Abstract
Purpose
We sought to determine the frequency and clinical characteristics of patients with lung cancer harboring NRAS mutations. We used preclinical models to identify targeted therapies likely to be of benefit against NRAS mutant lung cancer cells.
Patients and Methods
We reviewed clinical data from patients whose lung cancers were identified at 6 institutions or reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) to harbor NRAS mutations. 6 NRAS mutant cell lines were screened for sensitivity against inhibitors of multiple kinases (i.e. EGFR, ALK, MET, IGF-1R, BRAF, PI3K and MEK).
Results
Among 4562 patients with lung cancers tested, NRAS mutations were present in 30 (0.7%; 95% confidence interval, 0.45% to 0.94%); 28 of these had no other driver mutations. 83% had adenocarcinoma histology with no significant differences in gender. While 95% of patients were former or current smokers, smoking-related G:C>T:A transversions were significantly less frequent in NRAS mutated lung tumors compared to KRAS-mutant NSCLCs (NRAS: 13% (4/30), KRAS: 66% (1772/2733), p<0.00000001). 5 of 6 NRAS mutant cell lines were sensitive to the MEK inhibitors, selumetinib and trametinib, but not to other inhibitors tested.
Conclusion
NRAS mutations define a distinct subset of lung cancers (~1%) with potential sensitivity to MEK inhibitors. While NRAS mutations are more common in current/former smokers, the types of mutations are not those classically associated with smoking.
Keywords: NRAS mutation, EGFR mutation, KRAS mutation, lung cancer, non-small cell lung cancer, driver mutation, MEK inhibitor, erlotinib, gefitinib, crizotinib
Introduction
Recent advances have been made in targeting molecularly defined subsets of non-small cell lung-cancers (NSCLCs) that depend on specific molecular alterations for cell survival. Prime examples include tumors which harbor mutations in the gene encoding the epidermal growth factor receptor (EGFR) or translocations in the gene encoding the anaplastic lymphoma kinase (ALK). Patients with these tumors can derive substantial clinical benefit from EGFR (gefitinib, erlotinib) or ALK (crizotinib) tyrosine kinase inhibitors (TKIs), respectively (1–8).
To date, many other potential “driver mutations” occurring in genes encoding cellular signaling proteins have also been identified in NSCLCs. Genomic alterations include mutations in the GTPase KRAS (25%) (9, 10), the receptor tyrosine kinase ERBB2 (2–3%) (11, 12), the lipid kinase PIK3CA (2–4%) (10, 13, 14), the serine-threonine kinase BRAF (2–4%) (9, 10, 15), and the serine-threonine kinase MEK1 (1%) (16), as well as translocations in the tyrosine kinases ROS1 (1–2%) (17–19) and RET (1%) (19–21). A tumor with a mutation in one of these genes rarely harbors a mutation in another (22). Although targeted therapies have not yet been approved for all of these molecular subsets of lung cancer, pre-clinical and emerging clinical data suggest that molecular subtyping will allow for the rational prioritization of treatment options for lung cancer patients (23).
NRAS is a GTPase related to KRAS, originally identified in neuroblastoma cell lines as a third RAS family member following KRAS and HRAS (24). RAS GTPases regulate cell growth, proliferation and differentiation. Although the three family members share conserved sequences, their protein products generate distinct signal outputs (25, 26) and have distinct roles in development (27, 28) and tumorigenesis in mice (29, 30).
NRAS mutations have been reported to occur in lung cancers,(31) but as yet no comprehensive report has focused on the characteristics of patients whose tumors harbor NRAS mutations. Here, we used retrospective clinical data as well as preclinical models to define the clinical relevance of NRAS mutations in lung cancer.
Results
Characteristics of Patients Whose NSCLCs Harbor NRAS mutations
At multiple centers, NSCLCs undergo routine multiplexed mutational profiling for recurrent driver mutations. From 6 institutions (Memorial Sloan-Kettering Cancer Center (MSKCC), Massachusetts General Hospital (MGH), University of Colorado Cancer Center (UCCC), John Hopkins University (JHU), University of California at Los Angeles (UCLA), and Vanderbilt-Ingram Cancer Center (VICC)), we identified 18 NSCLC patients with NRAS mutations from a total of 3698 tested (0.5%; MSKCC:2, MGH:10, UCCC:1, JHU:2, UCLA:1, VICC:2). The spectrum of mutations (not including ALK fusions) from patients with NSCLC at VICC (Figure S1) shows a distribution of driver mutations consistent with the literature (EGFR 17%, ERBB2 1%, KRAS 21%, BRAF 3%, PIK3CA 3%, MEK1 0.5%, and NRAS 0.25%) (10). Another 12 NRAS mutant NSCLCs were listed in the COSMIC database, among 864 lung cancers reported (including small cell lung cancers) (1.4%); 83% of these were adenocarcinoma histology (Table S1). There was no overlap between the two datasets. Thus, in total, we identified 30 NRAS mutant cases among 4562 tested (0.7%; 95% confidence interval 0.45% to 0.94%) (Table 1). One of the tumors also had a KRAS G12A, while another had a MET amplification. Only NRAS mutations were found in the other 28 tumors (Table 2).
Table 1.
The frequency of NRAS mutations in lung cancers from 6 institutions and the COSMIC database.
| MSKCC | MGH | UCCC | JHU | UCLA | VICC | Subtotal | COSMIC | Total | |
|---|---|---|---|---|---|---|---|---|---|
| No. of pts | 1567 | 1397 | 145 | 95 | 43 | 451 | 3698 | 864 | 4562 |
| No. with NRAS mts | 2 | 10 | 1 | 2 | 1 | 2 | 18 | 12 | 30 |
| % NRAS mutant | 0.1 | 0.7 | 0.7 | 2.1 | 2.3 | 0.4 | 0.5 | 1.4 | 0.7 |
No – number, pts – patients, mts – mutations.
Table 2.
Characteristics of individual patients with NRAS mutant tumors.
| Unclassified stage | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | Age | Gender | Race | Histology | TNM | Stage | NRAS | EGFR | KRAS | ALK | MET amp | Smoking | Pack Year | Treatment | PFS (month) | OS (months) |
| 1* | na | na | A | Adeno | na | na | Q61K | na | na | na | na | na | na | na | na | na |
| 2* | na | na | na | Adeno | na | na | Q61L | WT | WT | na | na | na | na | na | na | na |
| 3 | 73 | M | C | na | na | na | Q61L | WT | WT | na | na | Former | 90 | na | na | na |
| 4 | 68 | F | AA | Adeno | na | na | Q61L | WT | WT | na | na | Current | 25 | na | na | na |
| 5* | na | na | A | na | na | na | Q61R | WT | WT | na | na | na | na | na | na | na |
| 6* | na | na | A | Adeno | na | na | G12A | na | na | na | na | na | na | na | na | na |
| 7* | na | na | na | Sq | na | na | G12D | na | na | na | na | Y | na | na | na | na |
| Early stage | ||||||||||||||||
| 8* | na | na | A | Adeno | na | I-III | Q61H | na | na | na | na | na | na | na | na | na |
| 9* | 51 | M | A | Adeno | T1N2M0 | IIIA | Q61L | na | na | na | na | na | na | Surgery | 113 | 113 |
| 10* | na | na | na | Adeno | na | I-III | Q61L | WT | WT | na | na | na | na | Surgery | na | na |
| 11* | na | na | na | Adeno | na | I-III | Q61L | WT | WT | na | na | na | na | Surgery | na | na |
| 12* | na | na | na | Adeno | na | I-III | Q61L | WT | WT | na | na | na | na | Surgery | na | na |
| 13 | 60 | M | C | Adeno | na | IIIA | Q61L | WT | WT | na | na | Former | 20 | Surgery/chemo | 33 | 38+ |
| 14 | 49 | F | C | Adeno | T1bN2M0 | IIIA | Q61L | WT | WT | WT | na | Current | 53 | CRT/surgery | 28 | 34 |
| 15 | 85 | F | C | Adeno | T1aN0M0 | IA | Q61L | WT | WT | WT | na | Current | 105 | RT | na | 14+ |
| 16 | 77 | F | C | Large | T1aN0M0 | IB | Q61L | WT | WT | WT | na | Former | 30 | Surgery | 1 | 1 |
| 17 | 50 | F | C | Adeno | T1bN2M0 | IIIA | Q61L | WT | WT | WT | na | Current | 40 | RT | na | 20 |
| 18 | 71 | M | C | Sq | T1aN2M0 | IIIA | Q61R | WT | WT | WT | na | Former | 15 | CRT/surgery | 20 | 20 |
| 19* | 59 | M | A | Adeno | T3N0M0 | IIB | G12C | WT | WT | na | na | Current | 38 | Surgery | 132 | 132 |
| 20 | 62 | M | C | Adeno | T1aN0M0 | IA | G12S | WT | WT | WT | na | Current | 100 | Surgery | 27 | 27 |
| Metastatic stage | ||||||||||||||||
| 21 | 91 | M | C | Sq | T1aN2M1 | IV | Q61K | WT | G12A | na | na | Former | 30 | Supportive care | na | 1+ |
| 22 | 48 | F | AA | Adeno | T4N2M1 | IV | Q61K | WT | WT | WT | N | Current | 15 | Carbo/paclitaxel | 2 | 23+ |
| 23* | na | na | na | Adeno | T3N0M1 | IV | Q61L | na | na | na | na | Y | na | na | na | na |
| 24 | 53 | M | C | Adeno | na | IV | Q61L | WT | WT | na | na | Former | 30 | Carbo/paclitaxel/bev | na | 26 |
| 25 | 79 | F | C | Adeno | T4N2M1 | IV | Q61L | WT | WT | WT | na | Current | 163 | Supportive care | na | 18 |
| 26 | 51 | M | C | NOS | T2aN3M1 | IV | Q61R | WT | WT | WT | na | Current | 55 | Carbo/pem | na | 7 |
| 27 | 69 | M | na | Adeno | T2bN3M1 | IV | Q61R | WT | WT | na | na | Current | na | Carbo/paclitaxel | 4 | 4 |
| 28 | 50 | F | C | Adeno | T4N3M1 | IV | Q61R | WT | WT | na | na | Former | 32 | Carbo/pem | 1 | 3+ |
| 29 | 58 | F | C | Adeno | T3N2M1 | IV | G12S | WT | WT | WT | na | Current | 75 | Pem | na | 3+ |
| 30 | 30 | M | AA | Adeno | T2bN2M1 | IV | G12R | WT | WT | WT | Y | Never | 0 | Carbo/pem | 2 | 8+ |
na- not available, Y- Yes, N- No, M- Male, F- Female, C- Caucasian, A- Asian, AA- African American, Adeno- Adenocarcinoma, Sq- Squamous carcinoma, Large- Large cell carcinoma, NOS- not otherwise specified histological type, WT- wild type, chemo- chemotherapy, CRT- chemoradiotherapy, carbo- carboplatin, pem- pemetrexed, bev- bevacizumab, PFS-progression free survival, OS- overall survival,
deceased,
COSMIC database. Case 18 had a mediastinal lymph node aspiration with cell block showing squamous cell carcinoma, with immunohistochemistry (IHC) positive for CK5/6 and p63 and negative for TTF1. This was the sample that was genotyped. The patient also had a surgical resection after neo-adjuvant chemoradiation that showed areas of residual squamous cell with IHC positive for p63, CK5/6, and CK903 and negative for CEA, TTF-1, synaptophysin, and chromogranin. Case 21 had metastatic disease with a couple of biopsies. Mediastinoscopy was performed with bronchoscopic biopsies and lymph node dissection - these were read as squamous cell carcinoma but no IHC was ordered. This is the sample that was genotyped. The patient also had a liver biopsy with IHC positive for CK7 and negative for CK20, positive for p63, and negative for TTF1. Case number 22: This patient received docetaxel, gemcitabine and pemetrexed as salvage chemotherapies. Case number 24: This patient received pemetrexed as second line treatment and gemcitabine as third line treatment.
Clinical characteristics of patients with NRAS mutations are summarized in Tables 2, 3 and S2. Among the 21 patients for whom smoking history was known, 20 were current or former smokers (95%) with a median smoking history of 34 pack years (Table 3). In a cohort of 3247 lung cancer patients (from MSKCC, MGH, UCCC, JHU, and UCLA) for which there was detailed clinical information, there was no significant correlation with NRAS mutations and gender, histology, or clinical stage, but there was a significant association of NRAS mutations with smoking history [current smoker (1.5%), former (0.3%), never smoker (0.1%) (Fisher’s exact test: never smoker vs current smoker (P=0.0065), former smoker vs current smoker (P=0.0043))] and race [Caucasian (0.5%), African American (4.1%), Asian (0%), Hispanic (0%) (Caucasian vs African American (P= 0.0274), African American vs Asian (P= 0.0603))] (Table S2).
Table 3.
Clinical Characteristics of patients with NRAS mutant lung cancers.
| Characteristic | No. | % |
|---|---|---|
| Stage | Total 30 | |
| Early (I-IIIA) | 14 | 61 |
| Advanced (IIIB/IV) | 9 | 39 |
| unknown | 7 | |
| Histology | Total 30 | |
| Adeno | 23 | 82 |
| Squamous | 3 | 11 |
| Large | 1 | 3.5 |
| NOS | 1 | 3.5 |
| Small | 0 | 0 |
| unknown | 2 | |
| Gender | Total 30 | |
| Male | 10 | 53 |
| Female | 9 | 47 |
| unknown | 11 | |
| Age (years) | Total 30 | |
| Median | 59.5 | |
| Range | 30–91 | |
| Race | Total 30 | |
| Caucasian | 13 | 64 |
| Asian | 5 | 27 |
| African American | 3 | 9 |
| unknown | 8 | |
| Smoking history | Total 30 | |
| Never | 1 | 5 |
| Former | 7 | 33 |
| Current | 11 | 52 |
| Yes | 2 | 10 |
| unknown | 9 | |
| Pack years | Total 19 | |
| Median | 34 | |
| Range | 0–163 | |
No. - number of patients, NOS - not otherwise specified histologic, Yes- smoking history positive but details were unknown.
NRAS Mutation Genotypes
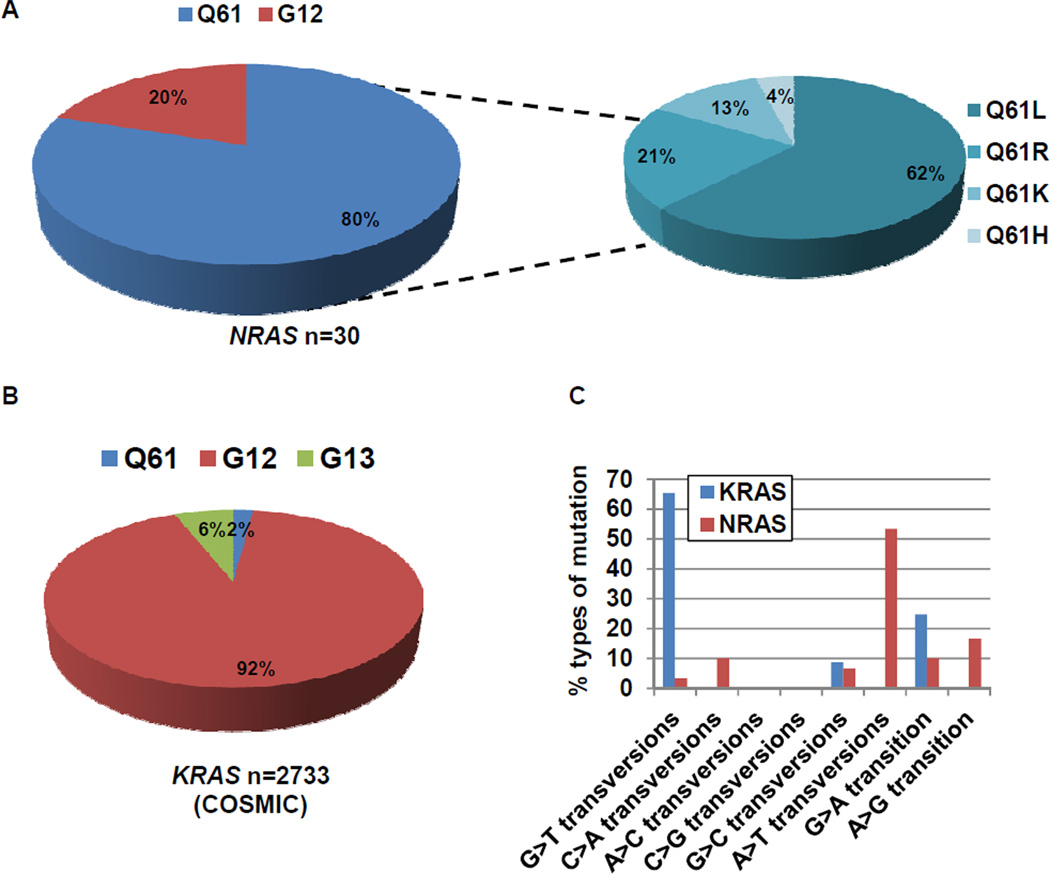
The 30 NRAS mutations corresponded to 9 different amino acid substitutions: Q61H/K/L/R (exon 3) and G12A/C/D/R/S (exon 2). Codon Q61 was the most frequently mutated (80%), and half of mutations were NRAS Q61L (Fig. 1A). Although NRAS and KRAS are related genes, the distribution of KRAS mutations (n=2733) in NSCLC as reported in COSMIC was completely different; more than 90% of KRAS mutations involved codons 12 or 13 (Fig. 1B). The types of mutations were also distinct. G:C>T:A transversions, thought to be associated with direct exposure to tobacco carcinogens,(32–34) were found in 1772 of 2733 (66%) KRAS mutant lung cancers. By contrast, among the 30 NRAS mutations, only 4 (13%) were G:C>T:A transversions (Chi-square test; p<0.00000001) (Fig. 1C). Even among the 21 patients with NRAS mutations and known smoking histories, only 3 of the 20 former/current smokers had such transversions.
Figure 1. Distribution of the types of mutations in NRAS and KRAS mutated lung cancers.
A. Q61 was the most frequently mutated codon in 30 NRAS mutated lung cancers (80%). B. The type of mutations in KRAS (COSMIC). 92% of mutations occurred at codon G12. C. Comparison of the types of mutations in KRAS (COSMIC) and NRAS. G:C >T:A transversions were significantly more common in KRAS (1772/2733, 66%) than NRAS (4/30, 13%) mutated lung cancers (Chi-square test; p<0.00000001).
Sensitivity profiles of 6 NRAS mutant lung cancer cell lines tested against various kinase inhibitors
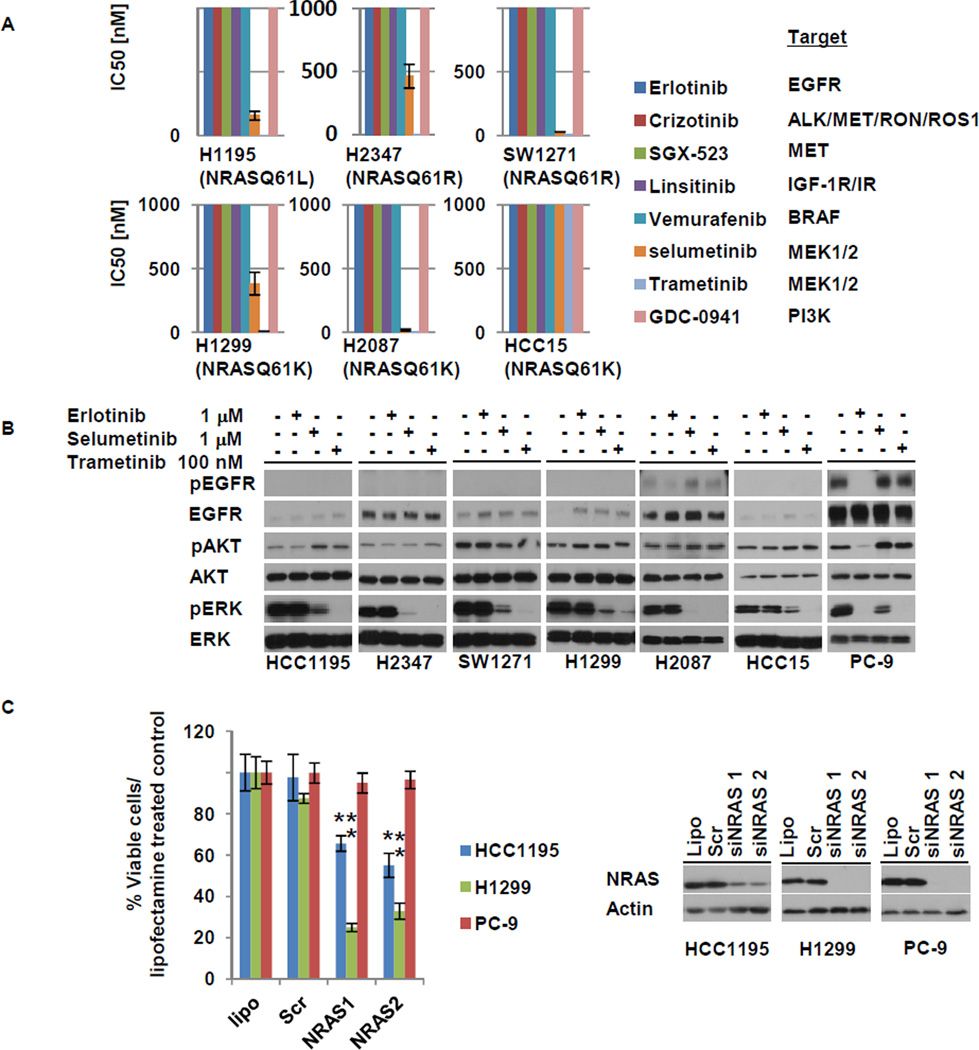
To identify potential therapies for patients with NRAS mutant tumors, we tested the sensitivity of 6 NRAS mutant NSCLC cell lines (Table S3) against a variety of kinase inhibitors in in vitro cell growth inhibition assays (Fig. 2A). None of the lines were sensitive (with lower than 1 micromolar IC50s) to the EGFR TKI, erlotinib, the ALK/MET/RON/ROS1 inhibitor, crizotinib, or the IGF-1R inhibitor, linsitinib. By contrast, 5 of 6 lines were sensitive to two different MEK inhibitors, selumetinib and trametinib. Consistent with these data, the MEK inhibitors inactivated ERK phosphorylation in the NRAS mutated cells while erlotinib did not (Fig. 2B). In order to verify further the dependency of these cells on NRAS, we performed siRNA-mediated knockdown experiments. As expected, NRAS knockdown led to growth inhibition in the NRAS mutant cell lines, H1299 and HCC1195, but not in PC-9 cells, which harbor an EGFR mutation (Fig. 2C).
Figure 2. Sensitivity profiles of 6 NRAS mutant lung cancer cell lines tested against various kinase inhibitors.
A. IC50 values derived from growth inhibition assays were plotted for each drug and each cell line. HCC15 cells were resistant to MEK inhibitors but sensitive to the combination of a MEK inhibitor plus linsitinib (see text and Figure S2 for details). B. MEK inhibitors but not erlotinib led to de-phosphorylation of ERK in NRAS mutated cells. Erlotinib inhibited phosphorylation of EGFR, AKT and ERK in PC-9 cells which harbor an EGFR mutation. C. siRNA-mediated knockdown of NRAS inhibits growth of the NRAS mutated HCC1195 and H1299 cells but not of PC-9 cells. Mean +- SD of three independent experiments performed in hextuplicate replicates is shown. *, **, P < 0.01 (Student’s t-test) for the comparison of siRNAs against NRAS versus scrambled controls in HCC1195 and H1299. Lipo – lipofectamine control; scr – scrambled siRNA control.
Like MEK, the PI3 kinase is reported to be a signaling protein activated downstream of RAS. We found that the selective PI3 kinase inhibitor, GDC0941, had little effect in the NRAS mutant lines. We also tested the efficacy a MET inhibitor, SGX-523, since a recent report showed that melanomas with mutant NRAS displayed activated MET (35). However, none of the NRAS mutant lung lines were sensitive to MET inhibition, either alone or in combination with MEK inhibitors (Fig S2A and data not shown).
HCC15 cells were the only NRAS mutant line insensitive to MEK inhibition alone. We previously reported that these cells displayed high levels of IGF-1R (36). Therefore, we assessed the effect of an IGF-1R inhibitor, linsitinib, together with trametinib. The combination showed a greater effect on cell growth than either drug alone (Fig S2B), suggesting that resistance to MEK inhibition could be overcome by linsitinib in these cells.
Discussion
To our knowledge, this is the largest study of NRAS mutant lung cancer to date, describing clinical characteristics associated with 30 unique patients among 4562 patients tested (0.7%). The actual frequency of NRAS mutations in NSCLC could be lower than in this study, because over 80% of tumors were adenocarcinomas in the cohorts examined. Although the frequency of NRAS mutations in NSCLC is relatively rare, NSCLC is a common disease with 230,000 new cases in the US. Thus, about 1,500 patients in the US would develop lung cancer harboring NRAS mutations every year. NRAS mutations were most significantly associated with smoking and potentially African American race, although the numbers for the latter association were too small to make meaningful conclusions. NRAS mutations were also, for the most part, mutually exclusive with other known driver mutations, including EGFR, KRAS, and ALK, etc. Of course, the probability has to be considered that these driver mutations could exist simultaneously in a single tumor at low frequency but, collectively, these data suggest that NRAS mutations in NSCLC define a distinct molecular subset.
NRAS and KRAS both encode GTPases involved in cell growth, proliferation, and differentiation. They share conserved sequences, but their protein products lead to differential downstream signaling events (25, 26) and have different roles in development (27, 28) and tumorigenesis in mice (29, 30). Recent data has suggested that oncogenic and wild-type RAS isoforms play independent and nonredundant roles within cancer cells. Oncogenic RAS regulates basal effector pathway signaling, whereas wild-type RAS mediates signaling downstream of activated receptor tyrosine kinases (37). Furthermore, oncogenic K-Ras promotes the activation of wild-type H- and N-Ras (38). Why certain lung tumors harbor NRAS vs KRAS mutations is unclear (39). One clue may involve the types of mutations that occur in each gene. Tobacco components, particularly benzo[a]pyrene, are believed to be strong carcinogens for KRAS mutated lung cancer (32, 33), and G:C >T:A transversions are found in 70–90% of KRAS mutations in smoking-related lung cancers (33, 34). This relationship has also been observed for TP53 mutations in lung cancers from smokers (40). By contrast, more than 50% of NRAS mutations involve A:T >T:A transversions (Fig. 1C). Carcinogens known to induce A:T >T:A transversions include 7,12-dimethylbenz[a]anthracene (DMBA), which is released into the environment through the combustion of fossil fuels (41). Perhaps the combination of smoking and such a carcinogen are associated with the etiology of NRAS mutated lung cancer.
The outcomes of NSCLC patients with early stage or metastatic disease remain poor (42). Here, we were able to determine relapse free survival after resection of early stage disease for 7 patients (33 months) and overall survival in the metastatic setting after treatment with systemic chemotherapy for 7 patients (8 months). Although the number of patients in each cohort was small, these preliminary data suggest at least for patients with advanced stage disease that NRAS mutations may be a poor prognostic marker, relative to EGFR and ALK alterations, which have been associated with better prognosis (9). These data will need to be verified in independent datasets.
Recent advances in lung cancer biology and molecular tumor profiling have allowed for rational prioritization of targeted therapies in patients with improved outcomes (5–8). Using preclinical models, we showed that 5 of 6 NRAS mutant NSCLC cell lines (83%) were sensitive to MEK inhibitors but not to other kinase inhibitors. These data are consistent with previous reports using some but not all related compounds (43). By contrast, KRAS mutant lines display much greater variability in sensitivity to this class of drugs (44, 45), suggesting that NRAS mutant lines display a greater dependence upon the MEK pathway for tumor maintenance in lung cancers. To our knowledge, no patient with NRAS mutant lung cancer has yet been treated with a MEK inhibitor, but our data would suggest such patients are likely to benefit from this class of agents.
In summary, NRAS mutations occur in about 1% of NSCLCs (mostly those with direct tobacco exposure), are mostly exclusive of other known driver mutations, have a nucleotide transversion profile different from that of KRAS mutations, and may be associated with sensitivity to MEK inhibitors. Such patients should be prospectively identified in order to prioritize targeted therapies most likely to be of maximal benefit.
Materials and Methods
Patient data
Patients with NSCLC who underwent molecular profiling were identified for review. Clinical characteristics including age, gender, race (reported by the patient), smoking history and clinical stage were recorded. All chart review/tissue collection was carried out under institutional review board/privacy board–approved protocols or waivers.
Genotype Analysis
Genomic DNA was extracted from patient samples (>70% tumor cells) and cell lines using standard procedures. Tumor specimens were obtained as standard of care for clinical management or with patients’ consent under Institutional Review Board–approved protocols. A mass spectrometry-based (Sequenom)(22) or SNapShot assay(46, 47) was performed for genotyping as described. Cell lines were genotyped using SNapShot and/or direct sequencing.
Statistical analysis
Fisher’s exact tests (for small sample size) were applied to test associations among NRAS mutations, smoking history and race. Chi-squared tests were applied to compare the frequency of tranversions in KRAS vs NRAS mutant cancers.
Cell culture
H1299, H2347, H2087 and SW1271 were purchased from ATCC. HCC15 were obtained as described before (36). HCC1195 was kindly provided by Dr. Roman Thomas. H1299, H2347, HCC15, and HCC1195 cells were cultured in RPMI 1640 media (Mediatech) supplemented with 10% heat inactivated fetal bovine serum (Atlanta Bio) and pen-strep solution (Mediatech; final concentration 100U/mL penicillin, 100μg/mL streptomycin). H2087 and SW1271 cells were cultured in DMEM (Mediatech) with the same supplements. Cells were grown in a humidified incubator with 5% CO2 at 37°C.
Growth inhibition assay
Cells were seeded in 96-well plates at a density of 500 to 5000 cells per well and exposed to drugs alone or in combination the following day. At 120 hours after drug addition, Cell Titer Blue Reagent (Promega) was added and fluorescence was measured on a Spectramax spectrophotometer (Molecular Devices), according to the manufacturer's instructions. All experimental points were set up in hextuplicate replicates and were performed at least 3 independent times. Erlotinib was synthesized by the MSKCC Organic Synthesis Core. Selumetinib, Trametinib, Vemurafenib, GDC-0941, Crizotinib, Linsitinib, and SGX-523 were purchased from Selleck Chemicals.
Antibodies and immunoblotting
The following antibodies were obtained from Cell Signaling Technology: phospho-EGFR, EGFR, MET, phospho-ERK, ERK, phospho-AKT, AKT, actin, HRP-conjugated anti-mouse, and HRP-conjugated anti-rabbit. NRAS antibody was purchased from Santa Cruz. For immunoblotting, cells were harvested, washed in PBS, and lysed in 50 mmol/L Tris-HCl, pH 8.0/150 mmol/L sodium chloride/5 mmol/L magnesium chloride/1% Triton X-100/0.5% sodium deoxycholate/0.1% SDS/40 mmol/L sodium fluoride/1 mmol/L sodium orthovanadate and complete protease inhibitors (Roche Diagnostics). Lysates were subjected to SDS-PAGE followed by blotting with the indicated antibodies and detection by Western Lightening ECL reagent (Perkin Elmer).
siRNA Experiment
NRAS and negative control oligos (Dharmacon) were used at a concentration of 10 nM and transfected with Lipofectamine RNAiMAX according to the manufacturer's protocol (Invitrogen).
Supplementary Material
Statement of translational relevance.
Recent advances in lung cancer biology and molecular tumor profiling have allowed for rational prioritization of targeted therapies in patients. NRAS mutations have been reported to occur in lung cancers, but as yet no comprehensive report has focused on the characteristics of patients whose tumors harbor NRAS mutations. Here, we describe clinical characteristics associated with 30 unique patients with NRAS mutated lung cancers among 4562 patients tested (0.7%). While 95% of patients were former or current smokers, smoking-related G:C>T:A transversions were significantly less frequent in NRAS mutated lung tumors compared to KRAS-mutant NSCLCs. NRAS mutations were for the most part, mutually exclusive with other known driver mutations, suggesting that NRAS mutations define a distinct molecular subset. In preclinical models, 5 of 6 NRAS mutant NSCLC cell lines were sensitive to MEK inhibitors. Our data suggests the possibility of personalized treatment in this subset of lung cancers.
Acknowledgements
This work was supported by National Institutes of Health (NIH) NCI grants RC2-CA148394, R01-CA121210, P01-CA129243, and U54-CA143798. WP received additional support from Vanderbilt’s Specialized Program of Research Excellence in Lung Cancer grant (CA90949) and the VICC Cancer Center Core grant (P30-CA68485).
CMR has received consulting fees from AVEO Pharmaceuticals, and Oncothyreon. SD has received consulting fee from Tragara Pharmaceuticals. GJR has received consulting fees from Chugai, Tragara, ARIAD, Daiichi and Abbott and research funding for other projects from Pfizer, Merck, GlaxoSmithKline and Bristol-Myers Squibb. DD received consulting fees from Bio-reference laboratories. MGK has received consulting fees from Pfizer, Genentech and Boehringer Ingelheim and research funding for other projects from Pfizer and Boehringer Ingelheim. DD received consulting fees from Bio-reference laboratories. PAB has received consulting fees from AMGEN, Bristol-Myers Squibb, Merck, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Roche, Pfizer, Eli Lilly, Sanofi Aventis, Astelas, AstraZeneca and Bayer. WP has received consulting fees from MolecularMD, AstraZeneca, Bristol-Myers Squibb, Symphony Evolution, Clovis Oncology and research funding for other projects from Enzon, Xcovery, AstraZeneca, and Symphogen. We also acknowledge that WP is a part of a patent regarding EGFRT790M mutation testing that was licensed by Memorial Sloan-Kettering Cancer Center to Molecular MD. Each patent holder received a total of $500.00 (five hundred dollars) and no royalties.
Footnotes
Conflict of interest statement
There are no other conflicts to report.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004 May 20;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004 Jun 4;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004 Sep 7;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010 Jun 24;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010 Feb;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007 Aug 2;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010 Oct 28;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011 Oct;12(11):1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011 May 20;29(15):2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011 Dec;22(12):2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005 Mar 1;65(5):1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Sun Y, Fang R, Han X, Luo X, Wang R, et al. Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol. 2012 Jan;7(1):85–89. doi: 10.1097/JTO.0b013e318234f0a2. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008 Sep 1;68(17):6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012 Feb;11(2):485–491. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011 Sep 10;29(26):3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 16.Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008 Jul 15;68(14):5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Fang R, Sun Y, Han X, Li F, Gao B, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One. 2011;6(11):e28204. doi: 10.1371/journal.pone.0028204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012 Mar 10;30(8):863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nature Medicine. 2012 Mar;18(3):378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 20.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nature Medicine. 2012 Mar;18(3):375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nature Medicine. 2012 Mar;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007 Mar;39(3):347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 23.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011 Feb;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu K, Goldfarb M, Suard Y, Perucho M, Li Y, Kamata T, et al. Three human transforming genes are related to the viral ras oncogenes. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2112–2116. doi: 10.1073/pnas.80.8.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003 May;4(5):373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 26.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011 Nov;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997 Oct 1;11(19):2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, et al. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001 Mar;21(5):1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008 May;40(5):600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol Cell Biol. 2008 Apr;28(8):2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008 Oct 23;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999 Jul 21;91(14):1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 33.Ahrendt SA, Decker PA, Alawi EA, Zhu Yr YR, Sanchez-Cespedes M, Yang SC, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001 Sep 15;92(6):1525–1530. doi: 10.1002/1097-0142(20010915)92:6<1525::aid-cncr1478>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 34.Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008 Sep 15;14(18):5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattopadhyay C, Ellerhorst JA, Ekmekcioglu S, Greene VR, Davies MA, Grimm EA. Association of activated c-Met with NRAS-mutated human melanomas. Int J Cancer. 2012 Jul 15;131(2):E56–E65. doi: 10.1002/ijc.26487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong Y, Yao E, Shen R, Goel A, Arcila M, Teruya-Feldstein J, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS One. 2009;4(10):e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young A, Lou D, McCormick F. Oncogenic and Wild-type Ras Play Divergent Roles in the Regulation of Mitogen-Activated Protein Kinase Signaling. Cancer Discov. 2013 Jan;3(1):112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 38.Jeng HH, Taylor LJ, Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat Commun. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prior IA, Lewis PD, Mattos C. A comprehensive survey of ras mutations in cancer. Cancer Res. 2012 May 15;72(10):2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hainaut P, Pfeifer GP. Patterns of p53 G-->T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001 Mar;22(3):367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 41.Osaka M, Matsuo S, Koh T, Liang P, Kinoshita H, Maeda S, et al. N-ras mutation in 7,12-dimethylbenz[a]anthracene (DMBA)-induced erythroleukemia in Long-Evans rats. Cancer Lett. 1995 May 4;91(1):25–31. doi: 10.1016/0304-3835(94)03714-t. [DOI] [PubMed] [Google Scholar]
- 42.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002 Jan 10;346(2):92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 43.Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008 Nov 15;68(22):9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009 Jun;119(6):1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010 May;2(5):146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011 Jan;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.