciences), topotecan (TPT) (Enzo Life Sciences), VP16 (Topogen) or 5-aza-dC (Calbiochem) at indicated concentrations. Cell survival was quantified using the CellTiter-Glo® assay (Promega). Transfection of GM639 cells was performed using Lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer’s protocol, and drug response was assayed at 48–72 h post-transfection. SLFN11 small interfering RNA (siRNA) (s40702) and Silencer® Select Negative Control No. 2 siRNA were purchased from Ambion/Life Technologies.
Cell lysis and DPCC isolation
A detailed protocol for DPCC isolation is available in Supplementary Material. In brief, 5 × 105 cells were cultured in 1.5 ml, treated with drug, then culture medium aspirated and cells immediately lysed on the plate by addition of 1 ml of lysis reagent (not to exceed 2 × 106 cells per 1 ml lysis solution). Several different solutions containing guanidinium isothiocyanate (GTC) proved satisfactory for cell lysis, DPCC isolation and immunodetection. One, M buffer (MB), was non-proprietary and contained 6 M GTC, 10 mM Tris–HCl (pH 6.5), 20 mM EDTA, 4% Triton X100, 1% Sarkosyl and 1% dithiothreitol; others were based on proprietary reagents, RLT Plus (Qiagen), or DNAzol® (Life Technologies/ Invitrogen), alone or in combination with 1% Sarkosyl. An aliquot of lysate (10–20%) was saved for analysis of the unfractionated extract, and nucleic acids and DPCC were precipitated from the remainder by addition of 0.5 volume of 100% ethanol followed by centrifugation. The precipitate was washed twice in 75% ethanol and immediately resuspended in 200 µl of freshly prepared 8 mM NaOH, to achieve final concentration of 20–30 µg/ml DNA. For complete solubilization, it was essential not to allow the pellet to dry. A solution of weak base has also been used by others to solubilize DNA following ethanol precipitation (17–19), and in our hands, it worked well. Most importantly, it preserved epitopes of Top1 and DNMT1 necessary for their recognition by antibodies. It is possible that other conditions might be better for preserving critical immuno-epitopes of other proteins.
Recovered DNA was quantified by measuring fluorescence of DNA-bound PicoGreen dye (Invitrogen), as recommended by the manufacturer. DNA recovery was 5–7 µg per 106 cells, consistent with the DNA content of 7.1 pg of DNA per human cell. DNA recovery was typically sufficiently uniform that blotting and immunodetection could proceed using identical volumes of similarly processed DNA samples, and DNA content independently determined for normalization of signal to DNA. At high DNA concentrations, solutions will be viscous and may clog membranes, which can be addressed by brief sonication (20 s at 50%). Protein assays using the BCA reagent (Pierce) showed that >98% of free protein was removed in the course of DNA adduct recovery. Significant amounts of RNA were recovered on precipitation, as evident on agarose gel electrophoresis or comparison of yields quantified by A260 and PicoGreen fluorescence. RNA was readily removed by treatment with RNase A.
DPCC immunodetection
Samples were diluted in 25 mM sodium phosphate (pH 6.5) or Tris-buffered saline [TBS; 10 mM Tris (pH 7.5), 150 mM NaCl] and applied to either a polyvinylidene difluoride (PVDF, Millipore) or nitrocellulose (Bio-Rad, Hercules CA) membrane using a vacuum slot-blot manifold (Bio-Rad). The membrane was then blocked for at least 1 h in 0.5% alkali-soluble casein (Novagen) dissolved in TBS containing 0.1% Tween 20 (TBST); incubated with primary antibodies diluted in TBST for at least 3 h at 25°C or overnight at 4°C; washed three times for 5 min with TBST; then incubated with horseradish peroxidase-conjugated secondary antibodies (1:10 000 dilution in TBST) for 1 h; washed thrice with TBST; and DPCC quantified by imaging on a Bio-Rad ChemiDoc XRS Plus Analyzer, which quantifies signal automatically subtracting surrounding background. For the 96-well plate-based immunoassay, Acrowell™ 96 filter plates with nitrocellulose membranes (Pall Corporation) were used and signal detected with a plate reader (FLUOstar Omega, BMG Labtech) in chemiluminescence mode. Each well of a 96-well plate has a 3.5-fold larger membrane area than a standard slot blot manifold (25 and 7 mm2, respectively) and requires a corresponding increase in amount of sample.
Sample loading was normalized based on DNA content as determined by PicoGreen fluorescence. Top1 was detected with rabbit polyclonal human anti-Top1 antibodies (ab28432, Abcam) at 1:2000 dilution. DNMT1 was detected with mouse monoclonal anti-human DNMT1 antibodies (ab13537, Abcam) at 1:2000 dilution. Specificity of each antibody and appropriate dilution was established in control experiments.
For assays of response to a range of drug doses, the ratio of signal from treated samples to the untreated control was determined and denoted as DPCC fold induction. In some cases, biological replicates of assays performed on different days were compared by normalization of weighted signals across immunoblots, using a procedure analogous to that used to compare results from different microarrays (20).
The appropriate range of the RADAR assay will depend on cell type, drug and antibody potency, and level of total DPCC that can form. This can be determined experimentally by an initial screen of 0.2–0.8 µg DNA/slot and subsequently refined. Antibody specificity should be independently verified by western blot. If good antibodies to an endogenous protein are not available, the RADAR assay can be used effectively on cells expressing a tagged version of the protein of interest (not shown).
RESULTS
Specific and sensitive detection of Top1–DPCC
Isolation of DPCCs from cells requires efficient cell lysis and clean separation of covalent DNA–protein adducts from free protein in conditions that preserve protein epitopes for immunodetection. We therefore tested solutions containing the chaotropic salt, GTC, which has been widely used for separation of both DNA and RNA from bulk protein in cell lysates, and permits nearly quantitative recovery of nucleic acid by ethanol precipitation (15,18). We assayed recovery of covalent Top1–DNA adducts from GM639 human fibroblasts treated with the Top1 poison, CPT, and found that a solution containing GTC, non-ionic detergents (Sarkosyl and/or Triton X-100) and dithiothreitol permitted recovery of Top1–DPCC, as did two proprietary reagents of apparently similar composition (Supplementary Figure S1).
We established specificity of DPCC recovery by assaying HCT116 cells, a colorectal line, treated for 30 min with either CPT or the related compound TPT, both of which trap Top1–DPCC; or etoposide (VP16), which traps Top2–DPCC. Cells were lysed in GTC/Sarkosyl/dithiothreitol (DTT), nucleic acids and DPCCs recovered by ethanol precipitation, applied to a membrane and probed with anti-Top1 antibodies. A clear signal was evident in samples from cells treated with the Top1 poisons CPT and TPT, but not from cells treated with VP16, and quantification confirmed that CPT and TPT specifically induced Top1–DPCC (Figure 1B).
To determine RADAR assay sensitivity for Top1–DPCC, we assayed serial dilutions of purified DPCC from CPT-treated or untreated HCT116 cells, corresponding to from 30 to 1000 ng of genomic DNA. Top1 signal increased with DNA concentration in samples from CPT-treated cells; there was essentially no background in untreated cells (Figure 1C). The assay was linear with amounts of DNA as low as 60 ng of DNA per well, corresponding to 104 cells per well. Thus, the RADAR assay is suitable for testing small numbers of cells, which will make it useful for analysis of primary tissues from animals or human tumours.
We tested technical reproducibility by comparing assays of DPCC induction as determined by blotting 0.4 or 0.8 µg of DNA. The curves were comparable (Supplementary Figure S2A); therefore, the technical reproducibility is very good. To determine whether cell density may influence DPCC induction, we compared Top1–DPCC induction in GM639 cells grown to either 40 or 80% confluence before drug treatment (Supplementary Figure S2B). The more confluent culture exhibited greater DPCC induction at lower drug dose and greater maximal induction. The possibility that relatively minor differences in growth conditions that affect cell density may affect DPCC induction must be borne in mind as a source of day-to-day quantitative variation.
Induction and quantification of Top1–DPCC
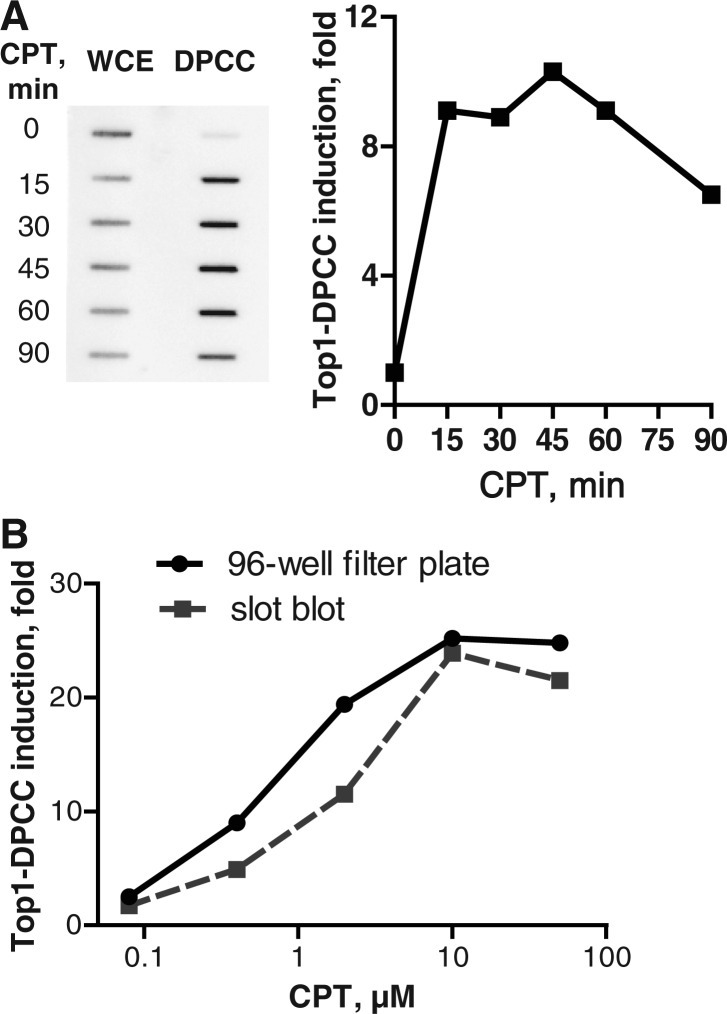
We analysed induction of Top1–DPCC in GM639 cells treated with 10 µM CPT, measuring fold increase relative to untreated cells. Top1–DPCCs were evident at 15 min and persisted at a fairly constant level through 60 min, then began to diminish (Figure 2A). Total Top1 in whole cell extracts of GM639 cells decreased in the course of CPT treatment, consistent with reported proteolysis of Top1 that occurs in response to transcriptional arrest in CPT-treated cells (21). However, this was not evident in all cell lines (not shown).
Figure 2.
Top1–DPCC immunoassay. (A) Kinetics of Top1–DPCC induction. Left, slot blot comparing Top1 signal in whole-cell extract (WCE) or DPCCs isolated from GM639 cells, which were treated with 10 µM CPT for indicated number of minutes. Right, quantification of fold induction of Top1–DPCC at indicated time, normalized to signal from untreated (t = 0) cells. (B) Comparison of Top1–DPCC induction as assayed by slot blotting or 96-well filter plate chemiluminescent detection. Assays were performed in parallel on samples from GM639 cells treated with indicated doses of CPT.
We then compared two different approaches to immobilizing and quantifying DPCC. DPCC isolated from GM639 cells treated for 30 min with CPT were applied to a slot blot and detected by imaging; or applied to a 96-well microtiter plate with nitrocellulose membrane forming the bottom of each well, then detected with a chemiluminescent reagent and read with an automated plate reader. Quantification by these two methods produced very similar results (Figure 2B). Use of 96-well filter plates permits convenient processing and data accumulation from a large number of samples.
Mechanism-based assay of 5-aza-dC
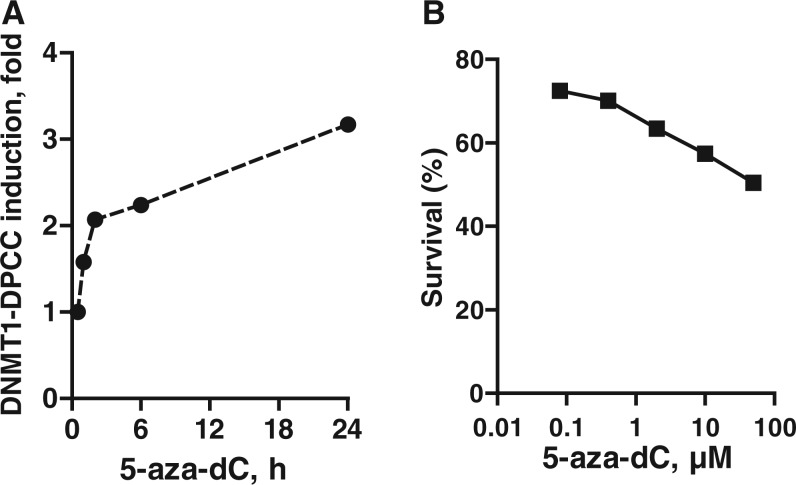
The nucleoside analogue 5-aza-dC (clinically known as decitabine or DAC) is incorporated into DNA and forms a stable covalent adduct with the DNMTs that catalyse methylation of cytosine to regulate gene expression. The 5-aza-dC is widely used to inhibit DNMTs and reactivate genes otherwise downregulated by cytosine methylation. However, there is no convenient mechanism-based assay for DNMT activity. Instead, its downstream effects on DNA methylation are typically assayed by measuring conversion of C, but not 5-methyl-C, to T following bisulfite treatment, a relatively cumbersome assay. To establish whether the RADAR assay could be used to monitor DNMT–DNA adduct formation, CCRF-CEM cells, derived from a T cell acute lymphoblastic leukaemia, were treated with 5-aza-dC and induction of covalent DNMT1–DNA complexes quantified using the RADAR assay. Rapid induction of DNMT1–DPCCs was clearly evident shortly after exposure to 10 µM DAC, and DNMT1–DPCC levels increased during the course of 24 h (Figure 3A). DAC also killed cells, with IC50 approximately 50 µM following 30 h continuous exposure (Figure 3B). This establishes utility of the RADAR assay for validating DNMT adduct formation resulting from 5-aza-dC treatment. The experiments shown queried only DNMT1, but they could be extended to discriminate effects of drug treatment on the maintenance methyltransferase, DNMT1, and the de novo methyltransferases, DNMT3A and 3B.
Figure 3.
Detection of DNMT1–DPCC generated on 5-aza-dC treatment. (A) Kinetic analysis of induction of DNMT1–DPCC in CCRF-CEM cells treated with 10 µM 5-aza-dC for indicated time. Data are normalized to maximal DNMT1–DPCC levels at 24 h. (B) Survival of CCRF-CEM cells following continuous exposure to indicated doses of 5-aza-dC.
DPCC induction correlates with CPT sensitivity
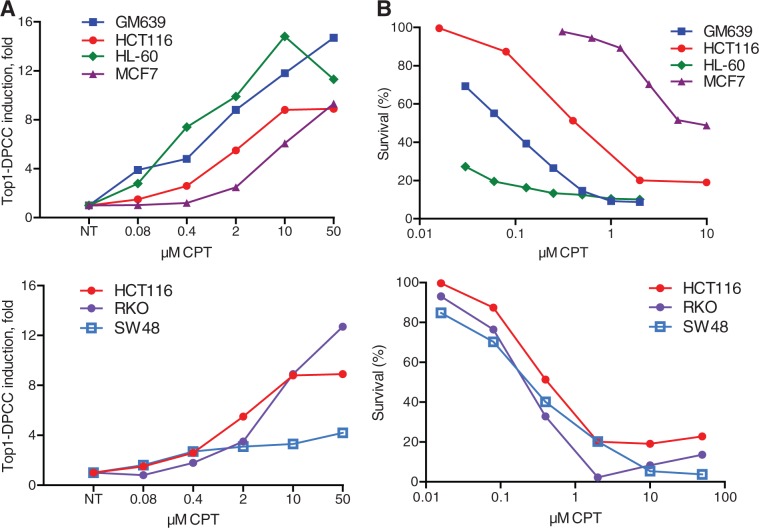
We asked whether Top1–DPCC induction correlated with CPT sensitivity in human cell lines derived from four different tissues: GM639 transformed fibroblasts; HCT116, RKO and SW48 colorectal carcinomas; HL-60 promyelocytic leukaemia, and MCF7 breast adenocarcinoma (Figure 4; colorectal lines in lower panel, and HCT116 shown in both upper and lower panels for reference). Induction of Top1–DPCC was readily evident following 30 min CPT treatment in all these lines, but different lines responded at very different doses (Figure 4A). Similarly, survival assays showed that IC50 values ranged from low (<0.1 µM for HL-60; 0.1 µM for GM639) to nearly 10 µM (MCF7). Sensitivities of the colorectal lines were in a fairly narrow range (IC50 = 0.6–0.8 µM; Figure 4B, below). In general, Top1–DPCC induction at low CPT doses correlated with CPT sensitivity, whereas higher CPT doses were necessary to induce Top1–DPCC in the more resistant lines. This validates the common assumption that the mechanism of drug action depends on accumulation of cytotoxic adducts and provides an assay that can be used to systematically test the contributions of specific factors to DNA adduct repair.
Figure 4.
Comparison of Top1–DPCC induction and cell survival in cells treated with CPT. (A) Top1–DPCC induction in indicated cell lines following 30 min exposure to indicated doses of CPT. Above, results for four cell lines from different tissues. Below, results for three colorectal cell lines. See text for cell descriptions. (B) Cell survival following 30 h continuous exposure to indicated concentrations of CPT, for cell lines tested in panel A.
For most cell lines tested, Top1–DPCC induction ranged from 6- to 15-fold at 10 µM CPT. SW48 cells were unusual, as they were comparably CPT sensitive as the other colorectal lines tested exhibited maximum induction of 2.5-fold, evident at much lower CPT concentrations (0.4 µM; Figure 4A, below). Comparison of cell extracts revealed that SW48 cells contained only 20% as much Top1 as RKO or HCT116 cells (not shown). It has been shown that, in some cell types, reductions in Top1 levels can diminish CPT sensitivity (22). This contrasts with results for SW48 and suggests that in other cell types, Top1 levels may limit DPCC induction at high CPT doses, but not limit drug sensitivity. This points to the utility of quantifying DPCC as one measure of drug response.
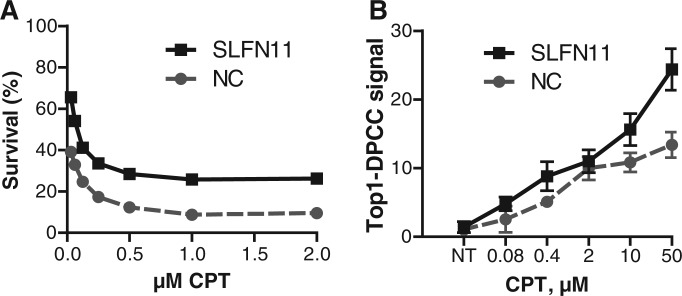
SLFN11 promotes CPT sensitivity but does not increase the burden of genotoxic DNA–protein adducts
SLFN11 is an interferon-responsive gene, and its expression has recently been shown to correlate with sensitivity to CPT and to other drugs that cause DNA damage (23,24). The mechanism by which this occurs has not been established. One possibility is that SLFN11 expression inhibits repair of DNA damage, to enhance genotoxicity owing to drug treatment. If so, then treatment of cells with siRNA to SLFN11 would be predicted to diminish both drug sensitivity and the level of Top1–DPCC. To test this, we compared survival and Top1–DPCC levels in GM639 cells treated with siRNA to SLFN11 or a control siRNA. Treatment with siRNA targeted to SLFN11 reduced CPT sensitivity (Figure 5A), as predicted. However, it did not decrease Top1–DPCC levels. Control experiments showed that SLFN11 siRNA treatment had no effect on total levels of Top1 or basal levels of Top1–DPCC (data not shown). Thus, altered Top1–DPCC levels do not explain the effect of SLFN11 on CPT sensitivity.
Figure 5.
SLFN11 siRNA treatment promotes CPT resistance but increases the burden of genotoxic DNA–protein adducts. (A) Survival of GM369 cells treated with siRNA to SLFN11 or a negative control (NC) siRNA, then exposed for 30 h to indicated doses of CPT. (B) Top1–DPCC signal in GM369 cells treated with siRNA to SLFN11 or a negative control (NC) siRNA, then exposed for 30 min to the indicated doses of CPT. Weighted signals were normalized across immunoblots to compare biological replicates of assays performed on different days, (20).
DISCUSSION
The RADAR assay permits rapid isolation of DPCCs and their quantification by immunodetection. It is sufficiently sensitive to detect CPT-induced Top1–DPCC in 20 000 human cells, conveniently cultured in microtiter format. Throughput is several hundred samples a day even without automation. The RADAR assay represents a substantial improvement over existing approaches in speed and throughput. The ICE assay (12), currently the standard for quantification of DPCC, involves many steps, with ultracentrifugation the most rate limiting. Relative to the ICE assay, the RADAR assay offers at least a 20-fold increase in throughput and 4-fold reduction in processing time, with further improvement possible with automation. Moreover, the RADAR assay requires 50-fold less material, making it suitable for experimental or clinical analysis of small numbers of cells (20 000 cells/well).
The RADAR assay should enable detailed genetic and biochemical analysis of the mechanism and kinetics of DPCC formation and resolution in living cells. For example, there is considerable interest in mechanisms that establish, maintain and eliminate cytosine methylation. Compounds like 5-aza-dC or 5-aza-C are commonly used to modulate methylation patterns, but it has not been possible to distinguish its effect on de novo methylation by DNMT3A or DNMT3B and maintenance of methylation marks by DNMT1 in specific cell types. This can now be done using the RADAR assay, distinguishing DPCCs formed by de novo and maintenance methylases with specific antibodies.
The RADAR assay can also be used to systematically test the role of specific factors in adduct repair. We have, for example, shown that although SLFN11 expression does correlate with CPT sensitivity, it does not inhibit repair of Top1–DNA adducts and thus must function by another mechanism.
The RADAR assay can also be used to establish whether formation of covalent adducts is an obligatory step in a catalytic pathway. More than 20 different human proteins are now known to form such intermediates, and it is likely that this activity is a property of other proteins, particularly in light of the fact that this activity of two intensively studied factors, PARP-1 and Ku, was demonstrated only recently (2,3). The RADAR assay need not be limited to proteins for which specific antibodies are available, as it can be used to quantify DPCC in cells expressing a tagged version of a protein of interest (not shown). The RADAR assay should also be useful for quantification of non-protein adducts for which specific detection reagents are available.
The RADAR assay has clear potential for clinical applications. For example, the RADAR assay could be used to measure DPCC induction in samples from biopsies or blood draws, thereby assessing whether a colon cancer is likely to respond to irinotecan or acute myeloid leukaemia blasts to VP16. This would identify patients who might benefit from treatment and spare others from treatment that is unlikely to be effective. The RADAR assay can also be applied to optimization of existing and newly discovered drugs to increase potency and selectivity and to drug discovery by high-throughput screening. The likely success of such mechanism-based drug discovery is highlighted by the fact that only a small fraction of the proteins known to form DPCC are currently drug targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–2 and Supplementary Methods.
FUNDING
US National Institutes of Health via National Cancer Institute [P01 CA77852 to N.M.]. Funding for open access charge: National Institutes of Health [NCI P01 CA77852 to N.M.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jim Champoux, our Program Project colleagues and the Maizels laboratory for their interest and suggestions. We are grateful to the US National Institutes of Health for supporting this research via National Cancer Institute.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Khodyreva SN, Prasad R, Ilina ES, Sukhanova MV, Kutuzov MM, Liu Y, Hou EW, Wilson SH, Lavrik OI. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1) Proc. Natl Acad. Sci. USA. 2010;107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ide H, Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol. Pharm. Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- 5.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair. 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, Maloney DJ, Dedkova LM, Hecht SM. Inhibitors of DNA polymerase beta: activity and mechanism. Bioorg. Med. Chem. 2008;16:4331–4340. doi: 10.1016/j.bmc.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 9.Haracska L, Prakash L, Prakash S. A mechanism for the exclusion of low-fidelity human Y-family DNA polymerases from base excision repair. Genes Dev. 2003;17:2777–2785. doi: 10.1101/gad.1146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gros C, Fahy J, Halby L, Dufau I, Erdmann A, Gregoire JM, Ausseil F, Vispe S, Arimondo PB. DNA methylation inhibitors in cancer: recent and future approaches. Biochimie. 2012;94:2280–2296. doi: 10.1016/j.biochi.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AM. Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 2011;711:113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Nitiss JL, Soans E, Rogojina A, Seth A, Mishina M. Topoisomerase assays. Curr. Protoc. Pharmacol. 2012 doi: 10.1002/0471141755.ph0303s57. Chapter 3, Unit 3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian D, Furbee CS, Muller MT. ICE bioassay. Isolating in vivo complexes of enzyme to DNA. Methods Mol. Biol. 2001;95:137–147. doi: 10.1385/1-59259-057-8:137. [DOI] [PubMed] [Google Scholar]
- 14.Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA. Etoposide targets topoisomerase IIalpha and IIbeta in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol. Pharmacol. 1998;54:78–85. doi: 10.1124/mol.54.1.78. [DOI] [PubMed] [Google Scholar]
- 15.Barker S, Murray D, Zheng J, Li L, Weinfeld M. A method for the isolation of covalent DNA-protein crosslinks suitable for proteomics analysis. Anal. Biochem. 2005;344:204–215. doi: 10.1016/j.ab.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 16.Shoulkamy MI, Nakano T, Ohshima M, Hirayama R, Uzawa A, Furusawa Y, Ide H. Detection of DNA-protein crosslinks (DPCs) by novel direct fluorescence labeling methods: distinct stabilities of aldehyde and radiation-induced DPCs. Nucleic Acids Res. 2012;40:e143. doi: 10.1093/nar/gks601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–534. 536–537. [PubMed] [Google Scholar]
- 18.Chomczynski P, Mackey K, Drews R, Wilfinger W. DNAzol: a reagent for the rapid isolation of genomic DNA. Biotechniques. 1997;22:550–553. doi: 10.2144/97223pf01. [DOI] [PubMed] [Google Scholar]
- 19.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 20.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 21.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol. Cell. Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491:125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.