Abstract
DNA mimic proteins are unique factors that control the DNA-binding activity of target proteins by directly occupying their DNA-binding sites. To date, only a few DNA mimic proteins have been reported and their functions analyzed. Here, we present evidence that the Neisseria conserved hypothetical protein DMP12 should be added to this list. Our gel filtration and analytical ultracentrifugation results showed that the DMP12 monomer interacts with the dimeric form of the bacterial histone-like protein HU. Subsequent structural analysis of DMP12 showed that the shape and electrostatic surface of the DMP12 monomer are similar to those of the straight portion of the bent HU-bound DNA and complementary to those of HU protein dimer. DMP12 also protects HU protein from limited digestion by trypsin and enhances the growth rate Escherichia coli. Functionally, HU proteins participate in bacterial nucleoid formation, as well as recombination, gene regulation and DNA replication. The interaction between DMP12 and HU protein might, therefore, play important roles in these DNA-related mechanisms.
INTRODUCTION
Neisseria species are a large family of commensal bacteria that include two human pathogens Neisseria meningitidis and Neisseria gonorrhoeae. (1,2). During the past decade, the complete genomes of many Neisseria species have been sequenced (3,4). However, in these sequenced genomes, a number of genes still lack functional annotations. Here, we present evidence that one of these uncharacterized Neisseria genes/proteins, N. meningitidis conserved hypothetical protein NMB2123, is a DNA mimic protein.
NMB2123 is a conserved protein that can be found in pathogenic N. meningitides and N. gonorrhoeae and non-pathogenic N. lactamica. The gene of this protein is located in an operon located on the 3′-end of the Neisseria maf-3 (N. meningitidis adhesion) gene. Microarray analysis showed the genes in this operon being significantly downregulated on Neisseria attachment to the epithelial cells, suggesting these genes may play a role during bacterial adhesion (5). NMB2123 was one of the candidate DNA mimic proteins selected for further study after a sequence analysis. As a small (118 amino acid) acidic (pI 4.39) protein, NMB2123 has two characteristics shared by the known DNA mimic proteins and, more importantly, a preliminary His–pull-down assay further showed that NMB2123 also interacted with a DNA-binding protein (bacterial histone-like HU protein). Subsequently in this article, we, therefore, refer to NMB2123 as DMP12 [DMP for DNA mimic protein; 12 (kDa) to indicate its SDS–PAGE location].
In this article, by using gel filtration and analytical ultracentrifugation, we found that DMP12 is in its monomeric form when it interacts with Neisseria HU protein dimer. We determined the crystal structure of DMP12, and we calculated the binding affinity between DMP12 and Neisseria HU using isothermal titration calorimetry (ITC). Finally, we used a limited trypsin digestion assay to show that DMP12 prevented Neisseria HU protein from being digested by trypsin. As HU proteins promote the assembly of higher-order DNA–protein structures (6–9), the interaction between DMP12 and HU protein may be instrumental in controlling the stability of the nucleoid in Neisseria.
MATERIALS AND METHODS
Preparation and purification of recombinant DMP12 and Neisseria HU protein
The methods used in this study were modifed from the standard research protocols developed in our laboratory for the DNA mimic protein DMP19 (10). The full-length DMP12 gene (NMB2123, amino acid residues 1–118) was ligated into pET16b expression vector (Novagen). After induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the recombinant DMP12 protein with its N-terminal His10-tag was expressed by Escherichia coli BL21 (DE3) cells at 37°C for 4 h. Soluble DMP12 was purified by immobilized metal-ion chromatography with an Ni-NTA (nitroloacetic acid) column, followed by gel filtration using Superdex 200 pg (GE Healthcare). The His10-tag was then removed by FactorXa cleavage (Novagen).
The full-length Neisseria HU gene (NMB1230, residues 1–89) with or without the stop codon were ligated into pET21b expression vector. After induction with 1 mM IPTG, the recombinant Neisseria HU proteins with or without C-terminal His6-tag were expressed by E. coli BL21 (DE3) cells at 16°C for 16 h. Soluble Neisseria HU protein with C-terminal His6-tag protein was purified by immobilized metal-ion chromatography with an Ni-NTA column, followed by gel filtration using Superdex 200 pg (GE Healthcare). Non-tagged Neisseria HU was purified by using a monoS ion exchange column and gel filtration with Superdex 200 pg (GE Healthcare). As Neisseria HU protein does not contain any Trp residues and its computed extinction coefficient is low (1 g/l = 0.159), we determined the concentration of this protein using a Coomassie blue dye-binding assay kit (Bio-Rad).
Bissulfosuccinimidyl suberate cross-linking and His–pull-down assays to confirm the interaction between Neisseria DMP12 and HU
For the cross-linking assay, 3 μM purified C-terminal His6-tagged Neisseria HU protein was mixed with 3 μM DMP12 in 1 ml of binding buffer [1× phosphate-buffered saline (PBS) with or without MgCl2], and the reaction mixture was incubated for 30 min at 25°C. After the addition of bissulfosuccinimidyl suberate (BS3) (Sigma-Aldrich) to a final concentration of 0.5 mM, incubation was then continued for 1 h. The reactions were quenched by 50 mM Tris–HCl. All reactions were analyzed by SDS–PAGE.
In a His–pull-down assay, which was to confirm the interaction between DMP12 and Neisseria HU, purified N-terminal His10-tagged DMP12 (final concentration: 1 μM) was mixed with Neisseria HU protein without any tag (final concentration: 3 μM) in 1 ml of binding buffer (1× PBS with 20 mM imidazole). The reaction mixture was incubated for 1 h at 25°C. Incubation was then continued with Ni-NTA beads (50 μl) for 1 h. After incubation, the beads were washed with 5 ml of the same binding buffer. The proteins that bound to the beads were eluted by the addition of buffer containing 500 mM imidazole and analyzed by SDS–PAGE.
Gel filtration to determine the native molecular weight of target proteins
To evaluate the approximate molecular weight of DMP12, as well as Neisseria HU and the DMP12/Neisseria HU complex, each protein was analyzed separately using a Superose 16 gel filtration column (Amersham Biosciences). The buffer used in this assay was 20 mM Tris, pH 7.4, and 5 mM MgCl2. Gel filtration standard proteins [ovalbumin (43 kDa), carbonic anhydrase (29 kDa) and RNase A (13.7 kDa)] were used to calibrate the column. For each protein, the logarithm of molecular weight (log MW) was plotted against Kav, which was calculated as follows: Kav = (Ve − Vo)/(Vt − Vo), where Ve is the elution volume, Vo is the column void volume using blue dextran 2000 and Vt is the total column bed volume (24 ml for the Superose 16 10/30 gel filtration column). For the sample loading, we used 100 µl of sample buffer containing 750 µM C-terminal His6-tagged Neisseria HU, or 1500 µM N-terminal His10-tagged DMP12, or else both 750 µM C-terminal His6-tagged Neisseria HU and 1500 µM N-terminal His10-tagged DMP12 together.
Recombinant DMP12 crystallization, data collection and structure determination
For crystallization, 2 µl of the DMP12 solution (20 mg/ml; 20 mM Tris, pH 8.0, and 100 mM NaCl) was mixed with 2 µl of a reservoir containing 0.2 M magnesium acetate, 0.1 M sodium cacodylate, pH 6.5, and 20% (w/v) PEG 8000 (polyethylene glycol 8000) as a precipitant. Equilibration with the reservoir was achieved by the sitting drop method. Before flash-cooling, ethylene glycol at a final concentration of 15% was added as a cryoprotectant. After soaking the DMP12 crystals with 1 mM AuCl3 (Sigma-Aldrich) for 30 min, both native and SAD (single-wavelength anomalous dispersion) X-ray diffraction data were collected on beamline 13B at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan. The data were processed using HKL2000 (11). The space group of the native and AuCl3 soaked-DMP12 crystals was tetragonal I4. Unit cell dimensions for the native DMP12 crystal and AuCl3 soaked-DMP12 are given in Table 1.
Table 1.
Data collection and refinement statistics of DMP12 crystals
| Data collection | AuCl3-DMP12 | Native-DMP12 |
|---|---|---|
| Wavelength (Å) | 1.02243 (high remote) | 1.00000 |
| Space group | I4 | I4 |
| Unit cell a, b, c (Å) | 116.5, 116.5, 54.9 | 112.7, 112.7, 57.3 |
| Resolution (Å) | 30–2.9 (3.0–2.9) | 50–1.85 (1.92–1.85) |
| Unique reflections | 8110 (792) | 30 304 (3012) |
| Redundancy | 4.8 (3.8) | 5.4 (5.5) |
| Completeness (%) | 97.5 (95.1) | 97.0 (97.6) |
| I/σ(I) | 21.5 (3.4) | 32.7 (2.7) |
| Rmerge (%) | 5.9 (42.4) | 4.7 (49.6) |
| Refinement | ||
| Rwork (%) | 18.6 | |
| Rfree (%) | 21.8 | |
| Bond RMSD (Å) | 0.018 | |
| Angle RMSD (°) | 1.7 | |
| Mean B value/no of atom | 30.1/2235 | |
| Ramachandran plot (%) | ||
| Most favored (%) | 98.67 | |
| Allowed (%) | 1.33 |
The DMP12 structure was determined using the SAD phasing method and the programs Shelx CDE and Phaser (12,13). High remote data in the range of 30–2.9 Å resolution collected at the wavelengths of 1.02243 Å were used. Four Au sites were located in an asymmetric unit (His 4 and His 117 from two monomers). The initial model was produced by the program BUCCANEER (14) and COOT (15) and revealed that each asymmetric unit contained two DMP12 molecules. The phase was further extended to 1.85 Å by using the native data set, and the programs COOT (15) and Refmac (16) were used for the subsequent refinement. Statistics for the data collection and refinement are shown in Table 1. The CCP4 package (17) and Pymol program (18) were used for the structural analyses and also for figure production.
Isothermal titration calorimetry to determine the binding affinity between Neisseria HU and DMP12/DNA
The binding affinity of Neisseria HU to DMP12 and dsDNA was determined by using an ITC200 (GE Healthcare) at 25°C (298 K). The 8mer dsDNA fragment used in this assay was prepared by incubating the complementary oligonucletide solution (5′-CTCGCGAG-3′) at 95°C for 3–5 min, and then lowering the temperature to room temperature (RT) for another 30 min. Before titration, both proteins and the DNA fragments were dialyzed into ITC buffer (20 mM Tris, pH 7.4, and 50 mM NaCl). For the Neisseria HU-DMP12 reaction, the syringe contained C-terminal His6-tagged Neisseria HU dimer at 400 μM, and the sample cell contained DMP12 monomer at 30 μM. Two microliters of the HU was injected every 3 min until the DMP12 was saturated (20 injections). The Neisseria HU-dsDNA reaction used 300 μM C-terminal His6-tagged Neisseria HU dimer in the syringe and 30 μM 8mer dsDNA in the sample cell, whereas the negative control reaction used 400 μM C-terminal His6-tagged Neisseria HU dimer in the syringe and 30 μM BSA (Bovine serum albumin; pI 4.7; Sigma-Aldrich) in the sample cell. Titration proceeded at the same rate in all assays, i.e. 2.0 μl every 3 min. Except for the first injection point, data from the entire titration curve were used to calculate the thermodynamic parameters. The program ORIGIN 7 (GE Healthcare) was used to calculate enthalpy (ΔH), binding entropy (ΔS), the equilibrium constant (1/Kd) and the stoichiometric ratio (N). Gibbs free energy (ΔG) was calculated using the equation ΔG = ΔH − TΔS.
DNA-binding assays (electrophoresis mobility shift assay assays)
For the HU/DNA-binding assays, 1, 2 and 4 µM C-terminal His6-tagged Neisseria HU protein was mixed with 2.5 nM plasmid DNA substrate (65 ng of the 2710-bp plasmid pUC57; GenScript) in 15 µl reaction buffer (20 mM Tris, pH 7.4, 100 mM NaCl and 5 mM MgCl2) and incubated at RT for 15 min. For the competitive binding assays, 4 µM C-terminal His6-tag protein HU protein was mixed with 50, 100 and 150 µM DMP12 in 14 µl of reaction buffer and pre-incubated at RT for 30 min. Subsequently, 1 μl of binding buffer containing the DNA substrate was added and incubation continued for 15 min. The reaction products were run on a 0.8% agarose gel and stained with SYBR Green I (Sigma-Aldrich).
Trypsin-limited digestion
The protocol used here followed that of Puvirajesinghe et al. (19), with minor modifications. The C-terminal His6-tagged Neisseria HU (200 µg) and DMP12 (300 µg) proteins were incubated in 200 µl of reaction buffer (20 mM Tris, pH 7.4) either alone or together in the presence of trypsin at a 1:500 ratio (w/w; trypsin:protein). Digestion started with the addition of trypsin, and the reaction was carried out at 4°C for 3 h. After incubation, uncleaved C-terminal His-tagged Neisseria HU was purified using Ni-NTA. The reaction products were analyzed on a 12% Bis–Tris SDS–PAGE and stained with Coomassie blue.
Cell growth assays
E. coli BL21 (DE3) cells were transformed with the expression vector pET21b that contains full-length DMP12 gene or an empty pET21b. When the bacterial cells grew to an absorbance at 600 nm of 0.5–0.8 at 37°C, the expression of DMP12 protein (or a null expression) was induced by addition of 1 mM IPTG, and the cell density was subsequently monitored by measuring OD600 each hour. To monitor the DMP12 expression at different times, cell lysate was extracted from the transformed E. coli cells using B-PER protein extraction reagent (Thermo Scientific). After centrifugation, the soluble fractions were analyzed by SDS–PAGE.
RESULTS
Observing the interaction between Neisseria HU protein and DMP12
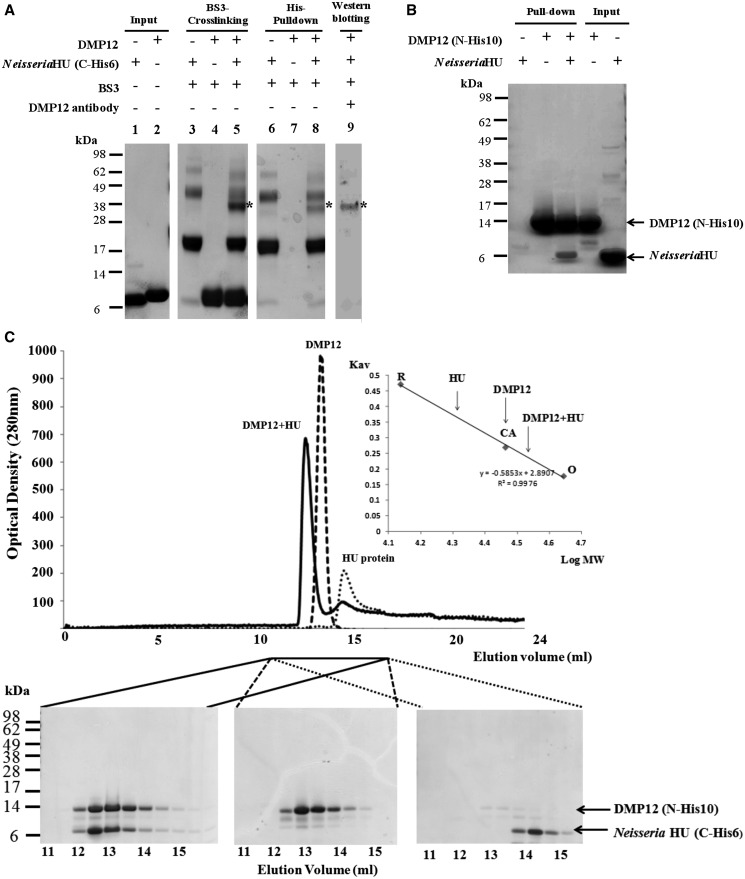
A preliminary pull-down assay to determine the targets of DMP12 showed that DMP12 may interact with two DNA-binding proteins: bacterial histone-like HU protein and integration host factor (IHF) (Supplementary Figure S1 and Supplementary Table S1). To further clarify these interactions, BS3 cross-linking and His–pull-down assays were used. The cross-linking results showed that only the Neisseria HU/DMP12 reaction produced signals that indicated cross-linking between the proteins (Figure 1A and Supplementary Figure S2). The His–pull-down assay also confirmed that Neisseria Hu without any tag could be pulled down by His10-DMP12 (Figure 1B), whereas IHF was not pulled down by His10-DMP12 (data not shown). These results clearly suggest a Neisseria HU protein/DMP12 interaction.
Figure 1.
BS3 cross-linking, His–pull-down and gel filtration assays all demonstrate an interaction between Neisseria DMP12 and HU. (A) A BS3 cross-linking assay demonstrates an interaction between DMP12 and C-terminal His6-tagged Neisseria HU. After the addition of BS3, shifted bands that indicated protein–protein cross-linking could be seen (lane 5); the shifted bands are marked with an asterisk. C-terminal His6-tagged Neisseria HU was pulled down using Ni-NTA beads, and cross-linked DMP12/ C-terminal His6-tagged Neisseria HU was detected (lane 8). The presence of DMP12 in the shifted bands was confirmed by using anti-DMP12 antibody (lane 9). (B) In this His–pull-down assay, HU without any tag was used as prey, and N-terminal His10-tagged DMP12 was used as bait. HU was pulled down by His10-tagged DMP12. (C) The molecular weights of DMP12, Neisseria HU and DMP12/HU complex were measured by gel filtration on a Superose 12 HR 10/30 column monitored at 280 nm. The standard proteins ovalbumin (OA; 43 kDa), carbonic anhydrase (CA; 29 kDa) and RNase A (RA; 13.7 kDa) were fractionated on the same column and used to generate a plot of Kav against log MW (inset). The Kav of each target protein (DMP12, HU and DMP12/HU complex) was then used to find its molecular weight (The DMP12 peak seems to correspond to its dimeric form in the low ionic buffer (20 mM Tris, pH 7.4, and 5 mM MgCl2) that was used in this gel filtration. However, in a higher ionic strength buffer, such as PBS (pH7.4), DMP12 only appears in its monomeric form; see Table 2, footnote C).
Next, gel filtration chromatography was used to investigate the native molecular weights of DMP12, Neisseria HU and the DMP12/Neisseria HU complex (Figure 1C). The theoretical molecular weights of tag-free DMP12 (13.8 kDa) and C-terminal His6-tagged Neisseria HU protein (10.4 kDa) are so close to each other that we used N-terminal His10-tagged DMP12 (16.4 kDa) in this gel filtration study to achieve better protein separation. Based on the protein standards that were fractionated in the same column (Figure 1C, inset), the apparent molecular weights of the target proteins were calculated to be 21, 29 and 35.7 kDa, respectively. The predicted molecular weights of N-terminal His10-tagged DMP12 and C-terminal His6-tagged Neisseria HU protein match well with their theoretical dimer molecular weights (32.8 and 20.8 kDa; Table 2). However, the apparent molecular weight of complex can only be matched with the sum of the theoretical molecular weights of DMP12 monomer and HU dimer (36.8 kDa; Table 2). Analytical ultracentrifugation experiments of this complex formation gave similar results (Supplementary Figure S3 and Table 2); therefore, we conclude that DMP12 forms a monomer-to-dimer complex with HU protein.
Table 2.
The results of gel filtration and analytical ultracentrifugation (AUC)
| Gel filtration |
Analytical ultracentrifugation |
|||
|---|---|---|---|---|
| Theoretical MWa | Calculated MW from Superose 12 column | Sb predicted by HYDROPRO | Observed S from AUC | |
| DMP12 (N-His10) | ||||
| DMP12 (N-His10) monomer | 16.4 kDa | not seenc | 2.0 | 2.01 |
| DMP12 (N-His10) dimer | 32.8 kDa | 29 kDa | 3.23 | not seenc |
| Neisseria HU (C-His6) | ||||
| Neisseria HU (C-His6) monomer | 10.4 kDa | not seen | 1.07 | not seen |
| Neisseria HU (C-His6) dimer | 20.8 kDa | 21 kDad | 1.9 | 2.11 |
| DMP12 (N-His10)/Neisseria HU (C-His6) complex | ||||
| DMP12 monomer to HU monomer | 26.8 kDa | not seen | 2.51 | not seen |
| DMP12 dimer to HU monomer | 43.1 kDa | not seen | 3.71 | not seen |
| DMP12 monomer to HU dimer | 36.8 kDa | 35.7 kDae | 3.19 | 3.02 |
| DMP12 dimer to HU dimer | 53.5 kDa | not seen | 4.3 | not seen |
aThe theoretical molecular weight is based on the amino acid sequence.
bHYDROPRO (20) predicted the S values based on the atomic coordinates model.
cIn a buffer of higher ionic strength (PBS), we found that DMP12 appeared only in monomeric form (data not shown), which suggests that the apparent presence of dimeric DMP12 might also be due to non-specific binding to the column material at low ionic strength.
dHigher ionic strength (PBS) had no effect on the Neisseria HU results (data not shown).
eAt Higher ionic strength (PBS), gel filtration results for the complex were broadened and slightly shifted (data not shown). This observation suggests that charge–charge interaction plays an important role in the DMP12/HU complex formation.
The crystal structure of DMP12
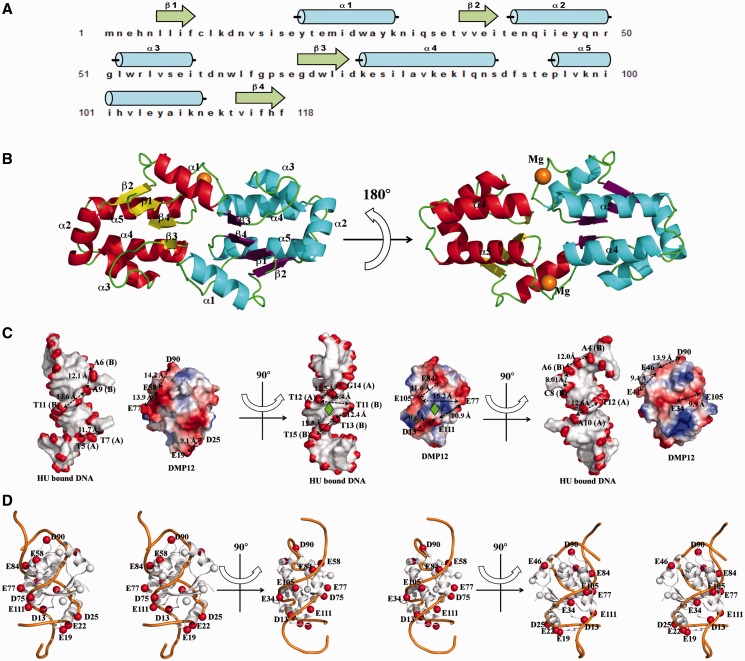
The protein structure of DMP12 provides further evidence of its DNA mimic properties. It contains five α-helices (α1 ∼ α5) and four β-strands in antiparallel configuration (β1 ∼ β4; Figure 2A). The refined crystal structure contains two DMP12 molecules in the asymmetric unit (Figure 2B), with two magnesium ions bound to the surface (Supplementary Figure S4). A structural homology search using the DALI website showed that DMP12 has no similarity to any other known structure, indicating a novel fold.
Figure 2.
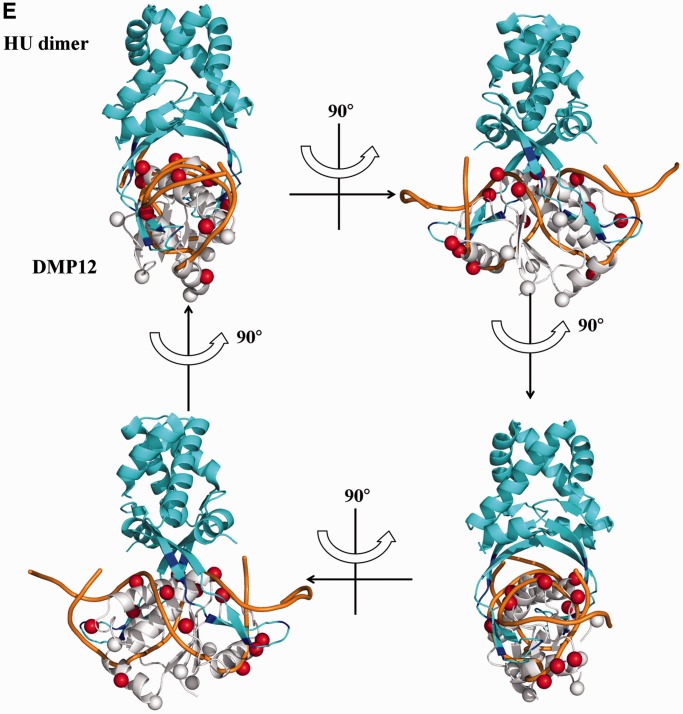
The crystal structure of DMP12 presents a DNA-like surface. (A) The secondary structural elements are shown above the amino acid sequence, with blue cylinders representing α-helices and green arrows representing β-sheets. (B) A ribbon diagram of two DMP12 monomers in an asymmetric unit. The α-helices and β-sheets were colored red and yellow, respectively, in one DMP12 monomer, and cyan and purple in the other, respectively. Two magnesium ions (orange) were found on the protein surface. (C) Comparison of the surface charge distribution of DMP12 and HU-bound dsDNA. DMP12 electrostatic potential surface is colored by Pymol, with red to blue representing the electrostatic potential from −77 to +77 kBT. The DNA surface is colored according to atomic charge by the same program. The distances between the β-carbons of neighboring acidic residues on DMP12 and the phosphate groups on HU-bound dsDNA were measured, and similar distances between the negative spots on DMP12 and HU-bound dsDNA were found. The pseudo 2-fold symmetries of the Hu-interacting surface of the HU/DNA complex and our proposed HU-DMP12-binding model are indicated by a green rhombus. (D) DMP12’s β-carbons were used to indicate the location of the negatively charged side chains of ASP and GLU on the DMP12 monomer. β-Carbons that were matched in the DNA of the HU-bound dsDNA are labeled as red spheres; non-matched carbons are white. This diagram is shown in stereo. (E) Proposed HU-DMP12–binding model. This model was produced by the alignment of the negatively charged spots and shapes of the DMP12 monomer and the phosphate backbone of HU-bound dsDNA in Figure 2C and D. In this model, the HU-bound dsDNA in the HU/DNA complex (PDB code: 1P78) is overlaid by DMP12.
On the surface of the DMP12 monomer, the negatively charged amino acids Asp-13, Glu-19, Glu-22, Asp-25, Glu-34, Glu-58, Asp-75 Glu-77 Glu-84, Asp-90, Glu-105 and Glu-111 are arranged into two rows with a dsDNA-like topology (Figure 2C and D; red spheres). The carboxyl groups on these two rows are a good match to the phosphate groups of the Anabaena HU-dimer–bound DNA backbone [from PDB 1P71; (8)] (Figure 2D; orange ribbon). A binding model was thus proposed based on the complementary charges and surface shape of the DMP12 monomer and HU dimer (Figure 2E). In this model, the DNA-binding β-ribbon arms of the HU protein dimer fit into the DNA groove-like surface on the DMP12 monomer. These results suggest that DMP12 mimics the DNA substrate of HU protein.
Binding affinity for the interaction between Niesseria HU and DMP12
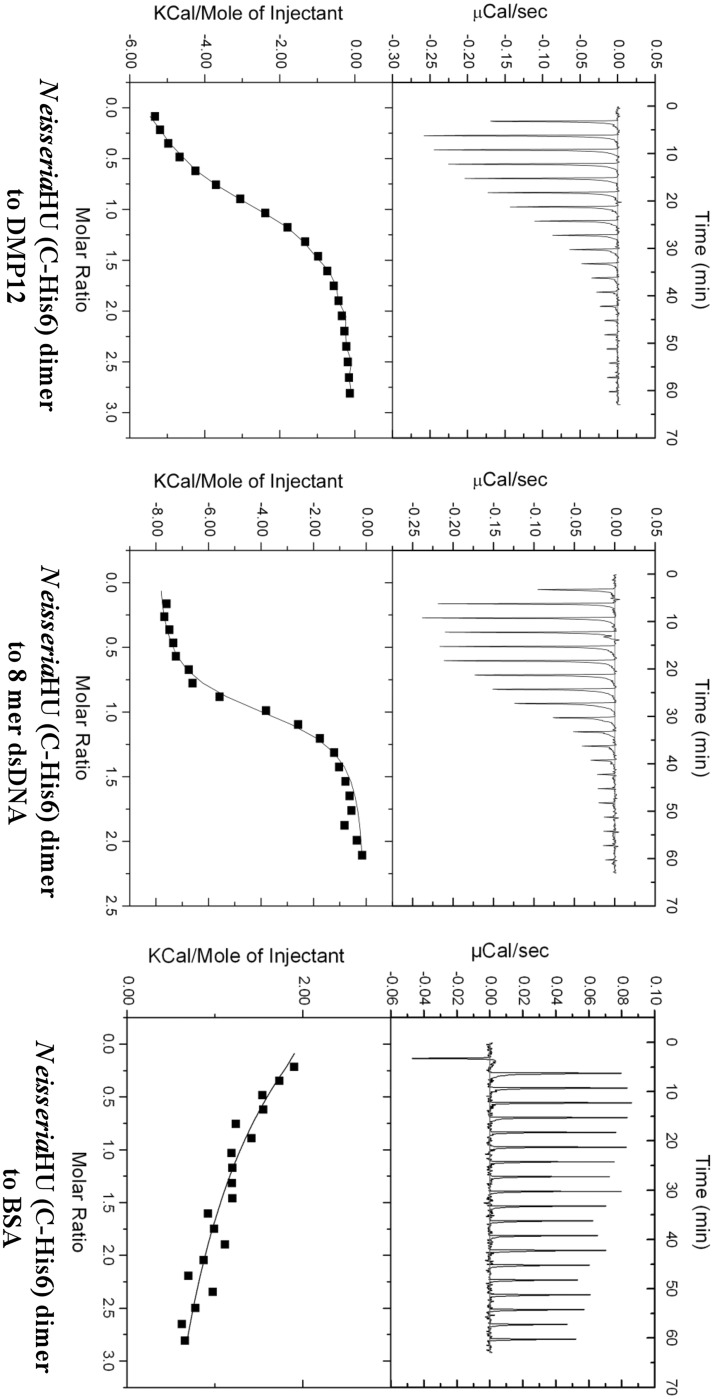
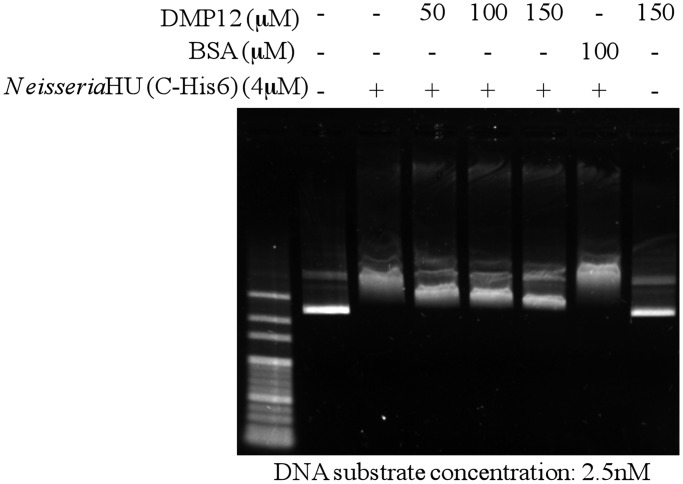
ITC was used to investigate the binding affinity of Niesseria HU and DMP12 (Figure 3). The calorimetric titration of Neisseria HU dimer into a solution of DMP12 showed that the proteins bind with a Kd of 2.81 ± 0.43 µM (Table 3). This is ∼4-fold lower than the 0.72 ± 0.06 µM for HU dimer binding to the 8mer dsDNA. A competitive electrophoresis mobility shift assay (EMSA) further showed that HU-plasmid DNA complexes were partially disrupted by the addition of DMP12 (Figure 4). The dosage-dependent decrease in the shifted bands when DMP12 was pre-incubated with Neisseria HU protein suggests that DMP12 prevented the binding of HU protein to the plasmid DNA. Also of note is that a high concentration of DMP12 did not completely return the plasmid DNA to its original unbound form. A similar finding has been previously reported for the HU-bound DNA mimic protein HI1450 (22). In the present case, the relatively weak affinity of DMP12 to Neisseria HU suggests that it may not fully block the HU protein–plasmid binding. These results suggest that although DMP12 and DNA evidently share the same binding sites on HU protein, DMP12 is more likely to act as a regulator than a competitive inhibitor.
Figure 3.
The binding affinity between of DMP12 and 8mer dsDNA to Neisseria HU protein was determined by an ITC assay. To obtain good ITC data, we tested the binding between Neisseria HU and DNA substrates with different lengths (35, 21, 16 and 8mer DNA). Only the Neisseria HU/8mer DNA reaction yielded a good binding curve, and these results are similar to previous ITC data obtained by using E. coli HU and different DNA lengths (21). The 8mer DNA was, therefore, used for the DMP12 comparison. The Kd of DMP12/Neisseria HU is 2.82 ± 0.43 µM and 8mer DNA/Neisseria HU is 0.72 ± 0.06 µM. Data are from three replicate experiments. No evidence of an interaction signal was seen in the negative control titration that used the acidic protein BSA (pI 4.7) in place of DMP12 protein or dsDNA.
Table 3.
Thermodynamic parameters for Neisseria HU/DMP12 and Neisseria HU/8mer dsDNA interactions
| ΔH (kcal/mol) | ΔG (kcal/mol) | Kd (µM) | N | |
|---|---|---|---|---|
| Neisseria HU dimer to DMP12 | −6.17 ± 0.55 | −7.58 ± 0.09 | 2.82 ± 0.43 | 0.96 ± 0.01 |
| Neisseria HU dimer to 8mer dsDNA | −7.82 ± 0.15 | −8.38 ± 0.05 | 0.72 ± 0.06 | 0.97 ± 0.01 |
| Haemophilus HI1450 to HUa | 3.00 ± 0.20 |
aThis Kd value of Haemophilus HI1450 and HU was measured by ITC in the presence of 200 mM NaCl (22).
Figure 4.
DMP12 partially prevents the DNA binding of Neisseria HU protein. A preliminary EMSA showed that the addition of 4 µM Neisseria HU protein produced a marked band shift of the 2.5 nM plasmid DNA substrate (Supplementary Figure S5), and this concentration was, therefore, used in this competitive EMSA assay. The results show that the DNA mimic DMP12 partially reduces the DNA shift of Neisseria HU in a dosage-dependent manner.
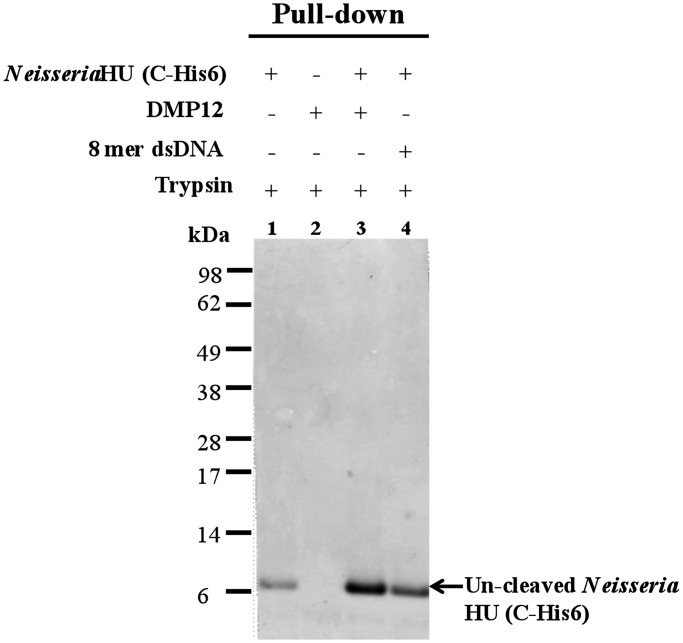
Neisseria HU protein is protected from trypsin digestion in the presence of DMP12
The DNA-binding β-ribbon arms of bacterial HU protein are disordered in the absence of DNA (23,24). A disordered region is expected to undergo protease digestion ∼105–107 times faster than an ordered one (25). To further provide evidence that DMP12 targets the DNA-binding region on Neisseria HU protein, we performed a limited trypsin digestion assay (Figure 5). The results showed Neisseria HU protein is sensitive to trypsin in the absence of DMP12 or dsDNA (Figure 5; lane 1), whereas DMP12 (lane 3) and dsDNA (lane 4) both reduced the rate of HU protein degradation. This protection effect is dose-dependent (Supplementary Figure S6). Taken together, these results suggest that DMP12 binding promotes and stabilizes the formation of DNA-binding β-ribbon arms on Neisseria HU in a similar way as does DNA binding. The biological significance of this protecting effect is not clear, but the growth rate of DMP12-expressing E. coli is noticeably higher (∼1.85-fold after the addition of IPTG for 4 h) than the control bacteria (Supplementary Figure S7). This suggests DMP12 protein may benefit bacterial replication.
Figure 5.
Neisseria HU protein is protected from trypsin digestion in the presence of DMP12 or DNA. Because C-terminal–tagged Neisseria HU protein and DMP12 both have similar molecular weight, His–pull-down was used to purify the uncleaved C-terminal–tagged Neisseria HU protein. The uncleaved C-terminal–tagged Neisseria HU proteins were analyzed using a 12% Bis-Tris SDS–PAGE.
DISCUSSION
DNA mimic proteins regulate the DNA-binding activity of proteins by mimicking the charge and/or shape of DNA (26,27). Functionally, they are involved in a wide range of cellular mechanisms for various different purposes. DNA mimics can act as protectors. For example, bacterial phage Ugi and OCR inhibit the uracil repair and type I restriction/modification systems to help the phage evade the bacterial defense system (28,29), whereas Mycobacterium MfpA protects bacterial gyrase from antibiotics (30). DNA mimic proteins can also be involved in the endogenous mechanisms that are important to cell survival. Examples include Escherichia ArdA, which inhibits the type I restriction/modification systems to allow a horizontal transfer of mobile genetic DNA elements (e.g plasmid DNA) between different bacterial cells (31). Two other mimics, Neisseria DMP19 and Myxococcus CarS interact with transcription factors to affect gene expression (10,32). Other functional roles are found in Haemophilus HI1450 and white spot syndrome virus ICP11, both of which are implicated in DNA packaging by interacting with bacterial HU and eukaryotic histone protein, respectively (22,33,34). Despite their evident importance to living cells, currently there are only a handful (<20) of known DNA mimic proteins. The main reason that so few DNA mimics have been identified is probably related to the fact that each reported DNA mimic protein has a unique amino acid sequence and structure.
We present here another DNA mimic protein, Neisseria DMP12, which interacts with a bacterial histone-like HU protein (Figure 1). The Neisseria DMP12 monomer has a molecular shape and charge distribution similar to the straight portion of the bent HU-bound DNA (Figure 2C–E). The proposed binding model also shows how this specialized surface complements the two DNA-binding β-ribbon arms of the HU dimer. Generally, bacterial histone-like HU proteins are critical for the maintenance of the nucleoid structure (9). HU homologs are highly conserved and usually function as homodimers, except for homologs from E. coli and other enterobacteria, which are heterodimeric (6). Structural studies of Anabaena HU-DNA complex revealed that HU protein induces DNA bending (∼105°–140°) and produces consistent negative supercoiling (8). In addition to their roles in regulating nucleoid structure, HU homologs also participate in many DNA-dependent functions, such as replication, repair, recombination and gene regulation (7). The binding between Neisseria DMP12 and HU protein may, therefore, have multiple effects in the bacteria. Interestingly, the binding affinities determined by the ITC assay showed that Neisseria HU’s affinity for DMP12 is four times lower than it is for dsDNA (Figure 3). In the competitive EMSA assay, excess DMP12 could not fully prevent Neisseria HU from binding to the plasmid DNA (Figure 4). This suggests DMP12 may not serve as a competitive inhibitor, but instead may have other regulatory roles in the bacteria.
One possible role for DMP12 is suggested by the limited trypsin digestion assay (Figure 5): it may serve to protect the HU protein. As the DNA-binding β-ribbon arms of bacterial HU protein become disordered in the absence of DNA substrate (23, 24), this region (∼20–25 residues, 25% of protein) is unstable when it is not incorporated into the bacterial nucleoid. However, the bacterial nucleoid is highly dynamic; disassembly and reassembly of the bacterial nucleoid are important for controlling gene transcription, as well as for recombination and DNA replication (7). Under these circumstances, a protector may be necessary to maintain the stability of the disordered regions of HU protein. We, therefore, propose that the role of DMP12 is to preserve the conformation of the HU protein in its DNA-bound form, and we note that its relatively weak binding affinity will not block the subsequent assembly or reassembly of DNA and HU protein. We also note that the abundance of HU protein can be regulated by protease (35,36). Thus, for example, Lon protease controls the homo- and heterodimeric forms of HU protein in E. coli by degrading the E. coli HU β-subunit. Although the effect of Neisseria Lon protease on HU protein is still unknown, an operon prediction by OperonDB (http://operondb.cbcb.umd.edu/cgi-bin/operondb/operons.cgi) suggests that the Neisseria Lon protease (NMB1230) and HU (NMB1231) genes belong to the same operon. This strongly suggests a relationship between these two proteins, and we intend to investigate the interactions between Neisseria Lon, HU and DMP12 in the future. As part of this research, the in vivo expression levels of DMP12 will also need to be determined. To date, there are only two transcriptomic microarray studies that include DMP12, one of which shows downregulation of DMP12 mRNA during Neisseria attachment to epithelial cells (5), whereas the other shows DMP12 mRNA downregulation during meningococcal biofilm growth (37).
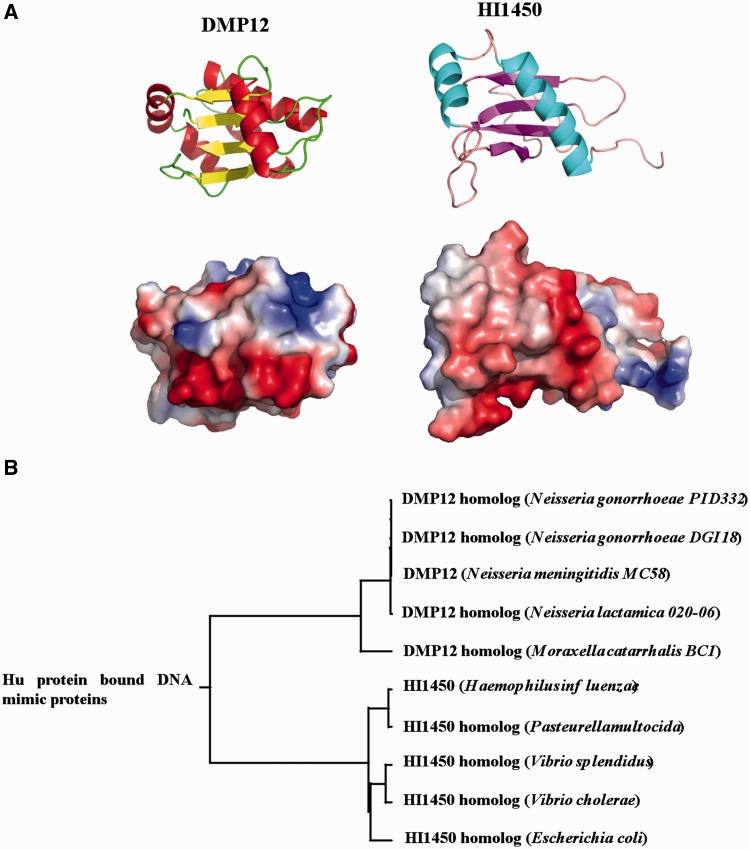
DMP12 is not the only DNA mimic protein that targets bacterial HU protein. Parsons et al. (22, 33) reported Haemophilus HI1450 as another DNA mimic that interacts with bacterial HU. However, these two proteins have many different features. We are currently working to extend our understanding of how new DNA mimics might be identified, but in this case, we note that although the DMP12 and HI1450 core structures fold into four matching β-strands in antiparallel configuration with an RMSD (root mean square deviation) fit of 1.05 Å for 21 Cα atoms (Supplementary Figure 8A), no other significant similarities can be found between these two structures: both their charge distributions and their shapes are also different (Figure 6A and Supplementary Figure 8A). Phylogenetic analysis also shows that these two proteins are classified into two distinct groups (Figure 6B; Supplementary Figure 8B and C). All of this suggests that HI1450 and DMP12 probably interact with bacterial HU protein for different purposes. Moreover, the biological roles of both HU-bound DNA mimic proteins in the bacteria need to be further clarified. Until now, no biological function of HI1450 is proposed. The HI1450 homolog (yciU) knockout E. coli strain still maintains the normal cell shape, and it suggests this protein is not necessary for E. coli survival [National BioResource Project (NBRP); http://www.shigen.nig.ac.jp]. However, the increasing growth rate of DMP12 expressing E. coli suggests this protein may manipulate expression of bacterial genome to favor replication (Supplementary Figure S7), and it encourages more studies to understand this issue.
Figure 6.
Comparison of two DNA mimic proteins that bind to HU. (A) Comparison of the structure and charge distribution of Neisseria DMP12 monomer and Haemophilus HI1450 (protein data bank accession number: 1NNV). (B) Rooted phylogenetic tree of DMP12 and HI1450 homologs with branch lengths.
In summary, we have shown that DMP12 is able to interact with Neisseria histone-like HU protein by mimicking the DNA substrate. The bacterial nucleoid is similar to the eukaryotic nucleosome in that it is highly dynamic, and its remodeling is believed to play an important role in gene regulation (7,38–41). DMP12 may support this mechanism by improving the stability of unbound HU protein. In eukaryotic cells, ‘histone chaperones’, such as Nap1 and Chz1, associate nearly synthesized histones, escort them into the nucleus and enhance their specific association with DNA in a variety of processes, such as DNA replication, repair or transcription (42,43). It is also possible DMP12 serves as an ‘HU chaperone’ in the Neisseria spps. Additionally, we note that as DMP12 homologs are only found in Neisseria and Moraxella species, this protein could be a potential target for specific anti-Neisseria drugs.
ACCESSION NUMBERS
The PDB ID of DMP12 is 3W1O.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–8.
FUNDING
Funding for open access charge: Academia Sinica and National Science Council grants [NSC100-2325-B-001-029 and NSC101-2319-B-001-003].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the staffs of the beamline BL13B1 at the Radiation Research Center (NSRRC) in Hsinchu, Taiwan; and the staffs of the beamline 44XU at the Spring-8 Synchrotron Radiation Research Center in Hyogo, Japan; for their help in X-ray crystal data collection. They also thank the Core Facilities for Proteomics Research located at the Institute of Biological Chemistry, Academia Sinica for their help in Proteomic mass spectrometry analyses.
REFERENCES
- 1.Ren J, Sainsbury S, Nettleship JE, Saunders NJ, Owens RJ. The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif. Proteins. 2010;78:1798–1802. doi: 10.1002/prot.22698. [DOI] [PubMed] [Google Scholar]
- 2.Snyder LA, Davies JK, Ryan CS, Saunders NJ. Comparative overview of the genomic and genetic differences between the pathogenic Neisseria strains and species. Plasmid. 2005;54:191–218. doi: 10.1016/j.plasmid.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 4.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 5.Joseph B, Schneiker-Bekel S, Schramm-Glück A, Blom J, Claus H, Linke B, Schwarz RF, Becker A, Goesmann A, Frosch M, et al. Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J. Bacteriol. 2010;192:5363–5377. doi: 10.1128/JB.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinson V, Takahashi M, Rouviere-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 7.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 8.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grove A. Functional evolution of bacterial histone-like HU proteins. Curr. Issues Mol. Biol. 2011;13:1–12. [PubMed] [Google Scholar]
- 10.Wang HC, Ko TP, Wu ML, Ku SC, Wu HJ, Wang AHJ. Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic Acids Res. 2012;40:5718–5730. doi: 10.1093/nar/gks177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Vagin AA, Steiner RS, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D. 2004;60:2284–2295. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative computational project number 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 18.DeLano WL. Palo Alto, CA: DeLano Scientific LLC; 2008. The PyMOL molecular graphics system. [Google Scholar]
- 19.Puvirajesinghe TM, Elantak L, Lignon S, Franche N, Ilbert M, Ansaldi M. DnaJ (Hsp40 protein) binding to folded substrate impacts KplE1 prophage excision efficiency. J. Biol. Chem. 2012;287:14169–14177. doi: 10.1074/jbc.M111.331462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortega A, Amorós D, García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh J, Saecker RM, Record MT., Jr DNA binding mode transitions of Escherichia coli HUαβ: evidence for formation of a bent DNA-protein complex on intact, linear duplex DNA. J. Mol. Biol. 2008;383:324–346. doi: 10.1016/j.jmb.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons LM, Liu F, Orban J. HU-alpha binds to the putative double-stranded DNA mimic HI1450 from Haemophilus influenzae. Protein Sci. 2005;14:1684–1687. doi: 10.1110/ps.041275705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White SW, Wilson KS, Appelt K, Tanaka I. The high-resolution structure of DNA-binding protein HU from Bacillus stearothermophilus. Acta. Crystallogr. D Biol. Crystallogr. 1999;55:801–809. doi: 10.1107/s0907444999000578. [DOI] [PubMed] [Google Scholar]
- 24.Christodoulou E, Rypniewski WR, Vorgias CR. High-resolution X-ray structure of the DNA-binding protein HU from the hyper-thermophilic Thermotoga maritima and the determinants of its thermostability. Extremophiles. 2003;7:111–122. doi: 10.1007/s00792-002-0302-7. [DOI] [PubMed] [Google Scholar]
- 25.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J. Mol. Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 26.Dryden DT. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotechnol. 2006;4:378–382. doi: 10.1016/j.tibtech.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair. 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 29.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DT. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 30.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 31.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.León E, Navarro-Avilés G, Santiveri CM, Flores-Flores C, Rico M, González C, Murillo FJ, Elías-Arnanz M, Jiménez MA, Padmanabhan S. A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix. Nucleic Acids Res. 2010;38:5226–5241. doi: 10.1093/nar/gkq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons LM, Yeh DC, Orban J. Solution structure of the highly acidic protein HI1450 from Haemophilus influenzae, a putative double-stranded DNA mimic. Proteins. 2004;54:375–383. doi: 10.1002/prot.10607. [DOI] [PubMed] [Google Scholar]
- 34.Wang HC, Wang HC, Ko TP, Lee YM, Leu JH, Ho CH, Huang WP, Lo CF, Wang AH. WSSV ICP11 is a multi-functional DNA mimic protein that disrupts nucleosome assembly. Proc. Natl Acad. Sci. USA. 2008;105:20758–20763. doi: 10.1073/pnas.0811233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnefoy E, Almeida A, Rouviere-Yaniv J. Lon-dependent regulation of the DNA binding protein HU in Escherichia coli. Proc. Natl Acad. Sci. USA. 1989;86:7691–7695. doi: 10.1073/pnas.86.20.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao JH, Lin YC, Hsu J, Lee AY, Chen TA, Hsu CH, Chir JL, Hua KF, Wu TH, Hong LJ, et al. Binding and cleavage of E coli HUbeta by the E. coli Lon protease. Biophys. J. 2010;98:129–137. doi: 10.1016/j.bpj.2009.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Dwyer CA, Li MS, Langford PR, Kroll JS. Meningococcal biofilm growth on an abiotic surface—a model for epithelial colonization? Microbiology. 2009;155:1940–1952. doi: 10.1099/mic.0.026559-0. [DOI] [PubMed] [Google Scholar]
- 38.Kar S, Edgar R, Adhya S. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl Acad. Sci. USA. 2005;102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanbichler M, Wang SC, Shapiro L. The bacterial nucleoid: a highly organized and dynamic structure. J. Cell Biochem. 2005;96:506–521. doi: 10.1002/jcb.20519. [DOI] [PubMed] [Google Scholar]
- 40.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 41.Sobetzko P, Travers A, Muskhelishvili G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl Acad. Sci. USA. 2012;109:42–50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avvakumov N, Nourani A, Côté J. Histone chaperones: modulators of chromatin marks. Mol. Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.