Abstract
The repair of DNA breaks by homologous recombination is a high-fidelity process, necessary for the maintenance of genome integrity. Thus, DNA synthesis associated with recombinational repair must be largely error-free. In this report, we show that human DNA polymerase delta (δ) is capable of robust DNA synthesis at RAD51-mediated recombination intermediates dependent on the processivity clamp PCNA. Translesion synthesis polymerase eta (η) also extends these substrates, albeit far less processively. The single-stranded DNA binding protein RPA facilitates recombination-mediated DNA synthesis by increasing the efficiency of primer utilization, preventing polymerase stalling at specific sequence contexts, and overcoming polymerase stalling caused by topological constraint allowing the transition to a migrating D-loop. Our results support a model whereby the high-fidelity replicative DNA polymerase δ performs recombination-associated DNA synthesis, with translesion synthesis polymerases providing a supportive role as in normal replication.
INTRODUCTION
Homology-directed repair of double-stranded DNA breaks (DSB) is essential to maintain genomic integrity. Homologous recombination (HR) is a high-fidelity repair process, and deficiencies in genes involved in recombination have been shown to predispose individuals to tumor formation (1). In HR-mediated repair, a DSB is processed to generate 3′-terminal single-stranded DNA (ssDNA), which is then bound by the ssDNA binding protein RPA (see Figure 1). Formation of the RAD51 nucleoprotein filament on such substrates involves RAD51 paralogs, BRCA2 and other factors, whereupon homology search takes place. The 3′-ssDNA invades the homologous donor double-stranded DNA (dsDNA) to initiate DNA strand exchange. At this stage, DNA synthesis primed by the invading strand occurs (first end synthesis), after which the recombination intermediate can be processed in a number of ways, leading to non-crossover or crossover outcomes. The precise mechanisms by which DNA synthesis takes place during HR are not fully understood. In humans, 15 different DNA polymerases have been identified (2). The primary replicative polymerases epsilon (ε) and delta (δ) are responsible for the bulk of genomic nuclear replication, performing synthesis on the leading and lagging strands, respectively. Pol ε and Pol δ are the most high-fidelity polymerases in mammals, generating less than one error per 105 base pairs synthesized; they also possess 3′–5′ proofreading exonuclease activities capable of excising misincorporated bases, rendering replicative synthesis a highly faithful process (2). On encountering a synthesis-blocking lesion, replicative polymerases dissociate and translesion synthesis (TLS) polymerases can be recruited to bypass these lesions (3). Unlike replicative polymerases, TLS polymerases lack proofreading exonuclease activity and synthesize DNA in both a distributive and low-fidelity manner (2). These properties of TLS polymerases allow them to bypass bulky or uninformative lesions in DNA, sacrificing fidelity to prevent DNA breaks that may lead to chromosomal rearrangements and tumorigenesis (2).
Figure 1.
Pathways of homologous recombination involve different modes of DNA synthesis. Homologous recombination (HR) initiates with the common steps of DSB processing, Rad51 nucleofilament assembly, homology search and DNA strand invasion. Following D-loop formation, three different HR pathways are recognized: double Holliday Junction (dHJ), Synthesis-Dependent Strand Annealing (SDSA) and BIR. The initial DNA synthesis is displacement synthesis primed from the 3′-OH end of the invading strand in the D-loop (first end DNA synthesis). There is evidence that this intermediate is already pathway-specific, but it is unclear, how this specification is achieved. In the dHJ pathway, the second end of the DSB is captured by the displaced strand of the D-loop. This second end DNA synthesis does not involve displacement synthesis per se, but after filling the gap may involve displacement synthesis once the extension reaches the 5′-resected end. Likewise in SDSA, after D-loop dissolution and annealing of the extended first strand, the second end synthesis is by a non-displacement mode at least until the gap is filled. In BIR, the requirements are more akin to elongation during replicative DNA synthesis.
As HR is a high-fidelity repair pathway, Pol ε or Pol δ might be expected to perform HR-mediated repair synthesis, and genetic evidence in Saccharomyces cerevisiae supports this hypothesis (4). Mutations in the catalytic subunit of Pol δ were shown to result in shorter gene conversion tract lengths (5), and HR events leading to chromosomal translocations were found to be dependent on Pol32, a subunit of Pol δ (6). The observation that Pol δ proofreading exonuclease activity is required to remove non-homology from DNA ends during HR-mediated DSB repair suggests that Pol δ can directly access the 3′-end of the invading strand (7). Additionally, it has been shown that mutations generated during gene conversion in S. cerevisiae were dependent on Pol ε and Pol δ, but not the TLS polymerases zeta (ζ) or eta (η) (8). Finally, biochemical experiments demonstrated that yeast Pol δ was far more efficient than Pol η at carrying out DNA synthesis at Rad51-mediated recombination intermediates (9,10). While these data demonstrate it is likely that replicative polymerases carry out the bulk of DNA synthesis during HR, genetic evidence in yeast and vertebrates provides compelling evidence for at least some role for TLS polymerases in recombination. In yeast, mutations at regions flanking a DSB repair site were found to be dependent on Rev3, which encodes the catalytic subunit of Pol ζ (11). In humans, recombination was significantly reduced in cell lines deficient in REV3, REV7 or REV1 (12). Pol η, a TLS polymerase involved in UV repair and antibody maturation, was shown to be required for efficient DSB-induced gene conversion in chicken DT40 cells (13). Additionally, biochemical experiments have suggested a role for human Pol η, but not Pol δ, in the extension of protein-free D-loops (14).
DNA synthesis during replication involves specific features that distinguish leading and lagging strand synthesis (15). Leading strand synthesis can be contiguous, extending over large distances and is executed by DNA Pol ε (16,17). Lagging strand synthesis is discontiguous, extending only to the 5′-terminus of the preceding Okazaki fragments that may only be a few hundred nucleotides long, and is catalyzed by DNA Pol δ (18–20). Lagging strand synthesis also involves displacement DNA synthesis to replace the RNA primer of the preceding Okazaki fragment with DNA (15,21,22). Both replicative DNA polymerases, ε and δ, are dependent on the processivity clamp PCNA, which endows processivity through tethering of the polymerase complex to the template DNA (23,24). As illustrated in Figure 1, recombination-associated DNA synthesis involves mechanistically different modes of DNA synthesis. Extension of the invading strand in the D-loop (first end synthesis) is akin to leading strand synthesis, as it can be contiguous. However, in leading strand synthesis, strand displacement is catalyzed by the replicative DNA helicase, whereas first end DNA synthesis from the invading strand in a D-loop during HR requires the displacement of one strand from the duplex template DNA without involvement of the replicative helicase (4). Mammalian DNA Pol ε is incapable of displacement synthesis (25), and hence an unlikely candidate for first end DNA synthesis during HR. DNA synthesis involving the second end is also leading strand type in that it can be contiguous but does not require displacement synthesis. HR is a high-fidelity DNA repair process involving extensive DNA synthesis. Given the complexity due to the presence of 15 different nuclear DNA polymerases in humans, it is critical to understand which DNA polymerases participate in HR. Biochemical reconstitution has been the approach of choice to decipher the mechanism of DNA replication and identify specific functions for DNA polymerases and its various cofactors at the replication fork (26).
Here, we reconstituted a homologous recombinational repair system with purified human proteins. While the fundamental function of many HR proteins is conserved. Some human HR proteins display different biochemical properties than their yeast counterparts. For example, the DNA binding properties of human RAD51 differ substantially from those of the yeast enzyme (27). Furthermore, human Pol δ lacks the processivity that characterizes the S. cerevisiae protein (28–30). Displacement synthesis at the lagging strand is different from displacement synthesis during first end synthesis in HR (Figure 1), involving displacement of a 5′-ending strand during lagging strand synthesis as opposed to displacing a contiguous strand during HR. Moreover, HR-associated DNA synthesis, excepting Break-Induced Replication (BIR), does not involve the replicative helicase and hence lacks the coordination of leading and lagging strand in the replication fork (4). Nevertheless, we show that human DNA Pol δ is capable of efficient and processive synthesis on D-loops formed by RAD51 and RPA, in a manner dependent on the replication clamp PCNA and the clamp loader RFC. RPA greatly stimulates recombination-associated DNA synthesis by binding to both the displaced and template strands of donor DNA at D-loops. RPA also helps to overcome polymerase stalling due to topological constraint. While human TLS Pol η is also capable of efficient D-loop synthesis stimulated by PCNA, its distributive activity does not allow for processive synthesis as compared with Pol δ. These results significantly extend previous data in yeast and vertebrates, supporting an important role for the replicative DNA polymerase δ in HR in humans and identifying novel roles of RPA during HR. The data allow for a supportive role for TLS polymerases in HR repair, in a manner similar to that performed during normal replication.
MATERIALS AND METHODS
Proteins
Native RAD51, native RPA, His6 C-terminally tagged DNA Pol η, DNA Pol δ (His-tagged at p58 and p13; this enzyme showed no difference to the untagged version), RFC with His-tagged p38 and N-terminally truncated RFA1 (this enzyme showed no difference to the full-length untagged version) and native PCNA were purified as described (29,31–35).
Substrates
pBluescript KS+ ssDNA and pUC19 plasmid DNA were purified as described (36). The 93mer oligonucleotide (olWDH1267, 5′-AAAGGCGGTAATACGGTTA TCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAA-3′) was obtained from Integrated DNA Technologies and purified by 16% denaturing acrylamide gel electrophoresis.
Reconstituted DSB repair assays
All reactions contained 2.4 μM (nt) ssDNA, 25.8 nM (molecule) supercoiled pUC19 and, where indicated, 0.8 μM RAD51, 20 nM DNA Pol δ or η, 20 nM RFC, 100 nM PCNA and the indicated concentrations of RPA. D-loops were formed by incubating RAD51 with ssDNA in buffer containing 25 mM Tris–acetate (pH 7.4), 0.1% ampholytes (pH 3–10), 0.1 mg/ml bovine serum albumin (BSA), 2 mM DTT, 2 mM Mg(OAc)2, 1 mM ATP, 0.03 U/μl creatine kinase, 20 mM phosphocreatine, 2 mM CaCl2 and 500 μM each dGTP, dATP, dTTP, dCTP for 15 min at 37°C, followed by addition of supercoiled pUC19 plasmid and further incubating for 10 min. After D-loop formation, Ca++ was chelated by addition of 2 mM EGTA, and additional 3 mM Mg(OAc)2 was added, followed by addition of RFC, PCNA, DNA polymerase and wheat germ Topoisomerase I and incubation at 37°C. Reactions were terminated by the addition of 1 mg/ml Proteinase K/1% SDS and incubation for 10 min at 37°C. D-loops were analyzed by 0.8% agarose gel electrophoresis in 0.5× TBE. DNA synthesis was analyzed by 1.2% alkaline agarose, 5.5% denaturing polyacrylamide and two-dimensional agarose gel electrophoresis. Two-dimensional gel electrophoresis was carried out by excising lanes from native 0.8% agarose gel electrophoresis (first dimension), rotating these 90° and electrophoresis through 1.2% alkaline agarose (12 × 14 cm2). Gels were dried, imaged with a Molecular Dynamics Phosphor Imager and quantified using ImageQuant software.
DNA synthesis on canonical replication substrates
Reactions used 10 nM 93mer annealed to pBKS+ ssDNA, 8 nm RFC, 40 nM PCNA, 8 nM DNA polymerase and indicated concentrations of RPA, in 25 mM Tris–acetate (pH 7.4), 0.1% ampholytes (pH 3–10), 0.1 mg/ml BSA, 2 mM DTT, 2 mM Mg(OAc)2, 1 mM ATP, 0.03 U/μl creatine kinase, 20 mM phosphocreatine and 500 μM each dGTP, dATP, dTTP, dCTP for 5 or 30 min at 37°C. Reactions were terminated by the addition of 1 mg/ml Proteinase K, 1% SDS and incubation for 10 min at 37°C, and then analysed on 1.2% alkaline agarose and 5.5% denaturing acrylamide gel electrophoresis.
RESULTS
Human Polymerase δ catalyzes robust DNA synthesis at RAD51-mediated D-loops in a PCNA-dependent manner
As human Pol δ requires the accessory factor PCNA and its clamp loader RFC for efficient synthesis of canonical primer-template substrates, we rationalized that these factors would also be necessary for efficient synthesis during recombination-associated DNA synthesis (23,24). To investigate the role of replication factors in DNA synthesis of recombination intermediates, we reconstituted an in vitro DSB repair system using human proteins (RAD51, RPA, Pol δ, PCNA, and RFC) in a plasmid-based D-loop assay (Figure 2A). The reaction was carried out by first incubating RAD51 and RPA with a 5′-endlabeled 93mer ssDNA in the presence of calcium ions to form competent nucleoprotein filaments (37). Negatively supercoiled plasmid (pUC19, 2686 bp) was added to initiate D-loop formation. Calcium is required for efficient filament formation by hRAD51, and chelation is necessary to allow ATP hydrolysis, necessary for maximum D-loop formation, and to allow for DNA polymerases to be active. RFC, PCNA and polymerase were added to initiate DNA synthesis. As previously demonstrated, in vitro D-loop formation and DNA synthesis using human proteins does not require RAD54 (38).
Figure 2.
D-loop formation and DNA synthesis. (A) Experimental scheme. The single-stranded 93mer is 32P-endlabeled (asterisk). See also Figure 4A legend for aspects of topology. (B) Analysis of D-loop formation and DNA synthesis by Pol δ and Pol η at D-loops as measured by 0.8 % native agarose gel. Percent D-loops were determined at 0 min. (C) Cartoon depicting analysis of products by two-dimensional gel electrophoresis. (D) Two-dimensional gel electrophoresis of D-loop reactions extended by Pol δ in the presence or absence of RFC, PCNA after 30 min extension time. (E) Same as in D, using Pol η.
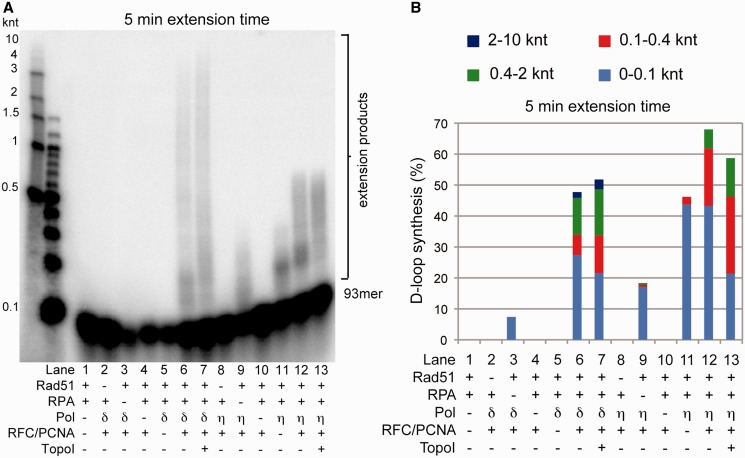
The reconstituted human D-loop formation/extension reactions were efficient, generating ∼30% D-loops (Figure 2B). Pol δ is capable of robust D-loop extension, and recombination-associated DNA synthesis is dependent on the presence of RAD51, PCNA and RFC (Figure 2B and D). In the presence of these factors and RPA, after 5 min incubation, Pol δ extended on average 48% of total primer termini engaged in D-loops, with 14% of products >400 nt in length, and 2% of products over 2 kb (Figure 3A and B, lane 6). We routinely observed products as long as 10 000 nt in these assays after 5 min incubation; longer products were predominant after 30 min incubation (Figure 2D and Supplementary Figure S1). This is significantly longer than the 2686 bp duplex DNA template and demonstrates that DNA synthesis in these reactions is processive and proceeds as a rolling circle.
Figure 3.
Efficient DNA synthesis by Pol δ. (A) Analysis of DNA synthesis products as measured by 1.2 % alkaline agarose gel electrophoresis with samples after 5 min extension time by polymerases. (B) Quantitation of DNA synthesis products shown in (A), as a function of size of products as well as percent of D-loops extended. Numerical data and errors are in Supplementary Table S1. knt: 1000 nucleotides.
While Pol δ is capable of limited DNA synthesis in the absence of PCNA and RFC, its efficiency has been shown to be low (39,40). Consistent with these observations and earlier studies using synthetic D-loops (14), we show that in the absence of these factors, Pol δ fails to catalyze significant DNA synthesis at D-loops (Figure 2D), and extended only 0.3% of substrate (Figure 3A and B, lane 5) with an average extension length of 5–10 nt (Figure 5D, lane 2). Single-stranded binding protein RPA is required both for recombination and efficient DNA replication (41). In the absence of RPA, D-loop formation was reduced by about one-third from ∼30 to ∼20% (Figure 2B), consistent with the previously identified role of RPA to bind to the displaced strand of the D-loop to stabilize this joint molecule (42). Omission of RPA also severely inhibited DNA synthesis by Pol δ. Pol δ extended only 7.4% of D-loops compared with 48% in the presence of RPA (Figure 3A and B, lanes 3 and 6; Figure 5D, lanes 3–10). In addition, DNA products formed were significantly shorter in the absence of RPA (Figures 2 and 3), owing to stalling induced by topological constraint and DNA sequence context, as discussed below. In the absence of RAD51, Pol δ generated a small amount of high molecular weight products that resulted from spontaneous annealing of the 93mer to the duplex DNA (Figure 2B). Pol δ contains a 3′–5′ nuclease and undergoes cycles of proofreading and DNA synthesis, a phenomenon termed idling (43), which causes loss of label in some reactions, in particular those lacking RPA, where idling is enhanced. In conclusion, we reconstituted a robust in vitro system with human proteins to analyze recombination-associated DNA synthesis from the invading strand of RAD51-mediated D-loops and show that human DNA polymerase δ efficiently extends the invading strand. The efficiency of using the invading 3′-end in the D-loop is similar to that observed with canonical primed templates (Figure 3B and Supplementary Figure S2) using the identical primer (93mer) and the same template sequence, as pBluescript and pUC19 share the same sequence for 1766 nt from the primer 3′-end. More importantly, the extension products displayed a highly similar length distribution, demonstrating that displacement synthesis is catalyzed with similar processivity as canonical primer extension by human Pol δ.
Figure 5.
RPA facilitates efficient D-loop extension by Pol δ. (A) Reactions were carried out as described, with RPA titrated at the following concentrations: 0.1, 0.25, 0.5, 1.0, 2.0, 4.0, 6.0 μM. Samples (30 min time point) were analyzed by 1.2 % alkaline agarose gel electrophoresis. (B) Quantitation of synthesis products in (A). Numerical data and errors are in Supplementary Table S2. (C) Quantitation of representative sites of replication stalling (0–2 µM: average ± SEM, n = 3). (D) Reactions in (A), analyzed by 5.5 % denaturing acrylamide gel electrophoresis. Arrows indicate examples of G-tracts on the newly synthesized strand that elicit polymerase stalling that is suppressed by RPA. The initial stall region due to topological constraints is suppressed by addition of topoisomerase I (see Figure 4D) and marked on the right hand side.
Human Polymerase η catalyzes non-processive DNA synthesis at RAD51-dependent D-loops
It has been shown previously that human Pol η was able to extend RAD51-dependent D-loops in a similar assay; however, the previous studies omitted deoxycytidine triphosphate from the reaction to limit DNA extension to ≤30 nt. Thus, processivity, polymerase stalling and the effects of DNA topology could not be examined (38). Here, we show that, like Pol δ, synthesis by Pol η is dependent on RAD51-mediated formation of D-loops (Figures 2B and 3A). However, unlike Pol δ, Pol η is able to extend D-loops in the absence of PCNA, extending an average of 46% of D-loops (Figure 3B). Pol η synthesis products were <400 nt in length, consistent with the more distributive nature of Pol η (Figures 2E and 3A and B, lane 11) (44). On addition of PCNA/RFC, an average of 68% of D-loops were extended and extension products increased ∼2-fold in length (Figure 3A and B, lane 12). The low processivity of Pol η synthesis at D-loops and the mild stimulation by PCNA is consistent with its polymerase activity on canonical primer-template substrates (Supplementary Figure S2) (45). As with Pol δ, RPA allowed more efficient usage of the invading 3′-end and led to longer extension products of up to 500 nt (Figure 3B). These data appear to show that human DNA polymerase η extends the invading strand of RAD51-mediated D-loops with equal if not greater efficiency than Pol δ. However, Pol η uses more primer because it is more distributive, catalyzing the addition of 7–10 nt per binding event (46), and dissociates/reassociates with the primer more readily than Pol δ. Surprisingly, the Pol η synthesis products displayed a similar length distribution in D-loop extension and canonical primed templates, showing that Pol η is capable of vigorous displacement synthesis. As expected, Pol η catalyzes recombination-associated displacement synthesis in a less processive manner than Pol δ, leading to significantly shorter extension products. Like Pol δ, Pol η extends the invading strand in D-loops with similar efficiency (Figure 3B) to a canonical primer:template with the same sequence context (Supplementary Figure S2).
DNA synthesis proceeds via a migrating D-loop
Extension of D-loops by DNA synthesis might proceed by one of two non-exclusive mechanisms. In the first, the D-loop size is directly correlated to the length of DNA synthesis, resulting in an expanding D-loop (Figure 4B). Alternatively, synthesis might proceed via a migrating D-loop bubble of relatively constant size (47). In our assay, the first mechanism would result in torsional strain caused by a DNA synthesis-dependent increase in D-loop size, due to unwinding of the negatively supercoiled plasmid, at one supercoil for every 10.4 bp of D-loop formed/extended (see also Figure 4A legend). The length of synthesis products would be determined by the D-loop size at which torsional strain caused by loss of negative supercoils and addition of positive supercoiling stalls the DNA polymerase. However, if synthesis proceeds via a migrating D-loop bubble of relatively constant size, synthesis length will not be inhibited, as the plasmid does not undergo significant changes in supercoiling during DNA synthesis. Eukaryotic chromosomes are topologically constrained by various mechanisms involving the telomeres and interstitial regions, so that in eukaryotes this topological question becomes mechanistically relevant for DNA replication and recombination-associated DNA synthesis.
Figure 4.
DNA synthesis proceeds via a migrating D-loop. (A) Experimental scheme. Single-stranded 93mer is 32P-endlabeled (asterisk). The dsDNA substrate contains ∼15 negative supercoils (−sc). On D-loop formation with the 93mer, this changes to ∼6 negative supercoils. For each 10.5 nt synthesized, one positive supercoil (+sc) is added, resulting in the accumulation of positive supercoils during D-loop extension. (B) Analysis of D-loop formation and DNA synthesis in the presence or absence of TopoI as measured by 0.8 % native agarose gel electrophoresis. (C) Cartoon depicting analysis of products by two-dimensional gel electrophoresis. (D) Two-dimensional gel electrophoresis of D-loops extended by Pol δ in the presence or absence of TopoI.
To distinguish between the two modes of DNA synthesis, we compared D-loop extension by Pol δ and Pol η in the presence or absence of wheat germ Topoisomerase I (TopoI) (Figure 3). Eukaryotic TopoI removes both positive and negative supercoils. In the expanding D-loop model, the torsional strain generated is expected to be relieved, resulting in an increase in synthesis product length. In the migrating D-loop model, addition of DNA topoisomerase is not expected to have an effect on the length of the extension products. In the presence of stoichiometrically saturating amounts of RPA and in the absence of TopoI, we observed significant transient stalling of both Pol δ and Pol η after ∼100 nt synthesis (Figure 3A and B, lanes 6 and 12; Supplementary Table S1; Figure 4). High-resolution analysis suggests that stalling occurs in a region of 50–150 nt synthesis (Figure 5D). Together with the change in supercoiling from the invading 93mer, this is equivalent to a change of ∼19 positive supercoils. The addition of TopoI significantly relieves stalling at this region, allowing synthesis by both polymerases to proceed past this block (Figure 3A and B, lanes 7 and 13, Figure 4). With the exception of this region, however, we observed no significant differences in product length distribution with either polymerase on addition of TopoI, indicating a lack of torsional constraint in the synthesis of longer DNA products. We also observed a slight decrease in the overall DNA synthesis in the presence of TopoI (Figure 3A and B, compare lanes 6 and 7, 12 and 13). This was expected, as addition of TopoI destabilizes D-loops and results in a decrease of available substrate for the DNA polymerases (Figure 4B and D).
We next analyzed D-loop synthesis products generated by Pol δ under native agarose gel conditions, in the presence or absence of TopoI. While supercoiled D-loops are stable over time, the relaxation of the plasmid by TopoI destabilizes D-loops (Figure 4B). Pol δ synthesis resulted in D-loops that migrated more slowly consistent with increased size, as well as dissociation of some fraction of extended primer. Under native conditions, little difference in D-loop synthesis products on addition of TopoI are detected (Figure 4B). We next separated these products in a second dimension by 2D gel electrophoresis under alkaline conditions. We observed three separate arcs: Arc 1 (Figure 4C and D, labeled 1 in c, see also Figure 2C and D) is composed of extended primers dissociated from the D-loop ranging in size from 100–2686 nt. Arc 2 (Figure 4C and D, labeled 2 in c, see also Figure 2C and D) is composed of extended primers on nicked plasmid, where the displaced strand has dissociated on reaching unit length of the 2686 bp pUC19 plasmid, enabling rolling circle synthesis. These structures range in size from 2.7 to >10 kb in length. Arc2 is only evident with dsDNA preparations that contain appreciable amounts of nicked DNA (Figures 2D and 4D). In the context of this product analysis, this was an advantage and allowed the differentiation of two-stranded structures (Arc2) from three-stranded structures (Arc3). Arc 3 (Figure 4C and D, labeled 3 in c, see also Figure 2C and D) is composed of extended D-loops where the displaced D-loop strand remains bound to the template strand, and synthesis proceeds via a migrating bubble. The species at ∼200 nt of this third arc corresponds to the stalled region observed under one-dimensional alkaline gel conditions (Figure 3A). This species, present in the absence of TopoI, disappears completely on TopoI addition as a result of the relief of torsional constraint. In our system, Pol δ synthesis generated products greater in length than the 2686 bp pUC19 plasmid used as the donor template (Figures 3A and 4B and D). If synthesis proceeded via an expanding D-loop mechanism, after completion of the first round of plasmid-length extension, further extension would necessarily proceed via rolling circle synthesis, as the displaced D-loop strand would then dissociate. In our assay, this would require that the displaced strand be nicked to allow dissociation. Analysis of these products on native agarose gels stained with ethidium bromide showed no nicking activity present in the reaction (data not shown).
Our results indicated that DNA synthesis proceeded in an expanding D-loop, until the size of the D-loop was ∼200 nt, at which time DNA synthesis transiently stalled because of the torsional strain caused by the buildup of positive supercoils ahead of the replication machinery. This strain was relieved by the dissociation of the 5′-end of the newly synthesized strand from the D-loop, and binding of RPA prevented reannealing, whereby synthesis continued via a migrating D-loop with a continuously dissociating 5′-end.
RPA stimulates DNA synthesis at D-loops via binding to the template strand of donor DNA
The ssDNA binding protein RPA is a crucial factor in DNA replication. It was previously demonstrated that DNA synthesis of canonical primer-template substrates is maximally stimulated by RPA addition to a stoichiometry of 1 RPA:30 nt ssDNA, the binding site size for human RPA (48). Extension of D-loops during recombinational repair synthesis requires displacement of the complementary donor strand by the invading DNA strand. The precise mechanism of this type of strand-displacement synthesis is not understood. It has been demonstrated that RPA binds the displaced strand during D-loop formation by Rad51, stabilizing the D-loop intermediate (42). This explains why we observe a reduction in D-loops levels in reactions lacking RPA in the absence of DNA synthesis (Figure 2B). On canonical templates, RPA binds both the primer-template junction of DNA, facilitating initiation of DNA synthesis, and to the template strand downstream of the 3′-primer terminus, facilitating synthesis by removing secondary structures (41,49,50). We hypothesized that during recombination-associated DNA synthesis from the invading strand of the D-loop, RPA would also bind the template strand in addition to the demonstrated binding of the displaced D-loop strand.
We observed maximum DNA synthesis for Pol δ at 2 μM RPA, a concentration that saturates the available RPA binding sites of both the displaced and template strands of the donor template, as normalized to the percent of D-loop products generated in these reactions. At concentrations above 2 μM, RPA inhibits DNA synthesis by Pol δ (Figure 5A and B, lanes 9–10; Supplementary Table S2). This is consistent with observations that supersaturating RPA is inhibitory to DNA synthesis on canonical templates (Supplementary Figure S2B and C). If, as in DNA replication, RPA binds to the template strand of D-loops, then subsaturating concentrations of RPA will be expected to limit the polymerase processivity in much the same manner as on canonical DNA substrates. This is exactly what we observe: at 0.1, 0.25, 0.5 and 1 μM RPA, where RPA fails to saturate the maximum number of binding sites, there is a marked decrease in processivity of synthesis by Pol δ (Figure 5A and B, lanes 3–7). In addition to lowered processivity of synthesis, we observe sites of replication stalling that become more prominent with decreasing RPA concentrations. We mapped these stall sites and found that several of the more prominent sites contain short repetitive guanine tracts, which have been shown to be prone to the formation of secondary structures (Figure 5C) (51,52). These stall sites at G tracts are also found in reactions with canonical primer-templates, which uses the same 93mer and ssDNA template that has identical sequence to pUC19 for 1776 nt from the 3′-terminus of the 93mer. Additionally, primer utilization was positively affected by increasing RPA concentrations (Figure 5B). At limiting concentrations of RPA, initiation of DNA synthesis was inhibited, as indicated by the decrease in primer utilization (Figure 5B). We interpret these results to indicate that RPA is bound on the template strand in the D-loop ahead of the DNA polymerase and suggests that the D-loop might be larger than estimated based on the length of the DNA synthesis products.
DISCUSSION
Here, we report the reconstitution of human recombination reactions involving DNA Pol δ, PCNA and RFC. This reconstituted system provides the first evidence of an involvement of human Pol δ in HR and allowed us to generate significant information about novel roles of RPA during HR. First, human Pol δ efficiently extends RAD51-mediated D-loops (Figure 6). Extension of the invading strand is dependent on the processivity clamp PCNA and displacement synthesis can extend over significant distances of several kb. The efficiency of D-loop extension by DNA Pol δ is surprisingly high and indistinguishable from that with canonical primer:templates. These findings are consistent with other in vitro data using human and yeast Pol δ (9,10,14) (L. Krejci, personal communication). Together with compelling genetic evidence in yeast (4–8), these data suggest that DNA Pol δ might be the primary polymerase that extends the invading strand present in a RAD51-mediated D-loop. Based on the reported biochemical properties for calf thymus DNA Pol ε (25), it appears unlikely that Pol ε is involved in first end synthesis during HR, due to its inability to perform strand displacement synthesis. The genetic requirement of HR for DNA Pol ε in yeast (4) may suggest that this DNA polymerase plays a role in second end synthesis during HR but there is currently no experimental evidence for this (Figure 1).
Figure 6.
Recombination-associated DNA synthesis. RAD51-mediated homology search and DNA strand invasion generates the D-loop intermediate and the invading 3′-OH end serves to initiate recombination-associated DNA synthesis. After dissociation of RAD51 from the heteroduplex DNA, RFC loads PCNA at the 3′-junction enabling binding of DNA polymerase. This process is aided by RPA binding to ssDNA. DNA polymerase initiates DNA synthesis and comes to an initial stall owing to the accumulation of positive supercoils. Extrusion of the 5′-end of the invading DNA alleviates the topological constraint and allows topologically unhindered extension via a migrating D-loop. RPA binding to the template strand in front of DNA polymerase to facilitate processive DNA synthesis.
Second, D-loop extension by human Pol δ proceeds as a migrating bubble as observed for T4 recombination (47) after an initial transient stall induced by topological constraint. D-loop extension creates significant topology problems. For every 10.4 bp of D-loop, formed either by strand invasion or DNA synthesis, one positive supercoil is induced, which ultimately leads to stalling of the DNA polymerases owing to topological constraints. In cells, this problem could be solved by DNA topoisomerases, but there is currently no evidence for such a mechanism. Biochemical studies with phage T4 recombination proteins illustrated another mechanism to overcome this problem by extrusion of the invading strand at the end opposite, where DNA synthesis takes place (47) (Figure 6). In our in vitro system, we observed by one- and two-dimensional analysis of reactions products that a fraction of DNA polymerases stalled in a region after 50–150 nt synthesis (Figure 4D and 5D). Importantly, this stalling was significantly suppressed by Topoisomerase I addition (Figure 4D, Supplementary Table S1). No other prominent stall sites were suppressed by topoisomerase addition, suggesting that only this initial stall is topological. Moreover, the majority of the extension products extend beyond this stall site, suggesting that this stall is only temporary. We propose that a significant fraction of the DNA polymerase stalls due to topological constraint after only 50–150 bp of synthesis (Figure 6). Extrusion of the 5′-end of the invading strand then switches the system to a migrating D-loop, which alleviates the topological problem and allows contiguous synthesis. This switch depends on saturating amounts of RPA. Insufficient RPA concentrations led to more significant stalling (see Figure 5), as previously observed in the yeast system (9), where subsaturating amounts of RPA were used. These results suggest a novel role of RPA in HR to bind the invading strand after extrusion during D-loop extension, which generates the substrate for strand annealing with the resected second end during Synthesis-Dependent Strand Annealing (SDSA) (Figure 6). The exact position of the initial stall is defined by the superhelicity of the template DNA. Presently, it is not known how DNA synthesis from the invading strand is regulated. A topology-limited D-loop extension could provide a mechanism to limit DNA synthesis in vivo during SDSA (Figure 1) or multiple invasion cycles (53–56).
Third, RPA is known to bind the displaced strand in the D-loop and stabilize D-loop formation (42) (see Figures 2B and 5). Here, we discovered a novel role of RPA during HR: binding to the opposite template strand in the D-loop (Figure 6). RPA suppresses polymerase stalling at certain sequence motifs, particularly tracts of G residues (Figure 5D). Importantly, the RPA-suppressed stall sites were the same in D-loop extension and extension of canonical primer:templates using substrates with the identical sequence context. It has been previously shown that during extension of standard primer:templates, RPA suppresses DNA sequence context-mediated polymerase stalling by binding to the template strand (41,57). During HR-associated DNA synthesis, the DNA polymerase encounters potential stall sites on the template within the confines of the D-loop, where the DNA is not under topological stress. While it has been demonstrated that PCNA can be loaded bidirectionally at a bubble (58), the structure of the D-loop is more complex and requires unidirectional loading. The architecture of the D-loop is poorly understood, in particular with respect to how PCNA can be loaded by RFC and how polymerase accesses the 3′-OH. Recent observations by Atomic Force Microscopy of human RAD51-mediated D-loops showed that the template duplex is sharply kinked, resulting in a 90° angle at the end of the D-loop opposite the invading 3′-end (59). This kink potentially induces the unwinding necessary to generate unpaired DNA that can be bound by RPA ahead of the invading 3′-end. The binding site size of human RPA has been estimated to be 30 nt, but RPA may also engage in binding modes with a smaller site size (41).
Fourth, RPA significantly increases the efficiency of primer extension by DNA polymerase (Figures 3A and B and 5D), which is a different role than preventing stalling after initial extension due to topological constraint and DNA sequence context discussed above. It appears likely that this role of RPA is a reflection of its role in DNA replication to enhance RFC-mediated loading of PCNA (41,50).
Fifth, consistent with other biochemical studies (14,38) (L. Krejci, personal communication), human Pol η extends RAD51-mediated D-loops, and we determined that the efficiency of primer utilization was similar to that of Pol δ (Figure 3B). Surprisingly, Pol η was able to perform strand displacement synthesis during D-loop extension of up to 400 nt after 5 min, and the length of the extension products was enhanced ∼2-fold by the addition of PCNA/RFC (Figure 3). This is significantly longer than expected from a translesion polymerase, whose primary function is to insert correct nucleotides opposite UV-induced pyrimidine dimers (46). Pol η has been implicated in gene conversion and HR-mediated DSB repair in chicken DT40 cells (13), but studies in human cells failed to identify a contribution of Pol η to HR (12,60). However, a possible role for other translesion polymerases in HR cannot be ruled out (11,61,62).
HR is a high-fidelity DNA repair pathway, and we provide evidence that this fidelity is achieved by engaging the high-fidelity replicative DNA Pol δ. The length of conversion tracts during HR in mammals has been estimated to be between 1 to >10 kb in length using different cell lines and recombination systems and provides an estimate for the amount of DNA synthesis required (63–66). This extent of synthesis strongly suggests the involvement of a processive replicative DNA polymerase. Similar to DNA replication, translesion polymerases may provide a supporting role. Establishment of a reconstituted in vitro system now allows identification of additional cofactors that affect D-loop extension and D-loop dissociation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–2.
FUNDING
National Institutes of Health (NIH) [GM38559 to J.H. and GM58015 to W.D.H.]. J.L.S. was a trainee on the T32 training grant in Oncogenic Signals and Chromosome Biology [CA108459]. Funding for open access charge: NIH [GM58015].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank X.-P. Zhang for providing materials, D. Meyer, W. Wright, E. Schwartz, C. Fasching, J. Liu and C. Ede for comments on the manuscript and L. Krejci for the exchange of unpublished results.
REFERENCES
- 1.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang XA, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz JF, Gomez-Gonzalez B, Aguilera A. Chromosomal translocations caused by either Pol32-dependent or Pol32-independent triparental break-induced replication. Mol. Cell. Biol. 2009;29:5441–5454. doi: 10.1128/MCB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pâques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiating recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebesta M, Burkovics P, Haracska L, Krejci L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair. 2011;10:567–576. doi: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res. 2012;40:682–691. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 14.McIlwraith MJ, Vaisman A, Liu YL, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyabe I, Kunkel TA, Carr AM. The Major Roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. Plos Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larrea AA, Lujan SA, McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl Acad. Sci. USA. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast - I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 22.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast - II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 23.Uhlmann F, Cai J, Flores-Rozas H, Dean FB, Finkelstein J, O'Donnell M, Hurwitz J. In vitro reconstitution of human replication factor C from its five subunits. Proc. Natl Acad. Sci. USA. 1996;93:6521–6526. doi: 10.1073/pnas.93.13.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 25.Podust VN, Hubscher U. Lagging strand DNA synthesis by Calf Thymus DNA polymerases-alpha, polymerase-beta, polymerase-delta and polymerase-epsilon in the presence of auxiliary proteins. Nucleic Acids Res. 1993;21:841–846. doi: 10.1093/nar/21.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornberg A, Baker TA. DNA Replication. New York: W.H.Freeman and Company; 1992. [Google Scholar]
- 27.Heyer WD. In: Molecular Genetics of Recombination. Aguilera A, Rothstein R, editors. Berlin, Heidelberg: Springer-Verlag; 2007. pp. 95–133. [Google Scholar]
- 28.Hu Z, Perumal SK, Yue H, Benkovic SJ. The human lagging strand DNA polymerase delta holoenzyme is distributive. J. Biol. Chem. 2012;287:38442–38448. doi: 10.1074/jbc.M112.404319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bermudez VP, Farina A, Raghavan V, Tappin I, Hurwitz J. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J. Biol. Chem. 2011;286:28963–28977. doi: 10.1074/jbc.M111.256289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langston LD, O'Donnell M. DNA Polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J. Biol. Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl Acad. Sci. USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 33.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee CG, Phillips B, Finkelstein J, Yao N, O'Donnell M, Hurwitz J. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc. Natl Acad. Sci. USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Sneeden J, Heyer WD. In vitro assays for DNA pairing and recombination-associated DNA synthesis. Methods Mol. Biol. 2011;745:363–383. doi: 10.1007/978-1-61779-129-1_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 39.Lee SH, Pan ZQ, Kwong AD, Burgers PM, Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J. Biol. Chem. 1991;266:22707–22717. [PubMed] [Google Scholar]
- 40.Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 41.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 42.Eggler AL, Inman RB, Cox MM. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J. Biol. Chem. 2002;277:39280–39288. doi: 10.1074/jbc.M204328200. [DOI] [PubMed] [Google Scholar]
- 43.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 45.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 46.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 47.Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 48.Kim CS, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 49.Bartos JD, Willmott LJ, Binz SK, Wold MS, Bambara RA. Catalysis of strand annealing by replication protein A derives from its strand melting properties. J. Biol. Chem. 2008;283:21758–21768. doi: 10.1074/jbc.M800856200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Weitzmann MN, Woodford KJ, Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 53.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 54.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 55.McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray A, Machin N, Stahl FW. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1989;86:6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutL alpha endonuclease in mismatch repair. Proc. Natl Acad. Sci. USA. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ristic D, Kanaar R, Wyman C. Visualizing RAD51-mediated joint molecules: implications for recombination mechanism and the effect of sequence heterology. Nucleic Acids Res. 2011;39:155–167. doi: 10.1093/nar/gkq766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cleaver JE, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe JP, Busch DB, Coleman DM, Ziffer DW, Yu Y, et al. Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- 61.Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 62.Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr. Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 63.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neuwirth EA, Honma M, Grosovsky AJ. Interchromosomal crossover in human cells is associated with long gene conversion tracts. Mol. Cell. Biol. 2007;27:5261–5274. doi: 10.1128/MCB.01852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruksc A, Bell-Rogers PL, Smith JD, Baker MD. Analysis of spontaneous gene conversion tracts within and between mammalian chromosomes. J. Mol. Biol. 2008;377:337–351. doi: 10.1016/j.jmb.2008.01.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.