Abstract
Polymerases belonging to the DinB class of the Y-family translesion synthesis DNA polymerases have a preference for accurately and efficiently bypassing damaged guanosines. These DinB polymerases also generate single-base (−1) deletions at high frequencies with most occurring on repetitive ‘deletion hotspot’ sequences. Human DNA polymerase kappa (hPolκ), the eukaryotic DinB homologue, displays an unusual efficiency for to extend from mispaired primer termini, either by extending directly from the mispair or by primer-template misalignment. This latter property explains how hPolκ creates single-base deletions in non-repetitive sequences, but does not address how deletions occur in repetitive deletion hotspots. Here, we show that hPolκ uses a classical Streisinger template-slippage mechanism to generate −1 deletions in repetitive sequences, as do the bacterial and archaeal homologues. After the first nucleotide is added by template slippage, however, hPolκ can efficiently realign the primer-template duplex before continuing DNA synthesis. Strand realignment results in a base-substitution mutation, minimizing generation of more deleterious frameshift mutations. On non-repetitive sequences, we find that nucleotide misincorporation is slower if the incoming nucleotide can correctly basepair with the nucleotide immediately 5′ to the templating base, thereby competing against the mispairing with the templating base.
INTRODUCTION
During replication, DNA polymerases frequently encounter unrepaired DNA lesions. Owing to their restrictive active sites and 3′→5′ exonuclease proofreading activities, replicative DNA polymerases tend to stall at sites of damage, which can lead to replication fork collapse and which, unless rescued, can eventually cause large-scale genomic rearrangements and cell death. As a preventative measure, most organisms have alternative polymerases that are capable of ‘damage tolerance’ and can synthesize past DNA lesions by a mechanism called translesion synthesis (TLS). However, these specialized polymerases are generally much more error-prone than replicative polymerases (1). Most polymerases involved in TLS belong to the Y-family of DNA polymerases. Phylogenetic data further subdivides this family into six classes: two UmuC subfamilies, found only in bacteria; Rad30A (pol eta), Rad30B (pol iota) and Rev1 proteins, found only in eukaryotes; and the DinB subfamily that is found in all domains of life (2).
The role of the DinB class of polymerases in cells has not been as clear as for some other Y-family polymerases, despite the fact that they are so widely conserved. However, bacterial, archaeal and eukaryotic DinB polymerases all have the ability to efficiently and accurately bypass N2-adducted guanosines in vitro (3–7), and their absence makes cells more sensitive to DNA damaging agents (3,8). Furthermore, all of these polymerases incorporate dCTP much more efficiently than other nucleotides, a property presumed to relate to a role in bypassing damaged guanosines (9–11).
A characteristic mutational feature of DinB polymerases is their ability to generate single-base deletions at high frequencies (∼10−2 to 10−4) on undamaged DNA sequences (11–15). Most deletions have been found to occur on ‘deletion hotspots’ containing a short run of identical pyrimidines followed by a 5′ G, but many deletions are also found in non-repetitive sequences where other polymerases rarely make deletions.
The human DinB ortholog, polymerase kappa (hpolκ), shares the lesion bypass and mutational characteristics with the bacterial (DinB) and archaeal (Dpo4 and Dbh) enzymes, but it also has some distinctive features. One key feature is its ability to efficiently extend from mispaired bases located at the primer terminus (16,17). Unlike other polymerases, hpolκ extends from mispairs at a higher frequency than it generates mispairs, with dC-C mispairs being extended most readily (16). Mispair extension can occur either by direct extension from the mispair or by a misalignment mechanism, where the misincorporated base at the primer terminus forms a correct pair with the next templating base. A single-base deletion results when DNA synthesis continues from this misaligned conformation (16,17). Efficient mispair extension, combined with an inability to incorporate nucleotides opposite the 3′ base of intra-strand cross-linked bases (cis-syn TT dimers and 6-4 photoproducts), led to the proposal that pol kappa works in concert with other TLS polymerases to extend DNA synthesis after another polymerase has copied the lesion itself (10,16).
Another characteristic that distinguishes hpolκ from the other DinB polymerases (and from other classes of Y-family polymerases) is that it has a moderate degree of processivity, ∼25 nucleotides incorporated per DNA binding event compared with at most a few nucleotides added by other Y-family polymerases (15). This moderate processivity is conferred on hpolκ by a C-terminal extension of the enzyme that is not found in other members of the polymerase family. Because of this processivity, hpolκ has a higher potential to introduce mutations in the genome. In vitro analysis shows that hpolκ residues 1–560 and full-length polymerase generate nearly identical mutational spectra at similar frequencies, with average base-substitution and deletion frequencies of 7 × 10−3 and 2 × 10−3, respectively (15).
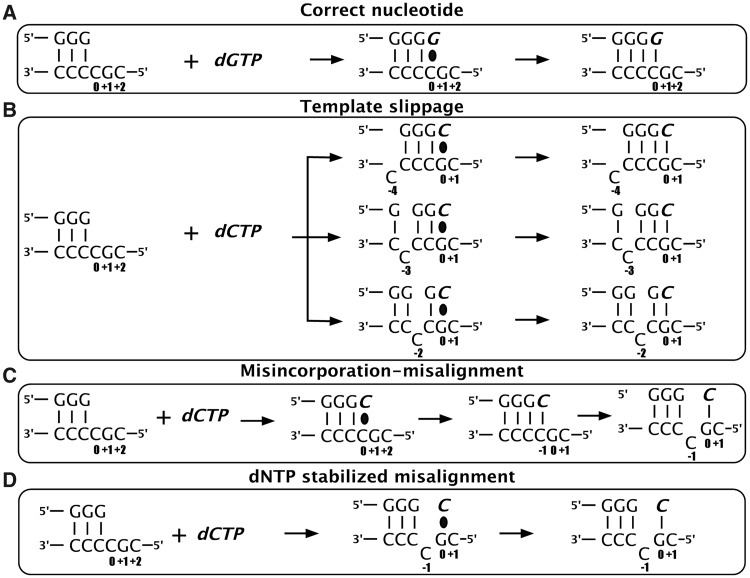
To date, three mechanisms have been suggested for generation of single-base deletions (Figure 1). On repetitive/iterative sequences, Streisinger slippage is thought to occur, where the intrinsic nature of the substrate allows DNA duplexes to readily misalign, leaving an extrahelical nucleotide in one strand (Figure 1B). After a round of replication, this can lead to addition or deletion of the unpaired nucleotide, depending on whether the unpaired nucleotide is in the primer or template strand. When this occurs in an open reading frame, Streisinger slippage results in +1 or −1 frameshift mutations (18,19). For deletions to occur by this mechanism, the polymerase must be able to tolerate the unpaired nucleotide that is skipped during replication in the template strand, causing the newly synthesized primer strand to be shorter than the template.
Figure 1.
Single-base deletion mechanisms. On a deletion ‘hotspot’ containing a homopolymeric run of pyrimidines followed by a 5′G, DinB polymerases could (A) add the correct nucleotide (dGTP) or generate single-base deletions in the presence of the incorrect nucleotide (dCTP) by any of three proposed mechanisms: (B) template slippage (C) misincorporation–misalignment and (D) dNTP-stabilized misalignment (see text for more details). The incoming nucleotide and newly added base are shown in italics (bold). The base positions assigned are shown with respect to the templating position defined as 0. The black oval represents base-pairing before phosphodiester bond formation.
For deletions on non-repetitive sequences, both misincorporation–misalignment and dNTP-stabilized misalignment mechanisms have been proposed (20–24). Misincorporation–misalignment requires incorporation of the incorrect nucleotide opposite the templating base (0 position), followed by isomerization of the template to allow base pairing between the newly added nucleotide and the next correct templating base (+1 position), resulting in the original templating base becoming unpaired at what becomes the −1 position (Figure 1C). The dNTP stabilized misalignment mechanism, however, directly uses the nucleotide next to the templating base (+1 position) for incorporation, again resulting in the templating nucleotide at the 0 position to become extrahelical in the −1 position (Figure 1D).
Mutational data show that hpolκ makes single-base deletions at higher rates in repetitive sequences (15). This is consistent with a template slippage mechanism, but this has not been tested directly. As the deletion rate on non-repetitive sequences is also high, hpolκ could use one of the other deletion mechanisms on both repetitive and non-repetitive sequences. The most likely alternative mechanism would be misincorporation–misalignment, as the misalignment part of this mechanism is used by hpolκ when extending from a mispaired primer terminus that is complementary to the next templating base (17).
Here, we investigate how hpolκ generates deletion mutations, finding that hpolκ does primarily use a template-slippage mechanism in repetitive sequences, as do the archaeal and bacterial DinB polymerases, but hpolκ is also highly proficient at realigning the slipped strands, a property that has not been investigated previously. Furthermore, the rate of nucleotide misincorporation is highly dependent on sequence context.
MATERIALS AND METHODS
Protein purification
The human DNA polymerase kappa (κ) construct encoding for amino acids 1–560 (hPol κ1–560) was cloned into pGEX vector with a N-terminal GST tag and was expressed in Rosetta2 Escherichia coli cells (EMD Millipore). For simplicity, we will refer to hPol κ1–560 as hPolκ, as earlier work has shown that constructs containing residues 1–560 and 1–526 have the same nucleotide incorporation activity and fidelity as the full-length polymerase (15,25,26). Cells expressing the construct were grown at 37°C until OD600 reached 0.7 and then induced with 0.5 mM isopropylthio-β-galactoside (IPTG) overnight at 20°C. All of the remaining steps were performed at 4°C. Cell pellets were resuspended in Buffer A [50 mM Tris–Cl (pH 8.0), 300 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 10 mM β-mercaptoethanol and 10%(w/v) sucrose] containing lysozyme. Resuspended cells were lysed by sonication followed by centrifugation at 20 000g for 1 h. Ammonium sulphate was added to the clear supernatant to a final concentration of 35% (0.208 g/ml) of supernatant to precipitate hPolκ. The precipitate was collected after centrifugation, resuspended in 35 ml of Buffer A and dialysed overnight against Buffer A. The dialysed protein was then loaded on (2 × 5 ml) GST columns (GE Healthcare) pre-equilibrated with Prescission protease buffer [50 mM Tris–Cl (pH 8.0), 100 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 10% glycerol]. Columns were washed again before treating overnight with 1 column volume of Prescission protease buffer containing 1 mg/ml of HRV3C protease. The untagged hPolκ was obtained the next day by eluting with Prescission protease buffer, and the eluate was immediately applied to (2 × 5 ml) HiTrap SP-columns (GE Healthcare), followed by gradient elution with 0.05–1 M NaCl in 20 mM HEPES (pH 7.25), 0.1 mM EDTA, 1 mM DTT and 10% glycerol. The eluted fractions were then dialysed overnight into storage buffer containing 25 mM HEPES (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, 5 mM DTT and 10% glycerol. After concentrating the protein using Amicon Ultra-15 filters with 10 kDa molecular weight cutoff (Milipore) to achieve a desired concentration (calculated using a theoretical extinction coefficient of 28 880 M−1 cm−1), the aliquots were frozen at −80°C.
Duplex DNA formation
All DNA oligonucleotides (shown in Table 1) were obtained from Integrated DNA Technologies, and primer oligonucleotides were synthesized with a 5′ 6- carboxyfluorescein (6FAM) label. Primer and template oligonucleotides were annealed in buffer containing 10 mM Tris–Cl (pH 7.5) and 50 mM NaCl by heating to 95°C followed by slow cooling to 25°C.
Table 1.
DNA substrate sequences
| Number | Name | Sequencea |
|---|---|---|
| 1 | 4C-G | 5′-(FAM)-AGG CAC TGA TC GGG -3′ |
| 3′-CC GTG ACT AG CCCCGC ATT -5′ | ||
| 2 | 1T-G | 5′-(FAM)-AGG CAC TGA TC GGG -3′ |
| 3′-CC GTG ACT AG CCCTGC ATT -5′ | ||
| 3 | 2T-G | 5′-(FAM)-AGG CAC TGA TC GG G -3′ |
| 3′-CC GTG ACT AG CCTCGC ATT -5′ | ||
| 4 | 3T-G | 5′-(FAM)-AGG CAC TGA TC G GG -3′ |
| 3′-CC GTG ACT AG CTCCGC ATT -5′ | ||
| 5 | 4T-G | 5′-(FAM)-AGG CAC TGA TC GGG -3′ |
| 3′-CC GTG ACT AG TCCCGC ATT -5′ | ||
| 6 | 4C-A | 5′-(FAM)-AGG CAC TGA TC GGG -3′ |
| 3′-CC GTG ACT AG CCCCAC ATT -5′ | ||
| 7 | 1T-A | 5′-(FAM)-AGG CAC TGA TC GGG -3′ |
| 3′-CC GTG ACT AG CCCTAC ATT -5′ | ||
| 8 | C-C mispair | 5′-(FAM)-AGG CAC TGA TC GGGC -3′ |
| 3′-CC GTG ACT AG CCCCGC ATT -5′ | ||
| 9 | 4C-GA | 5′-(FAM)-AGG CAC TGA TC GGG -3′ |
| 3′-CC GTG ACT AG CCCCGA ATT -5′ | ||
| 10 | 4G | 5′-(FAM)-AGG CAC TGA TC GGGG -3′ |
| 3′-CC GTG ACT AG CCCCGC ATT -5′ |
aPrimers are labelled on the 5′ end with 6-carboxyfluorescein (FAM).
Nucleotides in bold vary between substrates.
Pre-steady-state primer extension assays
All assays were done using a KinTek RQF-3 rapid quench instrument (KinTek Corp.). To achieve single turnover conditions, where all of the DNA substrate is pre-bound by polymerase, an excess of hPolκ (30 µM) was preincubated with 100 nM DNA at room temperature (25°C) in reaction buffer [25 mM MES–Tris (pH 7.5), 25 mM NaCl, 8 mM MgCl2, 2 mM DTT and 10% glycerol]. Reactions were initiated by mixing with an equal volume of 2 mM deoxynucleoside-triphosphate (dNTP) in the same buffer (final concentrations were 15 µM hPolκ, 50 nM DNA substrate and 1 mM dNTP) and quenched with 250 mM EDTA at appropriate time points (0.02–60 s). The extended product was separated from unextended primer on a 17.5% (19:1) acrylamide-(1×)Tris-Borate-EDTA (TBE) gel containing 7.5 M urea. The gel was imaged using a Typhoon 9400 scanner (GE Healthcare), and bands were quantitated using ImageQuant software (GE Healthcare). Percentage of primer extension was determined by measuring the relative intensity of the band corresponding to the extended primer with respect to the total labelled DNA (i.e. both extended and unextended primer strands) [Equation (1)].
| (1) |
Where applicable, the amount of extended primer includes sum of primer extension seen as multiple bands when extended by more than one nucleotide.
Most data were fit to an exponential equation [Equation (2)] unless a biphasic nature was observed in which case the burst equation [Equation (3)] was used:
| (2) |
where A is amplitude, kobs is the observed rate of the reaction, t is the time after which reaction was quenched and c is a constant.
| (3) |
where A is the amplitude of the fast phase, k1 is rate of the fast phase of the reaction and k2 is the rate of the slow phase, t is the time after which reaction was quenched and c is a constant.
Experiments were performed in duplicate with two different protein preparations, and error bars represent the standard deviation of the data collected. All of the graphs and non-linear regressions were done using GraphPad Prism, version 6.0a (GraphPad Software Inc.). Observed nucleotide incorporation rates for all experiments are summarized in Table 2.
Table 2.
Summary of observed nucleotide incorporation rates
| Substrate | Observed rate (s−1) |
|||||
|---|---|---|---|---|---|---|
| dCTP addition |
dGTP addition |
|||||
| First | Second | Overall | First | Second | Overall | |
| 4C-G | 2.0 ± 0.5 | 0.15 ± 0.08 | 0.72 ± 0.04 | 1.6 ± 0.3 | 0.09 ± 0.15 | 2.62 ± 0.39 |
| 1T-G | 0.44 ± 0.03 | |||||
| 2T-G | 0.38 ± 0.04 | |||||
| 3T-G | 3.47 ± 0.34 | |||||
| 4T-G | 1.59 ± 0.14 | |||||
| 4C-A | 0.12 ± 0.01 | |||||
| 1T-A | 1.14 ± 0.09 | |||||
| C-C mispair | 0.33 ± 0.02 | |||||
| 4C-GA | 2.5 ± 0.5 | 0.15 ± 0.02 | 0.79 ± 0.03 | |||
| 4G | 0.11 ± 0.36 | |||||
RESULTS
The template sequence 3′-CCCCG-5′ is a deletion hotspot for hPolκ
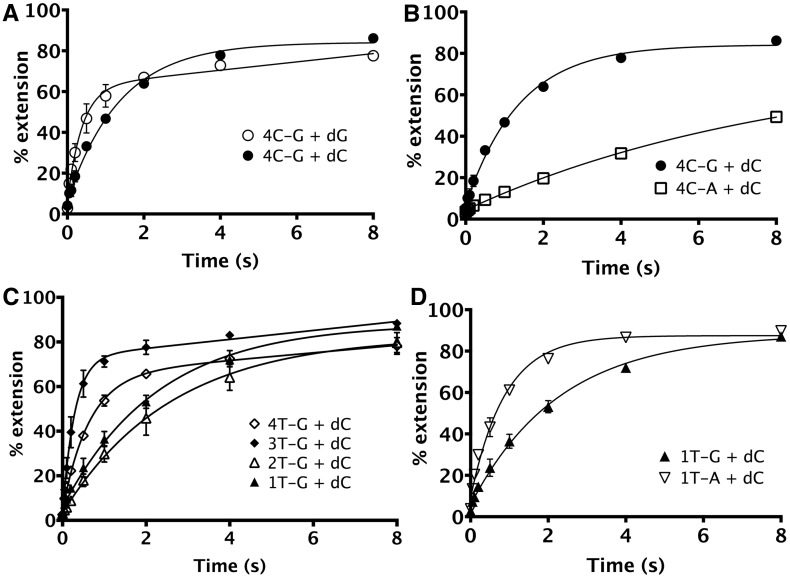
To determine whether hPolκ is error prone on the ‘deletion hotspot’ sequence characterized for other DinB polymerases (11,12,14,27,28), we used DNA substrate 4C-G, which contains a homopolymeric run of four C’s followed by a 5′ G on the template strand (3′-CCCCG-5′, Table 1). On this substrate, hPolκ adds the correct incoming nucleotide, dGTP, at a rate that is only 3.6-fold faster (kobs of 2.62 ± 0.39 s−1) than the rate of incorrect nucleotide, dCTP (kobs of 0.72 ± 0.04 s−1) addition (Figure 2A), indicating that deletions can be initiated on this substrate at a very high frequency.
Figure 2.
Nucleotide incorporation by hPolκ on repetitive sequence containing deletion hotspot. In all, 15 µM of hPolκ was pre-incubated with 50 nM DNA before initiating the reaction with addition of 1 mM dNTP (final concentrations). (A) On 4C-G (substrate 1), containing a run of four C’s followed by a 5′ G, hPolκ adds dGTP ∼4-fold faster than dCTP. (B) Modification of the nucleotide 5′ to the templating position (i.e. +1 position) from G to A [4C-A, substrate 6] results in reduction in overall kobs for incorporation of dCTP by 6-fold from 0.72 ± 0.04 s−1 to 0.12 ± 0.01 s−1. (C) Pre-steady-state assays were performed under single turnover conditions using DNA templates containing unpaired T’s at every C position in the homopolymeric run of 4 C-G (substrates 2–5) to test whether hPolκ can tolerate the presence of an extrahelical nucleotide at these positions. The dCTP incorporation by hPolκ on 1T-G, 2T-G, 3T-G and 4T-G substrates is shown. The hPolκ extends the various bulged templates with the -3T bulge (3T-G, substrate 4) being used most efficiently. (D) Modification of the +1 G position of the 1T-G substrate to an A [1T-A, substrate 7] resulted in a 3-fold increase in kobs from 0.44 ± 0.03 s−1 to 1.14 ± 0.09 s−1.
Interestingly, the dGTP (correct) addition follows biphasic behaviour with a burst amplitude of 56.2 ±3.2 nM and a slow second phase rate of 0.036 ±0.012 s−1. This was unexpected because the assay was designed to be performed under single turnover conditions, where all of the DNA is initially bound by polymerase so that the DNA would all be extended in one cycle without polymerase having to rebind. Under these identical conditions, dCTP (incorrect nucleotide) addition by hPolκ displays only a single phase of product formation, which is consistent with single turnover conditions and accordingly fits well to the single exponential equation [Equation (2)] with an amplitude of 77.41 ± 1.63 nM.
Hence, the biphasic nature of product formation for dGTP addition cannot be explained simply as a result of the presence of initial unbound DNA. Because the two different reactions used the same primer-template (p/t) DNA and polymerase preparation, the difference in amplitudes (56 nM for dG versus 77 nM for dC) is dependent on the identity of the incoming dNTP, which must influence the fraction of active enzyme complex. Previously, the active fraction of hPolκ was found to be ∼8-fold larger when extending from a mispair than from a perfectly base paired p/t DNA junction (29). It was suggested that this characteristic might imply the preference for mispaired rather than correctly paired DNA substrates. Here, we find that even when supplied with a perfectly basepaired p/t DNA, the active enzyme concentration of hPolκ varies based on the incoming dNTP. When dCTP is added to initiate a deletion, the nucleotide skipped is expected to adopt an extra-helical conformation, which could thereby disrupt the B-form conformation of the DNA. This alteration of DNA conformation may be the key difference that could favourably influence the active enzyme concentration of hPolκ.
Human Polκ predominantly uses a template-slippage deletion mechanism on iterative sequences
As hPolκ has been reported to use a misalignment mechanism to make deletions on non-repetitive sequences (17), we first investigated if the incorporation of dC on the deletion hotspot sequence also occurred by this mechanism. Of the three mechanisms for single-base deletion (Figure 1), only misincorporation–misalignment does not depend on the nucleotide on the 5′ side of the templating position (+1 position) for templating dC addition, as in this mechanism, the first nucleotide would be added by mispairing with the C at position 0 (Figure 1C). In the 4C-G substrate, a G occupies the +1 position. To determine whether hPolκ uses this +1G when generating deletions, we changed it from G to A (Table 1, 4C-A). On testing for addition of dCTP on this template, we observed that modifying the base at the +1 position reduced the kobs 6-fold, from 0.72 ± 0.04 s−1 to 0.12 ± 0.01 s−1 (Figure 2B). This suggested a clear dependence on the +1G, thus ruling out misincorporation–misalignment as the major deletion mechanism used by hPolκ on this repetitive sequence.
DNA templates with a repetitive sequence can exist in multiple conformations with the same primer, resulting in alternate base pairing situations that could lead to deletions or additions after a round a replication (18,19). In an attempt to localize and stabilize an extrahelical nucleotide at specific positions in the homopolymeric run of C’s in the 4C-G substrate (3′-CCCCG-5′), we systematically modified each of the four C’s on the template strand to a T one at a time (Table 1, substrates 2–5). The higher energy penalty of forming a G-T mispair compared to a G-C pair makes the former less favoured, biasing the DNA to adopt a conformation with an unpaired nucleotide at the altered position (27). We used this assay to test if hPolκ is able to tolerate and extend p/t DNA with an extrahelical nucleotide at positions −4, −3, −2 and −1 of the template (Figure 1B and D; Figure 2C). Efficient use of the templates with T at positions −4, −3 and −2 would indicate use of template slippage. Efficient use of the template with the T at the −1 position would indicate use of either dNTP-stabilized misalignment or misincorporation-misalignment deletion mechanism. Having already ruled out misincorporation-misalignment for the 4C-G template, we used these altered substrates to distinguish template slippage from dNTP-stabilized misalignment. Similar DNA substrates have been used to assess deletion mechanisms for other DinB polymerases (27,28,30).
We found that hPolκ showed a strong preference for unpaired nucleotides present at positions −3 and −4 (substrates 3T-G and 4T-G) with kobs of 3.47 ± 0.34 s−1 and 1.59 ± 0.14 s−1, respectively (Figure 2C). Interestingly, these data showed a biphasic behaviour and so were fit to a burst equation [Equation (3)], as was done for the dGTP (correct) addition on the 4C-G substrate. An unpaired base at the −2 position is used ∼10-fold less efficiently than at the −3 position, with a kobs of 0.38 ± 0.04 s−1, suggesting that proximity to the active site reduces tolerance for an extrahelical nucleotide. This preference for an unpaired base further away from the active site has been observed for other DinB polymerases as well (27,28,30).
The ability to efficiently use DNA substrates containing an unpaired nucleotide at the −4, −3 and −2 positions (substrates 3–5, Table 1) indicates that hPolκ predominantly uses a template slippage mechanism to generate single-base deletions. Compared with the 4C-G substrate, the dC incorporation rate is faster on the 4T-G and 3T-G substrates but slower on the 2T-G substrate. This suggests that the 4C-G template adopts multiple conformations, and the observed nucleotide incorporation rate results from a combination of different rates on substrates with different conformations.
Although we found that hPolκ is able to use slipped template DNA sequences efficiently, it is also able to add dCTP on 1T-G (Table 1), which has a T at the 0 position, with a kobs of 0.44 ± 0.03 s−1 (Figure 2C), comparable with the rate of addition on the 2T-G substrate and only ∼50% more slowly than on the 4C-G substrate. This left open the possibility that hpolκ could potentially use dNTP-stabilized misalignment or misinsertion–misalignment on this substrate, even though the latter had been ruled out on the original deletion hotspot sequence.
Human Polκ readily misincorporates dC when the deletion hotspot is changed to 3′-CCCTG-5′
We then considered the possibility that hPolκ used entirely different nucleotide incorporation mechanisms on substrates 4C-G and 1T-G, despite differing by only a single nucleotide. To test this possibility, we altered the template sequence on the 1T-G substrate such that the +1G was modified to A (1T-A, substrate 7). If dCTP addition on this substrate is through dNTP-stabilized misalignment, then the +1G on the template would prove crucial for extension, and a considerable reduction in rate of addition may be expected. However, if incorrect nucleotide addition was independent of the +1G, no such decrease would occur, suggesting that dCTP addition on the 4C-G substrate occurs by a different mechanism than on the 1T-G substrate.
Surprisingly, on 1T-A (i.e. 1T-G with +1G modified to A) a 2.6-fold increase in kobs from 0.44 ± 0.03 s−1 to 1.14 ± 0.09 s−1 was observed (Figure 2D), in contrast to the 6-fold decrease observed on the 4C-A substrate compared with the 4C-G substrate (Figure 2B). Absence of a rate decrease when the +1G is altered excludes the possibility that dNTP-stabilized misalignment occurs in the sequence context found in the 1T-G substrate. Earlier genetic evidence has shown that hPolκ produces T-dCMP mispairs at a very high frequency of 8.2 × 10−3 (15). On the 1T-G substrate, therefore, dCTP addition most likely occurs by misincorporation opposite the templating T. We conclude that dC addition occurs by misincorporation on both the 1T-G and 1T-A substrates, but the sequence context influences the efficiency of misincorporation, as evidenced by the 1T-A sequence context stimulating incorporation. The mechanism for the increased rate of dC incorporation on the 1T-A substrate is discussed later.
Human Polκ efficiently realigns primer-template DNA after initiating a deletion
We observed that hPolκ performs multiple additions on most DNA substrates examined, even when provided with just a single nucleotide. On the 4C-G substrate, this was true for both dGTP (correct) and dCTP (incorrect) incorporations [Figures 3A, panel (i), and 4B, panel (i)], but to differing extents. Interestingly, we found that multiple addition of dGTP followed a very different pattern than that observed for dCTP, indicating dissimilarity in mechanism. To better understand the relevance of this observation and ascertain its mechanistic implications, we decided to investigate how hPolκ performs multiple nucleotide additions on the 4C-G substrate.
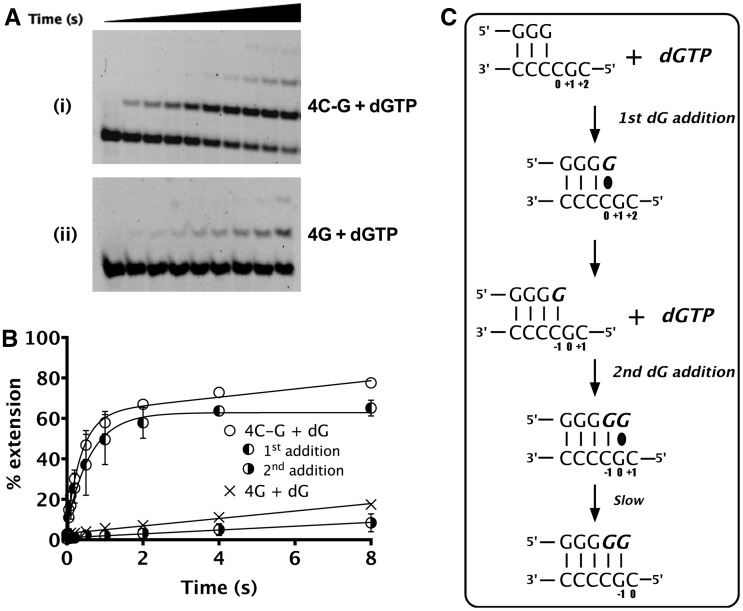
Figure 3.
Mechanism of second dGTP incorporation on 4C-G substrate. (A) Pre-steady-state assays under single turnover conditions were performed. (i) and (ii) show sequencing gels depicting multiple dG additions on substrates 4C-G and 4G, respectively. (B) Plot of percentage extension versus time for both 4C-G and 4G substrates. Addition of first dG plateaus at ∼60% followed by slow misincorporation of the second dGTP. Rate of second dG incorporation on the 4C-G corresponds well with misincorporation of dGTP on the 4G substrate that has the first dGTP already added. (C) Schematic showing the probable mechanism of second dGTP addition on 4C-G substrate. The incoming nucleotide and newly added base are shown in italics (bold). The base positions assigned are shown with respect to the templating position defined as 0. The black oval represents base-pairing before bond formation.
We found that for dGTP addition on the 4C-G substrate, the second dG is added slowly (kobs ∼0.09 s−1), after rapid addition of the first dG (kobs of 1.6 ± 0.3 s−1) [Figure 3A (i) and 3B], suggesting a gradual misincorporation opposite the +1G after the correct nucleotide at position 0 had been added (Figure 3C). To test this possibility, we designed substrates that already had the first nucleotide incorporated as a G (Table 1, substrate 4G) and tested the rate of insertion of the next nucleotide. The rate for the second dG addition (0.09 s−1) corresponds well with the misincorporation rate (kobs ∼0.11 s−1) [Figure 3A (ii) and 3B], confirming this interpretation.
In contrast to the slow rate of addition of a second dG, we found that addition of a second dC is considerably faster and followed an entirely different pattern (Figure 3A and B versus Figure 4B and C). As the aforementioned data indicate, the first addition of dC occurs predominantly by template slippage, allowing the proper base pairing of the dCTP opposite the +1G. We can envision two possibilities for the addition of the second dC (Figure 4A). One possibility is that the next dCTP is misincorporated opposite the templating C at +2 to generate a C-dCMP ‘mispair’. A second possibility is that the extrahelical nucleotide generated during the first dCTP addition is ‘rearranged’ such that now there is a C-C mispair generated at the p/t junction. As hPolκ is an efficient extender of mispairs (16), it could tolerate the mispair to add the second dCTP correctly paired opposite the +1G.
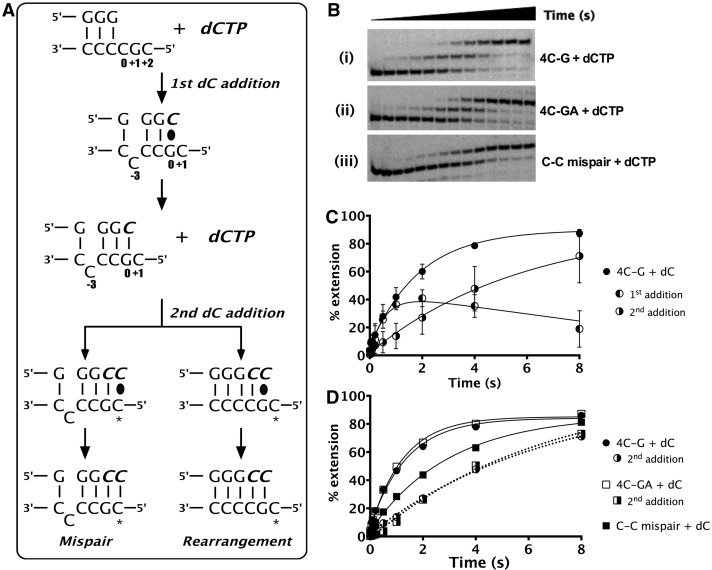
Figure 4.
Mechanism of multiple dCTP incorporations. (A) Schematic representation of the two possible ways in which hPolκ can efficiently add a second dCTP on 4C-G (substrate 1). The incoming nucleotide and newly added base are shown in italics (bold). The base positions assigned are shown with respect to the templating position defined as 0. The black oval represents base-pairing before bond formation. ‘*’ indicates the +2 templating position. (B) Gels showing the unique pattern of multiple incorporations of dCTP on (i) 4C-G (substrate 1) and (ii) 4C-GA (substrate 9) to test for mispair formation. (iii) Gel showing the incorporation of dC to extend from a C-C mispair (substrate 8) suggests hPolκ’s ability to realign. (C) Plot shows that mechanism of multiple dC additions is distinct from dG additions on 4C-G substrate. First dCTP addition reaches a maximum of ∼40% extension, after which increase in second dCTP incorporation occurs exponentially, implying that most of the primer extended by one nucleotide is rapidly extended by two. (D) Overall dCTP incorporation on the 4C-G, 4C-GA and C-C mispair substrates shown with respect to time along with percent of second dCTP addition as seen for the 4C-G and 4C-GA substrates. Second dCTP incorporation traces are shown as dotted lines.
To distinguish between these mechanisms, two different substrates were designed: one with the +2C modified to an A (Table 1, 4C-GA) and another where the primer contained a 3′-C to generate a mispair at the p/t junction (Table 1, C-C mispair). Modification of the +2C to an A (4C-GA) did not alter the overall rate of nucleotide addition; kobs for 4C-G and 4C-GA were 0.72 ± 0.04 s−1 and 0.79 ± 0.03 s−1, respectively (Figure 4B and D). Under steady-state conditions, dC-A incorporation is 3.8-fold less efficient than dC-C (comparing Vmax/Km of 8 × 10−4 for dC-A and 3 × 10−3 for dC-C) (10). Thus, the unchanged incorporation rate on substituting the +2 C to an A indicates that the base in the +2 position is not used to template the second dC addition.
Dissecting this data further, the rates of second dC incorporation on 4C-G and 4C-GA substrates were also found to be essentially identical (4C-G: 0.146 s−1 and 4C-GA: 0.153 s−1), but dCTP incorporation from a C-C mispair was 2-fold faster (0.33 ± 0.02 s−1) (Figure 4D). This observation can be explained, because time taken for rearrangement after the first nucleotide addition might slow the rate of second nucleotide addition when compared with a substrate with a preformed C-C mispair at the p/t junction. These data strongly suggest that after the first dCTP addition, rearrangement at the p/t junction occurs and allows extension from a C-C mispair for the second incorporation. From these experiments, we conclude that after initiating a deletion by incorporation of dCTP on the deletion hotspot sequence 3′-CCCCG-5′, hPolκ can efficiently realign the slipped p/t junction to correctly incorporate a second dC from a C-C mispaired p/t junction. Notably, this is the mispair that previously was found to be most efficiently extended by hpolκ (10).
Sequence context influences the rate of nucleotide incorporation
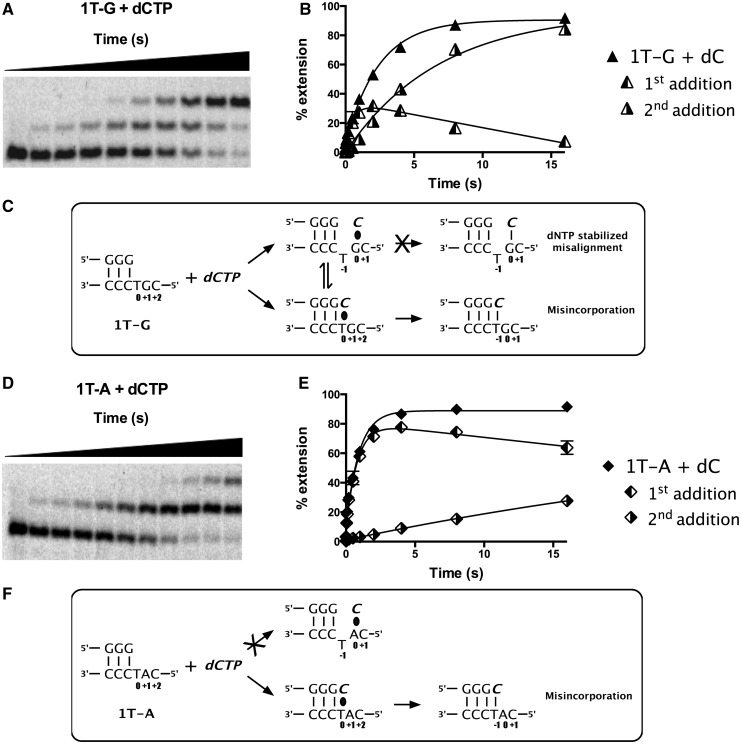
The ability to incorporate a single nucleotide multiple times was also observed on the 1T-G and 1T-A substrates (Figure 5A and D). As demonstrated earlier in the text, the addition of the first dCTP on 1T-G substrate occurred by misincorporation, as modification of the +1G to A in the 1T-A substrate resulted in ∼3-fold increase in overall rate. Although this clearly indicated that dNTP-stabilized misalignment was not the predominant incorporation mechanism, the increase in rate was puzzling. To understand this better, we calculated the amount of extension for the first and second additions separately (Figure 5B and E) and found that different patterns and rates of incorporation on the two substrates provided further information to decipher the mechanism.
Figure 5.
The hPolκ misincorporates efficiently on 1T-G substrate. (A) Gel and (B) graph showing multiple incorporations of dCTP on the 1T-G substrate. First addition, second addition and overall addition. (C) Schematic showing possible mechanisms of dCTP addition on the 1T-G substrate. (D) Gel and (E) graph showing multiple incorporations of dCTP on the 1T-A substrate. (F) Schematic showing possible mechanisms of dCTP addition on the 1T-A substrate. In (C) and (F), the incoming nucleotide and newly added base are shown in italics (bold). The base positions assigned are shown with respect to the templating position defined as 0. The black oval represents base pairing before bond formation.
On the 1T-G substrate, the amount of primer extended by just one nucleotide reached a maximum level of ∼30%, at which point the level decreased as the second dC was being added rapidly (Figure 5A and B). On the 1T-A substrate, in contrast, the amount of primer extended by one nucleotide reached a maximum level of ∼80% (Figure 5D and F) before dropping, as the second dC was being added slowly. For each substrate, the overall rate of primer extension (1.14 s−1 for 1T-A; 0.44 s−1 for 1T-G) was dominated by the rate of the first nucleotide incorporation.
We interpret these results as shown in Figure 5C and F. On both substrates, the first dC addition is templated by the T in position 0. On the 1T-G substrate (Figure 5C), the second dC is templated by the G at the +1 position, and the rate of this incorporation is fast because hpolκ can efficiently add the next correct nucleotide even from a mispaired primer terminus (16). On the 1T-A substrate (Figure 5F), the second dC is templated by the A at the +1 position, but the rate is slow because dC-A is a mispair, and previous steady-state data show that it is the slowest mispair formed by hpolκ (10). The fast rate of second nucleotide addition on 1T-G would lower the amplitude of the primer extended by one nucleotide, as this serves as the substrate for the second nucleotide addition. On the 1T-A substrate, the amplitude of the first addition would be higher because the primer extended by one nucleotide would only slowly be extended by a second nucleotide.
The lower overall rate of nucleotide incorporation (the sum of the first and second additions) on the 1T-G substrate can be explained if the dCTP can stably pair both with the T at the 0 position and with the G at the +1 position, and if the rate of addition is negligible for the nucleotide paired at the +1 position. In this case, only a fraction of the substrate initially has the nucleotide paired at the 0 position where it can be incorporated efficiently. Because ∼80% of the primer eventually becomes extended, the nucleotide paired with the +1G must slowly shift to mispair with the T at position 0, thus lowering the overall rate of addition. For the 1T-A substrate, the dCTP is unlikely to be able to pair stably with the +1A and not compete for binding at position 0. Thus, more of the substrate will initially have the nucleotide paired with the T at position 0 where it can be incorporated efficiently, increasing the overall rate of nucleotide addition on the 1T-A substrate.
DISCUSSION
As hpolκ is the most processive of the human Y-family polymerases, it has the greatest potential to introduce mutations into the genome (15). Investigating the mutational mechanisms of hpolκ is therefore important for understanding the mutagenic consequences of DNA damage tolerance by translesion synthesis. Through the studies reported here, we have found that template slippage is strongly preferred as the mechanism of deletion by hpolκ on a repetitive deletion hotspot 3′-CCCCG-5′ template sequence. Strikingly, however, our results indicate that hpolκ is able to efficiently realign the DNA strands, resulting in a mispaired primer terminus rather than an extrahelical template base. Extension from the mispaired primer terminus, which hpolκ is able to do proficiently (16), would result in a less deleterious base-substitution mutation being generated rather than a frameshift mutation.
A distinctive feature of the mutation spectrum produced by hpolκ when replicating undamaged DNA is that deletions are formed readily, with only a 2- to 3-fold difference existing between single-base deletion error rates observed on iterative (1.6 − 3.1 × 10−3) versus non-iterative (1.0 × 10−3) sequences (15). Our findings help explain why there is such a small difference in single-base deletion rates. On repetitive sequences, hPolκ can convert some of the deletions that are initiated into base substitution by realigning the slipped sequence and extending from the resulting mispaired primer terminus. Additionally, on non-iterative sequences, hPolκ has been found to extend from mispaired primer-template termini by initiating template-primer misalignment when direct extension does not occur (17). This would result in an increase in single-base deletions on non-repetitive sequences. Taken together, these biochemical experiments corroborate the earlier mutational studies showing high rates of T•dC misincorporation and of deletion of Ts that are located in a 3′-TG-5′ sequence context (15).
We have not found any evidence that hpolκ is able to use a dNTP-stabilized misalignment mechanism to create deletions, even on non-repetitive sequences. In fact, the preponderance of data indicates that none of the DinB polymerases is able to efficiently use this mechanism (27,28,30–32). The first suggestion that Y-family polymerases might use a dNTP-stabilized misalignment deletion mechanism came from one of the first two Y-family structures bound to DNA: the Dpo4 ‘type II’ complex, which was crystallized in an attempt to determine the structure of a mispair in the active site. Instead of forming a mispair, however, the incoming nucleotide was found paired with the next templating base, which happened to be complementary to the dNTP used (33). This pairing configuration is the hallmark of dNTP-stabilized misalignment; however, it was puzzling that the skipped template base was stacked within the DNA duplex instead of adopting an extrahelical conformation (33). Fluorescence measurements on both Dbh and DinB have shown that a skipped template base becomes unstacked from the surrounding DNA duplex when a single-base deletion is made (31,32). An extrahelical conformation has also been observed in crystal structures of both Dbh and Dpo4 with unpaired bases at positions −3 and −4 in templates that contain deletion hotspots (27,28).
Our data demonstrate that misincorporation is slower when the incoming nucleotide can pair correctly with the base in the +1 position rather than pairing incorrectly with the base in the 0 position (Figure 5). Unless the skipped base adopts an extrahelical conformation, the primer terminus and alpha phosphate would have difficulty approaching each other closely enough for efficient catalysis. Thus, rather than showing a deletion being formed by dNTP-stabilized misalignment, we believe that the Dpo4 type II structure shows instead how the rate of nucleotide misincorporation can be slowed depending on the sequence context. Consistent with this, nucleotide addition did not occur in the type II structure, even though neither the primer terminus nor incoming nucleotide were altered to prevent catalysis during crystallization.
Most polymerases show a strong dependence of deletion frequency on homopolymeric run length. Y-family polymerases differ from other polymerase families in not having a very strong dependence of deletion frequency on run length, with a run of just two nucleotides already resulting in a high deletion frequency. The structure of hpolκ (Figure 6) shows that nucleotides 3′ of the templating base are adjacent to a large solvent-accessible gap between the polymerase and polymerase-associated domains of the enzyme, suggesting that bulged bases can readily be accommodated in this area. Crystal structures of the archaeal DinB polymerases Dbh and Dpo4 show how extrahelical nucleotides at positions −3 and −4 interact with the protein in this region (27,28). In contrast, polymerases from other families have much tighter constraints around the DNA duplex, which would suppress nucleotides from adopting extrahelical conformations.
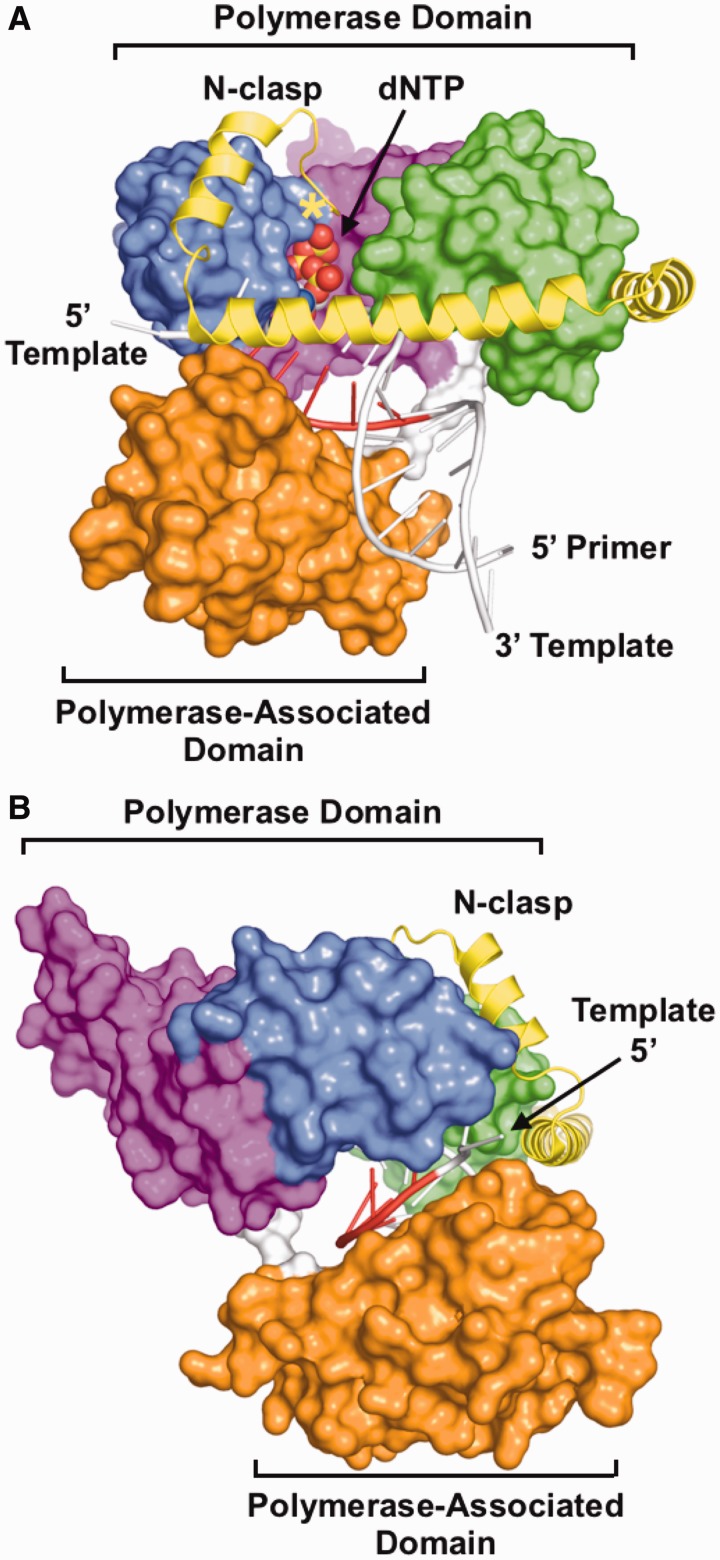
Figure 6.
Structure of an hPolκ ternary complex. (A) View looking into the active site of the polymerase. (B) View looking at the template strand of DNA entering the active site, between the polymerase and polymerase-associated domains. The polymerase [PDB code 3IN5 (34)] is shown in surface representation, except for the N-clasp (yellow), which is shown in ribbons representation. The polymerase domain is composed of fingers (blue), palm (magenta) and thumb (green) subdomains and is connected to the polymerase-associated domain (orange) by a relatively unstructured polypeptide linker (white). DNA is coloured white, except for the templating base and nucleotides at positions −1 through −4 on the 3′ side of the templating base. The first residue visible in the structure (amino acid 25) is marked with a yellow asterisk.
Recent data indicate that the overall polymerase conformation strongly influences differences in Y-family polymerase fidelity. Structural and biochemical comparisons of Dbh, Dpo4 and chimeric constructs of the two polymerases demonstrated that close contacts between the catalytic and polymerase-associated domains next to the templating base led to a greater rate of nucleotide misincorporation and a higher propensity for adding multiple nucleotides (35). In the case of hpolκ (Figure 6), the polymerase-associated domain does not directly contact the fingers subdomain, but the two domains are bridged by the N-clasp as aforementioned, which could stabilize the protein in a conformation that favours nucleotide misincorporation. The N-clasp is only found in the eukaryotic DinB polymerases. Deletion of the first 19 amino acids of the N-clasp reduces the ability of hpolκ to extend from mispaired primer termini without significanly reducing overall polymerase activity (25). The precise structural role of these residues remains to be determined, as they were not included in any of the constructs of hpolκ that have yielded crystals, but they are likely to reach into the polymerase active site, on the major groove side of the nascent basepair and primer terminus where they could directly contact and stabilize mispairs (Figure 6A).
Overall, hpolκ displays a remarkable ability to create deletion and base substitution mutations using both template slippage and nucleotide misincorporation mechanisms. As for other DinB polymerases, hpolκ has a strong preference for creating deletions in repetitive sequences by template slippage but is exceptionally proficient at realigning slipped DNA strands and extending DNA synthesis from mispaired primer termini. These unique properties give hpolκ a mechanism for suppressing deletion mutations at the expense of increasing base-substitution mutations.
FUNDING
National Institutes of Health (NIH) [GM080573 to J.D.P.] Funding for open access charge: NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Xiaojun Huang for constructing the hPol κ1-560 clone, Zhigang Wang for providing the full-length clone of hPol κ and Ryan Wilson for careful proofreading of the manuscript.
REFERENCES
- 1.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 3.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 4.Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J. Biol. Chem. 2008;283:27433–27437. doi: 10.1074/jbc.M801237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minko IG, Yamanaka K, Kozekov ID, Kozekova A, Indiani C, O'Donnell ME, Jiang Q, Goodman MF, Rizzo CJ, Lloyd RS. Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem. Res. Toxicol. 2008;21:1983–1990. doi: 10.1021/tx800174a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Repair (Amst.) 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Shen X, Sayer JM, Kroth H, Ponten I, O'Donnell M, Woodgate R, Jerina DM, Goodman MF. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J. Biol. Chem. 2002;277:5265–5274. doi: 10.1074/jbc.M109575200. [DOI] [PubMed] [Google Scholar]
- 8.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl Acad. Sci. USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc. Natl Acad. Sci. USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potapova O, Grindley ND, Joyce CM. The mutational specificity of the Dbh lesion bypass polymerase and its implications. J. Biol. Chem. 2002;277:28157–28166. doi: 10.1074/jbc.M202607200. [DOI] [PubMed] [Google Scholar]
- 12.Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 13.Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl Acad. Sci. USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 16.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfle WT, Washington MT, Prakash L, Prakash S. Human DNA polymerase kappa uses template-primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. Genes Dev. 2003;17:2191–2199. doi: 10.1101/gad.1108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J. Biol. Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 21.Bebenek K, Kunkel TA. Frameshift errors initiated by nucleotide misincorporation. Proc. Natl Acad. Sci. USA. 1990;87:4946–4950. doi: 10.1073/pnas.87.13.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom LB, Chen X, Fygenson DK, Turner J, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- 23.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase beta violates the “A-rule” Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel TA. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J. Biol. Chem. 1986;261:13581–13587. [PubMed] [Google Scholar]
- 25.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol. Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Wilson RC, Pata JD. The Y-family DNA polymerase Dpo4 uses a template slippage mechanism to create single-base deletions. J. Bacteriol. 2011;193:2630–2636. doi: 10.1128/JB.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlson KD, Johnson RE, Prakash L, Prakash S, Washington MT. Human DNA polymerase kappa forms nonproductive complexes with matched primer termini but not with mismatched primer termini. Proc. Natl Acad. Sci. USA. 2006;103:15776–15781. doi: 10.1073/pnas.0605785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foti JJ, Walker GC. Efficient extension of slipped DNA intermediates by DinB is required to escape primer template realignment by DnaQ. J. Bacteriol. 2011;193:2637–2641. doi: 10.1128/JB.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLucia AM, Grindley ND, Joyce CM. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry. 2007;46:10790–10803. doi: 10.1021/bi7006756. [DOI] [PubMed] [Google Scholar]
- 32.Foti JJ, Delucia AM, Joyce CM, Walker GC. UmuD(2) inhibits a non-covalent step during DinB-mediated template slippage on homopolymeric nucleotide runs. J. Biol. Chem. 2010;285:23086–23095. doi: 10.1074/jbc.M110.115774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 34.Vasquez-Del Carpio R, Silverstein TD, Lone S, Swan MK, Choudhury JR, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Structure of human DNA polymerase kappa inserting dATP opposite an 8-OxoG DNA lesion. PLoS One. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson RC, Jackson MA, Pata JD. Y-family polymerase conformation is a major determinant of fidelity and translesion specificity. Structure. 2013;21:20–31. doi: 10.1016/j.str.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]