Abstract
PARN, Nocturnin and Angel are three of the multiple deadenylases that have been described in eukaryotic cells. While each of these enzymes appear to target poly(A) tails for shortening and influence RNA gene expression levels and quality control, the enzymes differ in terms of enzymatic mechanisms, regulation and biological impact. The goal of this review is to provide an in depth biochemical and biological perspective of the PARN, Nocturnin and Angel deadenylases. Understanding the shared and unique roles of these enzymes in cell biology will provide important insights into numerous aspects of the post-transcriptional control of gene expression.
Keywords: poly(A) shortening, deadenylase, mRNA turnover
1. Introduction
This review covers three of an unexpectedly large contingent of deadenylase enzymes in eukaryotic cells: PARN, Nocturnin and Angel. Understanding the unique and shared roles of these enzymes in the cell is a major challenge to our working knowledge of the mechanistic details of the post-transcriptional regulation of gene expression. While each of these enzymes is generally widely conserved among eukaryotes, there are organisms that lack one or more these enzymes, providing evidence for functional redundancy among deadenylases.
From an enzymatic perspective, PARN is by far the best studied of the group. This versatile homodimeric enzyme plays a role in mRNA stability, the quality control of gene expression and the maturation of a class of small RNAs. Biochemical and structural studies have provided insight into the intriguing property of cap-mediated regulation of the activity of this enzyme, suggesting a potential mechanistic connection between deadenylation and translation. Nocturnin, on the other hand, may also participate in functions beyond poly(A) tail shortening. These include chaperoning the movement of factors to the nucleus by protein-protein interaction. Thus one needs to keep an open mind about the biochemical functions of these enzymes and consider them in the broader context of the networking of gene expression in the cell.
From a biological perspective, these three deadenylases have been associated with a variety of important developmental and metabolic responses. PARN-mediated deadenylation has been implicated in early development decisions in a variety of organisms. Furthermore, the enzyme has been linked with cancerous growth and DNA repair activities. Nocturnin is not only a key protein in the circadian rhythm of the cell, but also has been shown to have an interesting impact on adipogenic differentiation, metabolic processes related to obesity, and regulation of the inflammatory response. While less is known regarding the biological implication of the Angel deadenylase, it has been implicated as a regulator of cell cycle-specific genes. Therefore not only is this group of enzymes interesting from a molecular perspective, they also have significant impact on fundamental cell processes as well. Therefore the goal of this review is to provide an overview of work performed to date on these deadenylases to allow an in depth appreciation of their molecular and biochemical properties in an interdisciplinary context.
2 PARN (poly (A)-specific ribonuclease)
2.1 Overview
An activity in HeLa cells having the properties of PARN was first described by the Virtanen lab [1,2] and PARN was molecularly cloned in 1998 by Wahle and coworkers [3,4]. The enzyme is conserved and active in eukaryotic organisms from trypanosomes, through mosquitoes, plants and mammals [5,6]. However analysis of the sequenced genomes of Saccharomyces cerevisiae and Drosophila melanogaster has revealed that they lack PARN [7,8]. Similarly, PARN function appears to be largely dispensable in Caenorhabditis elegans and Schizosaccharomyces pombe [9] and PARN sequence is less conserved in these organisms compared to vertebrates (Figure 1). Thus although PARN appears to play an important role in the biology of many organisms, presumably other redundant deadenylases fulfill the role of PARN in the handful of eukaryotes that to date have been shown to lack the enzyme. Furthermore, it is important to note that some presumed PARN proteins may not be true homologs. The AtPARN, CePARN and TbPARN polypeptides, for example, all appear to lack conserved R3H and RRM domains (Figure 1).
Figure 1. Boxshade2 analysis of ClustalW alignment of PARN from various species.
Black shading exactly conserved amino acids; Gray shading conservative substitutions of amino acids. Genbank accession numbers are as cited. Mm: Mus musculus NP_083037; Hs: Homo sapiens AAH50029; Gg: Gallus gallus NP_001025800; Xl: Xenopus laevis NP_001081143; Dr: Danio rerio NP_957382; At: Arabidopsis thaliana Genbank 60390594; Ce: Caenorhabditis elegans from [7]; Ano Anopheles gambiae from [4]; Aed Aedes aegypti from [4] ; Tb1 Trypanosome brucei Genbank AAZ13051.
In addition to recognizing the poly(A) tail as would be expected of an adenosine-specific 3’-to-5’ exonuclease, interestingly PARN can also simultaneously bind the 7-methyl-guanosine cap on the 5’ end of mRNAs [10-12]. This interaction influences the processivity of the enzyme, increasing the rate of poly(A) tail removal. Thus cap binding proteins such as eIF-4E may possibly serve to regulate PARN activity via competition for binding the cap structure. This cap-binding property may help to specifically target PARN to RNA Polymerase II-generated transcripts and coordinate PARN-mediated deadenylase activity with the translational competence of the mRNA. Interestingly, directing cells towards a quiescent state by serum-starvation results in a modest increase in phosphorylation of PARN and a modest increase in cap occupancy relative to eIF-4E in the cell, suggesting a dynamic interplay between PARN and other cap binding factors [13]. Furthermore, as our understanding of the regulation of cap-dependent and cap-independent translation evolves through sensitive global approaches such as ribosome profiling [14], the interplay between the cap-binding activity of PARN and the coordinate regulation of translation and mRNA deadenylation/decay should become clearer.
Enzymatically speaking, PARN is a member of the DEDD class of exonucleases that contain conserved Asp-Glu-Asp-Asp residues in their active site [15]. Mutation of any of these four amino acids substantially reduces the overall enzymatic activity of PARN. PARN exists as a dimer with the exonuclease domain acting to coordinate the two subunits [16]. The dimeric structure of PARN has been determined by X-ray crystallography of partial proteins [17], analytical ultracentrifugation [16], and in solution for the entire protein by atomic force microscopy and dynamic light scattering [18]. PARN requires divalent cations, particularly Mg2+, for optimal enzymatic activity [19]. Two divalent cations specifically interact with the active site amino acids Asp28, Asp292, and Asp382 (numbers correspond to the human protein),with Asp28 being the most important for the binding of Mg2+ or other divalent cations like Fe2+ [19-21]. Mg2+ also plays a role in the structural stability of the PARN enzyme. Divalent cations protect the active site of PARN from thermal inactivation, causing retention of up to 90% of activity when the protein is incubated at 45°C [22].
PARN appears to act both in the nucleus and in the cytoplasm of eukaryotic cells. In addition to the standard trimming of poly(A) tails of messenger RNAs, PARN has two interesting functions in the nucleus. First, PARN has recently been shown to be involved in the maturation of the 3’ end of snoRNAs [23]. Second, the deadenylase plays a role in the directed decay of transcripts following DNA damage [24]. PARN also plays a role in the quality control of gene expression in addition to assisting in the regulation of poly(A)+ mRNA levels. The deadenylase is active in nonsense mediated mRNA decay (NMD) and can be immunoprecipitated with NMD factors Upf1, Upf2, and Upf3X. Knockdown of PARN by siRNAs to about 15% of wild-type levels led to the stabilization of a reporter transcript containing a premature nonsense codon [25].
PARN appears to play important roles in early development [4] as well as perhaps in certain cancers. A recent study found that PARN was 2.2-fold overexpressed in acute lymphocytic leukemia (ALL) and 2-fold overexpressed in acute myeloid leukemia (AML) compared to control patients [26]. In addition, PARN was phosphorylated to a greater extent in ALL patients compared to controls [26]. Similarly, PARN mRNA is expressed at significantly higher levels in growing follicles of the bovine oocyte compared to persistent follicles [27]. As PARN causes poly(A) tail removal at germinal vesicle breakdown, its expression silences maternal transcripts during oocyte maturation and embryogenesis. These studies are both consistent with a role for PARN in promoting cell growth.
PARN also plays a role in the regulation of mRNA turnover of a set of transcripts in mammalian cells. A recent study determined the effects of shRNA-mediated PARN knock-down in mouse C2C12 myoblasts using global mRNA expression and half-life analysis [28]. Forty stabilized mRNAs and 24 destabilized mRNAs were observed as a result of the PARN knock down. While this may appear to be a rather small subset of mRNAs in the cells, a similar small number of mRNAs (2%) was affected in cells depleted of other deadenylases (the CCR-like and CAF1-like enzymes) using RNAi technologies [29]. The effects of PARN on mRNA stability, at least on some mRNAs, is direct as analysis of the Zfp36l2 transcript indicated an elongated poly(A) tail up to 2-3 fold the length in PARN knock down compared to control cells. Interestingly, many of the destabilized mRNAs in PARN knockdown cells encoded factors involved in cell migration and adhesion. This may represent a biologically relevant regulon as PARN KD cells migrated faster in a wound-healing assay consistent with the identities of the destabilized mRNAs.
The PARN deadenylase, therefore, is a multifunctional enzyme with interesting structural, subcellular localization and enzymatic properties that plays a role in a number of important biological phenomena. The sections that follow go into more depth into several of the aspects of this enzyme to provide further perspectives on its biochemical and biological properties.
2.2 Structural studies of PARN
PARN consists of three domains: a RNA recognition motif domain (RRM), a nuclease domain, and an R3H domain (a protein domain that contains an invariant arginine residue and a highly conserved histidine residue four amino acids downstream [28]). Crystal structures of PARN are available [14, 29] and based on these crystal structures a structural model of PARN that visualizes the positions of the nuclease, R3H, and RRM domains has been reconstructed [29].
In addition to its expected role in single-stranded RNA binding, the RRM domain also plays a role in cap recognition. The m7GpppG cap binds to the RRM domain in one subunit of the PARN homodimer and to a pocket formed by the RRM domain and the nuclease domain in the other subunit [31]. The relative position of the RRM domains differs by 30° in the two units, resulting in the two alternative modes of cap interaction. Crystal structures of the human and mouse PARN enzymes indicate that the RRM domain binds the 7-methylguanosine cap (m7G) via a tryptophan residue (human Trp475 and mouse Trp468) [17,32]. The m7GTP is bound by coplanar stacking with human PARN Trp475 and by Trp456 hydrogen bonding with the m7G N1 atom [17]. PARN was found to bind GTP with at least 100-fold less affinity than m7GTP [17].
The R3H domain of one PARN subunit interacts with the active site of the other subunit as well as also being involved in RNA binding [16]. Deletion of either the R3H domain or the RRM domain revealed that both singly are able to bind RNA, but that both are required to be present for full enzymatic activity [33]. Crystallographic studies of a C-terminal truncation of human PARN (residues 1-430), lacking the cap binding domain, revealed that each subunit in the homodimer binds three adenosine residues of the poly(A) tail of the RNA substrate and that Glu30 specifically interacts with the 3’OH group [16]. Assays of a human PARN lacking the R3H domain revealed that 103 more protein was needed to achieve the same deadenylation activity as the C-terminal truncation (1-430 aa), further emphasizing the importance of the R3H domain [16].
In summary, significant progress has been made to date on the fundamental structure and interactions of this enzyme based on crystallographic analyses of PARN derivatives with cap analog or with short oligo-adenosine transcripts. Future co-crystallization of a capped and polyadenylated RNA substrate with PARN might reveal additional structural alterations in the position of amino acid side chains to further our knowledge of the structure-function of the overall interaction.
2.3 Recruiters of PARN and their role in general and developmentally-controlled regulation of gene expression
The activity of a deadenylase such as PARN must be tightly regulated in the cell. This control is likely mediated by positive and negative regulators of PARN activity. The interaction of the poly(A) tail with poly(A) binding proteins (PABPs) has been shown to repress PARN activity and protect RNA substrates [10]. Cap-binding proteins (CBPs) such as eIF4E and CBP80 have also been shown to repress PARN [10, 34]. Thus activities that increase the off rate of PABPs or CBPSs may serve to coordinate PARN activity. However the regulation of PARN by cap binding proteins appears to be more complex than that. Inhibition of PARN by CBP80 occurs in a cap-independent and RNase A-resistant manner suggesting direct interaction of PARN and CBP80. Presumably, this may help protect mRNAs being exported to the cytoplasm before the nuclear cap binding complex is replaced by translation initiation factors. Post-translational modification of PARN by phosphorylation may also play a role in regulating PARN interactions with capped RNA substrates [13].
In addition, several RNA binding proteins have been found to date that recruit PARN to specific mRNAs. In general, these proteins interact with elements that regulate cytoplasmic poly(A) tail length or RNA stability elements such as AU-rich sequences or GU-rich sequences to direct PARN to help in poly(A) remodeling or removal. The proteins that influence transcript-specific directed action of PARN are the focus of the remainder of this section.
CUG-binding protein, CUG-BP (or more recently renamed CELF1), specifically interacts with a variety of RNAs through AU-rich and GU-rich elements both in vitro and in living cells [35,36]. Interestingly, PARN can be co-immunoprecipitated with CUG-BP in an RNA-independent fashion and purified GST-CUG-BP beads pulled down recombinant PARN [35]. These data strongly suggest that PARN and CUG-BP directly interact. The proposed working model is that CUG-BP may assist in anchoring PARN to some mRNA targets to decrease its off rate and increase the processivity of deadenylation.
Human tristetraproline (TTP), a zinc-finger rich protein that binds AU-rich elements, or a rat and a Xenopus ortholog (ZFP36L1 or ZFP36L2, respectively) also can activate PARN when transfected into HEK293 cell extracts [37]. TTP binding to the RNA substrate is crucial for this activation, as TTP mutants that cannot interact with RNA failed to activate PARN activity. Whether or not TTP directly recruits PARN to RNA substrates is unclear, however, since attempts to demonstrate an association between the two factors were unsuccessful. In addition, recent experiments have shown that TTP can recruit other deadenylases such as hCcr4a, hCcr4b, hCaf1a, hCaf1b, and hPan2 through direct protein-protein interactions [38]. Marchese et al. have more recently demonstrated that TTP interacts with CAF1 deadenylase [39] similar to the situation in Drosophila melanogaster [40,41]. Thus TTP may be a relatively non-selective recruiter of deadenylases to mRNAs targeted for decay. Finally, the activation of deadenylation by TTP appears to be regulated by post-translational modification. Hyperphosphorylation of TTP inhibits the ability of TTP to recruit deadenylases and is associated with binding the phospho-serine/threonine binding protein 14-3-3 [37].
PARN plays a major role in regulating poly(A) tail length in the oocyte development [4]. PARN is active in degrading poly(A) tails in immature oocytes to keep the tail short and block translation. As oocytes mature, PARN dissociates from mRNAs to generate longer tail lengths and activate translation of stored mRNAs [39]. Three RNA binding proteins - Pumilio1, Pumilio2, and cis-acting cytoplasmic polyadenylation element binding (CPEB) protein - have been shown to interact with PARN in Xenopus oocyte extracts. Pumilio proteins bind a cytoplasmic polyadenylation element (CPE) in the 3’ untranslated region of many mRNAs [40]. Immunoprecipitation assays suggest a direct interaction between pumilio1 and pumilio2 proteins with PARN. Recent interesting experiments show CPEB protein recruits both the deadenylase PARN and a poly(A) polymerase, GLD2, to transcripts in oocytes [41]. In immature oocytes, PARN activity is dominant to the GLD2 poly(A) polymerase, leading to poly(A) tail shortening of CPE-containing mRNAs [41]. Stimulation of oocytes to reenter meiosis leads to phosphorylation of CPEB protein and the subsequent expulsion of PARN from the complex at the 3’ end of the transcript. This results in poly(A) tail elongation by GLD2 and the activation of translation of these stored mRNAs. However it is important to note that a study has challenged the association of PARN with CPEB complexes [42]. Nevertheless, these experiments illustrate the potential importance of auxiliary factors in PARN recruitment and the dynamic nature of PARN-containing complexes in the regulation of gene expression.
The use of PARN to counteract a co-recruited poly(A) polymerase on the same transcript to regulate poly(A) tail length is not unique to oocytes. In the mouse hippocampus, N-methyl-D-aspartate receptor (NMDAR) activation leads to CPEB protein phosphorylation and expulsion of PARN from a pentameric CPEB, symplekin, Gld2, PARN and neuroguidin (Ngd) complex [42]. This complex appears to control Gld2-mediated polyadenylation of at least 100 different hippocampal mRNAs, 27 of which have been implicated in synaptic plasticity or nervous disorders. Lentiviral knockdown of PARN levels by about 50% was not sufficient to alter synaptic plasticity. Thus a clear picture of the involvement of PARN in synaptic plasticity will require additional experimentation.
Given the ability of PARN to reshape poly(A) tails and its direct or indirect recruitment by several regulators of RNA stability, future studies are required to (i) identify the full complement of PARN protein-protein interactions, (ii) the range of mRNAs in additional tissues whose poly(A) tail and translatability are regulated by PARN, and (iii) catalog the direct and indirect effects of PARN on the regulation of both nuclear and cytoplasmic RNA stability. This work will more effectively place the role of PARN in the cell in the context of other deadenylases as well as help to better understand the role of the apparent functional redundancy of poly(A) shortening enzymes in various cellular and developmental contexts.
2.4 A role for PARN in development in Arabidopsis and trypanosomes
In addition to mammalian and frog development, PARN has also been shown to influence gene expression at specific developmental stages in other organisms. However, the “PARN” orthologs in these organisms as well as C. elegans lack the canonical R3H [28] and R3M domains. The overexpression of the active PARN1 homolog of Trypanosoma brucei was demonstrated to influence the expression of a set of stage specific coat proteins [43]. The Arabidopsis PARN protein is less conserved than seen with vertebrates (Figure 1), yet appears to fulfill a similar role as the vertebrate PARN in controlling poly(A) tail length of a defined subset of mRNAs [44]. Northern blots showed AtPARN expression in stem, leaves, root and flower with highest expression in root and flower [44]. The nuclear and cytoplasmic subcellular localization of PARN in plants also appears to be generally consistent with observations made in other systems [9,44].
The importance of PARN to plant biology has been well established using a variety of genetic and molecular approaches. PARN is an essential protein in Arabidopsis as no homozygous progeny could be recovered and aborted homozygous seeds were found using multiple different mutation alleles in the PARN gene [9,45,46]. PARN mutants were described to be defective in their responses to abscisic acid, salicylic acid and stress responses [45,46]. Culturing of PARN mutant embryos using in vitro culture media showed a profound retardation of development [9]. Importantly, the use of the ligation-mediated polyA tail assay (LM-PAT) revealed that the PROLIFERA (PRL1) mRNA encoding the MCM7 protein showed almost a doubling in the average length of its poly(A) tail in PARN mutants. Poly(A) tail length in this PARN mutant, however, was not affected on several other mRNAs that were tested [9]. These results are similar to the specificity shown in the knock-down of murine PARN to poly(A) tail lengthening of only a subset of transcripts [28].
2.5 Small Molecule Inhibitors of PARN
Since the expression of PARN is dysregulated in ALL and AML tumors, inhibitors of the deadenylase may be useful for interventions [26]. Small molecule regulators of PARN can currently be divided into three groups. First, mono-and divalent cations have been shown to significantly affect PARN activity in a variety of ways. PARN can be activated by K+ in an allosteric manner by binding the RRM domain [47]. Second, PARN can be inhibited by aminoglycoside antibiotics, including neomycin B [48]. Interestingly, the ability of aminoglycosides to modulate PARN activity is strongly influenced by divalent cation (Mg2+) concentrations [48]. Finally, several studies have found nucleotide analogs that inhibit PARN to varying degrees. PARN is activated by low concentrations (e.g. 0.2 μM) of cap analog m7G(5’)ppp(5’)G, but can be inhibited by high concentrations (e.g. 2 μM) in an allosteric manner by binding the RRM domain [47]. Early work showed PARN is inhibited by all purine nucleotides, either deoxynucleotides or ribonucleotides, in the low mM range [49]. Purine rNTPs showed non-competitive inhibition likely as simple feedback inhibition, while purine rNMPs and rNDPs displayed competitive competition [49]. Effective use of unsaturated fluoro-ketopyranosyl nucleotides as antiviral and antitumor agents [50,51] led researchers to try them as PARN inhibitors. PARN is competitively inhibited by binding in the active site of the enzyme by nucleotides with a fluoro-pyranosyl sugar moiety and a benzoyl-modified cytosine or adenine base and Mg2+ ions could not compete with the inhibition [52]. In silico analysis revealed that the sugar moiety stabilizes the interaction in the active site through interactions with catalytic amino acid residues suggesting these compounds could act therapeutically by lowering mRNA turnover rates [52]. Based on these results, improved synthetic uracil, 5-fluorouracil or thymine base analogs using glucopyranosyl rings were prepared that showed a Ki in the low μM range [53].
3. Nocturnin and Angel
3.1 Overview
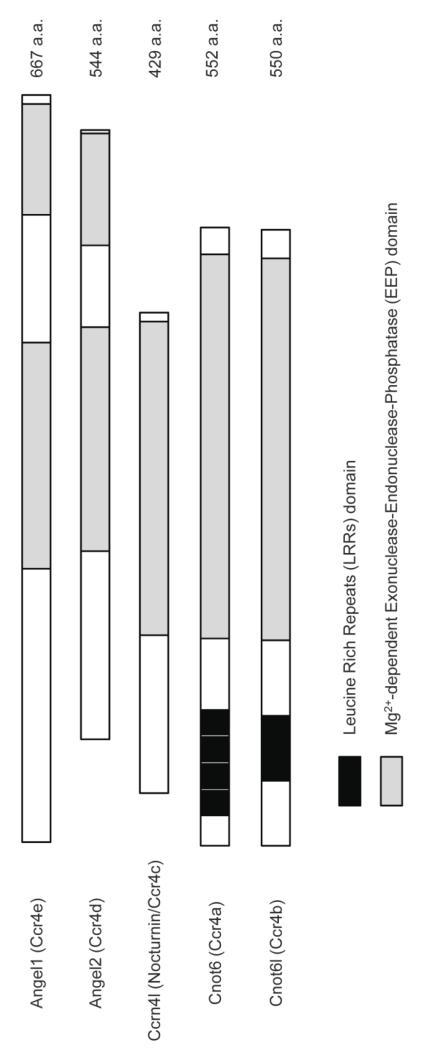
Nocturnin (NOC; Ccrn4l; CCR4c) and Angel1 (CCR4e) and Angel2 (CCR4d) are members of the CCR4 protein family with conserved Mg2+-dependent nuclease domains shared by all members of the Exonuclease, Endonuclease, and Phosphatase (EEP) superfamily to which the CCR4 proteins belong (Figure 2) [58-61]. Based on this strong conservation of the catalytic domain NOC, Angel1 and Angel2 are predicted to function as deadenylases, but there are some key differences between these proteins and the other CCR4 family members and many mysteries remain concerning their precise in vivo roles. Although the C-terminal catalytic domains are conserved in these proteins, NOC, Angel1 and Angel2 all have significantly divergent N-termini and lack the leucine-rich repeat region required for yeast Ccr4p and mammalian CCR4a and CCR4b to interact with Caf1 and other proteins in the major CCR4-NOT complex [61-63] (Figure 2). NOC exhibits deadenylase activity in vitro, as shown first in Xenopus and later in mouse and Suberites domuncula [64-66]. Deadenylase activity of Angel2 is an unsettled question [62, 67] and the activity of Angel1 has not been reported. Below we will review what is known about these proteins and discuss their possible in vivo functions.
Figure 2. Schematic representation of the mouse Ccr4 homologs.
Each has a conserved nuclease domain (shown in gray) related to a class of Mg2+-dependent Exonucleases-Endonucleases-Phosphatases, known as the EEP superfamily. Cnot6 and Cnot6l possess N-terminal Leucine Rich Repeats (LRRs) (shown in black), which are conserved in yeast Ccr4p. GenBank accession numbers are as given: Ccrn4l (Nocturnin/Ccr4c), NP_033964; Angel1 (Ccr4e), NP_653107; Angel2 (Ccr4d), NP_067396; Cnot6 (Ccr4a), NP_997649; Cnot6l(Ccr4b), NP_659159.
3.2 Nocturnin
Noc was first discovered in a differential display screen designed to detect mRNAs that cycled in abundance under the control of the circadian clock in retinal photoreceptors of Xenopus laevis [68, 69]. At the time of its discovery, this mRNA encoded a novel protein and was therefore named “Nocturnin’ based on its expression pattern, which was robustly rhythmic, peaking in the early night [68]. Noc is conserved among eukaryotes and its mRNA is widely expressed in many tissues throughout the body in various organisms [70-73]. In addition to the retina, in mammals Noc mRNA has been found in many parts of the brain, and in many metabolically-relevant tissues including liver, lung, pancreas, intestine, muscle, kidney, brown and white adipose and mesenchymal stem cells in bone marrow, as well as during all stages of the Drosophila life cycle, especially in metabolically active tissues such as the ring gland, salivary gland, and the proventriculus [58, 70-72, 74-76].
Noc is classified as a ‘circadian’ (circa-about, and dies-day) gene, one of the genes whose expression is driven by the biological clock, and this rhythmic expression pattern distinguishes it from other deadenylases. In Xenopus photoreceptors, the rhythmic mRNA levels of Noc likely result from transcriptional regulation by the circadian clock via rhythms in phosphorylated CREB (cyclic adenosine monophosphate response element-binding) [77, 78], whereas the regulatory mechanism in mammals appears to be more complicated. Noc expression can be activated directly by the CLOCK/BMAL1 heterodimeric transcription factor that is the core component of the circadian clock and that drives circadian transcription of thousands of genes [79-81], however, the expression of Noc remains rhythmic in Clock mutant animals, although with damped amplitude [82]. Similarly, Noc was among a small group of genes that remained rhythmic in the liver when the liver clock was conditionally inactivated while the remainder of the clocks throughout the body remained unaffected [83]. This suggests that non-cell autonomous signals can also control Noc’s rhythmic expression. Furthermore, the rhythmic profile of Noc mRNA is regulated by a post-transcriptional mechanism, as the liver-specific microRNA (miRNA) miR-122 represses Noc mRNA expression thus modulating the waveform of its rhythmic expression pattern [84].
In addition to its circadian expression, Noc is also unique among deadenylase genes in that it is an immediate early gene (IEG) showing acute responses to several stimuli including serum shock, phorbol ester, lipopolysaccharide (LPS) and rosiglitazone, a peroxisome proliferator-activated receptorγ (PPARγ) agonist [65, 76, 85, 86]. The corresponding in vivo correlate for this acute response has not been fully elucidated, however, it appears that the expression of Noc can be controlled by nutrient/metabolic cues. For example, a daytime bolus of lipid acutely induces Noc mRNA expression in the small intestine [87]. In contrast, its expression levels increase rapidly in some tissues upon food deprivation in mice and flies [82, 85], suggesting that other cues of nutrient status can also control its expression. The regulatory mechanisms for the acute induction of Noc are not established, however, there are conserved NF-κB sites in mammalian Noc promoter [86] that could be responsible for at least some acute responses. In addition, the Noc promoter has FoxO1 binding elements, and it has been shown that FoxO1 binds to these elements to regulate Noc expression [88]. It is possible that Noc’s regulation by food deprivation could be through this pathway, since FoxO transcriptional factors respond to nutrient signals to orchestrate transcriptional cascades such as those regulating glucose metabolism upon fasting.
Noc knock-out (Noc−/−) mice exhibit no obvious abnormalities in development or reproduction, however, they exhibit striking metabolic phenotypes when fed a High-Fat Diet (HFD). This diet causes wild-type mice to become obese, but the Noc−/− mice remain lean although they do not exhibit increased activity or reduced food intake [83]. This ‘leanness’ can be explained, at least in part, by a deficiency in the trafficking of dietary lipid in the intestinal enterocytes, resulting in inefficient absorption into the circulation [87]. These phenoytpes were accompanied by changes in mRNA levels that are associated with lipid metabolism. For example, hepatic expression of Srebp-1c, Scd1 and L-Fabp was decreased, and a normally robust diurnal rhythm of Pparγ mRNA was attenuated in the Noc−/− mice [89]. In addition, intestinal expression of adipophilin (Adrp/Plin2), adipocyte triglyceride lipase (Atgl/Pnpla2) and diacylglycerol acetyltransferase 2 (Dgat2), genes essential for regulating the intracellular trafficking and packaging of triglycerides, were also decreased, indicating the potential role of NOC in regulating the expression of these mRNAs. Nonetheless, it is not clear whether the deadenylase activity of NOC is responsible for the changes of expression of these mRNAs. If NOC removes poly(A) tails from these lipid-associated transcripts directly, one would expect that these transcripts would be more stable in the absence of deadenylation (i.e. in the Noc−/− mice), leading to higher mRNA levels. However, the opposite is observed, suggesting that NOC may act indirectly on other targets that in turn mediate the expression of these genes, or that it may have deadenylase-independent functions.
The proadipogenic function of NOC was also observed in 3T3-L1 cells undergoing adipogenesis with Noc mRNA levels rising as they differentiate into adipocytes [76]. Furthermore, Noc−/− mice exhibited a decreased marrow adiposity and an increased bone-mass [76]. Osteoblasts and adipocytes differentiate from a common pluripotent precursor, the mesenchymal stem cell (MSC), and lineage determination in these cells is therefore important because they can differentiate into multiple cell types [90]. Studies have identified numerous transcription factors and signaling pathways that determine the fate of MSC [90], and PPARγ is a prime inducer of adipogenesis that inhibits osteoblastogenesis [90, 91]. Interestingly, NOC interacts with PPARγ and promotes its nuclear translocation, and this nuclear translocation is promoted even with a catalytically-dead mutant of NOC [70]. Since PPARγ is a nuclear receptor that regulates the expression of genes important for fatty acid storage, glucose metabolism and osteogenesis [91], NOC perhaps regulates PPARγ target gene expression indirectly by promoting nuclear translocation of PPARγ [76]. These findings support the possibility that NOC can contribute to the regulation of gene expression independent of its deadenylase activity.
NOC has also been implicated in the inflammatory response, with Noc−/− mice showing increased survivability to the administration of a sub-lethal dose of LPS, a bacterial endotoxin that elicits strong immune responses in animals [86]. Mouse embryo fibroblasts (MEFs) harvested from Noc−/− mice showed drastically decreased levels of the proinflammatory transcript iNOS mRNA following LPS administration. Furthermore, the half-life of the induced iNOS mRNA is significantly shorter in the Noc−/− MEFs. The destabilization of this mRNA, in the cells lacking NOC, suggest that this change in iNOS mRNA half-life is either indirect, or is through some non-conventional activity for a deadenylase.
Despite NOC’s robust rhythmicity, it appears that NOC is an “output” of the clock and not part of the central clock mechanism itself in mammals. Noc−/− mice do not have overt circadian phenotypes either at the behavioral or molecular level [89]. Nevertheless, a recent finding demonstrated that NOC plays a more dominant role in controlling the circadian clock in Drosophila, as both mRNA and protein expression of noc are rhythmic in a subgroup of clock neurons (DN3s) in Drosophila, and noc knockdown flies have abnormal rhythmic behavior in constant light [73], indicating that NOC contributes to the light-mediated circadian behavioral response. It is not known whether this phenotype in Drosophila is due to its deadenylase activity. The nuclease domain of NOC in the C-teminus is highly conserved, however, the N-terminal domain is more divergent among organisms. Therefore, this difference in the phenotype might arise from the divergent portion of NOC, which does not encode the nuclease domain, or due to the different organization of the circadian system between flies and mice. This awaits further investigation.
Another interesting story of noc in Drosophila came from a study that identified noc, after a near-century-long gene hunt, as the gene that is responsible for upwardly bent and curled wing phenotype mutants widely used as markers in fruit fly genetics. Curled (Cu), one of the first fly mutants described in 1915 by C.B. Bridges and T.H. Morgan encodes Drosophila noc and this gene is required for proper wing morphogenesis during the pupal stage [92]. The nuclease domain is important for this phenotype as a truncated NOC/CU protein that lacks part of the predicted nuclease domain fails to rescue the wing phenotype [72]. However, noc/cu is not expressed in the developing wing, and noc/cu knockdown in various different individual organs such as the wing, nervous system, fat body, and gut, including each of the tissues in which endogenous noc/cu is expressed, was insufficient to result in the curled wing phenotype [72]. Noc/cu must be functioning in tissue-non-autonomous manner, and identification of mRNAs that are responsible for this phenotype and have altered poly(A) tail lengths would be of interest.
The biological clock controls approximately 10% of the transcriptome and the set of mRNAs that are rhythmic vary significantly between tissues [93]. NOC’s deadenylase activity likely contributes to circadian control of gene expression by rhythmically controlling transcript degradation to influence various circadian physiologic processes. Since Noc is expressed rhythmically in many tissues and organs [64, 70-72], it is likely that it may have cell-specific targets, but bona fide target mRNAs that are directly deadenylated by NOC in vivo have yet to be identified. The coordination of Noc expression with rhythmic food intake, as well as its acute responsiveness to various stimuli including lipids, supports the idea that NOC might function to regulate circadian nutrient metabolism, in other words the ingestion, trafficking and distribution of nutrients, with possible specificity for lipids.
3.3 Angel
The Angel proteins are thought to function as deadenylases, based on their sequence similarity to the other members of CCR4 protein family (Figure 2). Evidence supporting a deadenylase role for the Angel proteins came from the demonstration that Angel proteins interact with Caf1 proteins that possess deadenylase activity [62], analogous to the interactions between the CCR4a/b and Caf1 proteins in the major deadenylase complex. However, in vitro measurements of Angel1’s deadenylase activity have not been reported and attempts to demonstrate Angel2’s deadenylase activity were not successful [62, 67]. It is possible that the conditions used in these studies were not suitable for Angel proteins, or it could be due to the intracellular localization. Angel2 resides in the nucleus when exogenously expressed, while the majority of deadenylation reactions are thought to occur in the cytoplasm [94]. Indeed, other deadenylases such as CCR4a/b and NOC localize in the cytoplasm [63, 64, 67]. If Angel2 protein functions as a deadenylase, what are its substrates? One possibility is that Angel2 targets nuclear mRNAs or pre-mRNAs and promotes their decay as a nuclear RNA quality control mechanism. For example, mRNAs that fail to undergo splicing are degraded in the nucleus because the accumulation of these aberrant transcripts can be toxic [95, 96]. Another possibility is that Angel2 helps to maintain the poly(A) tail length of mRNAs to ~200-250 nt in the nucleus. Nuclear polyadenylation machinery adds the poly(A) tail to newly synthesized mRNAs, and the size of the poly(A) tail in the nucleus appears to be determined by the physical constraints imposed by the spherical protein-RNA complexes, which includes nuclear poly(A) binding proteins bound to the poly(A) tails [94, 97]. It has been proposed in yeast that a nuclear deadenylation process restricts newly added mRNA poly(A) tails to their proper lengths [98, 99] so that mRNA can be readily transported into cytoplasm.
Another interesting possibility came to light by the discovery of long non-coding RNAs (lncRNAs). These lncRNAs, defined as transcribed RNA molecules greater than 200 nt in length, biochemically resemble conventional mRNAs (they are 5’-capped, spliced and polyadenylated), yet do not template protein synthesis. Numerous high-throughput genomic analyses have provided evidence that the genome encodes at least as many lncRNAs as the known protein coding genes [100, 101], and approximately 30% of lncRNAs are highly enriched in the nucleus [102]. It would be interesting to pursue the possibility that Angel2 is responsible for deadenylating these lncRNAs that localize in the nucleus. Furthermore, despite its primarily nuclear localization, a role for the Angel2/Caf1z complex in the cytoplasm can also not be excluded, as both Caf1z and Angel2 are nucleocytoplasmic shuttling proteins [62]. Perhaps Angel2/Caf1z complex is normally sequestered in the nucleus but can be exported to the cytoplasm under certain conditions. Future studies should reveal whether such conditions exist and whether it targets specific mRNA substrates for degradation under those conditions.
The functions of the Angel proteins in vivo await further investigation, however, a cell-based study suggests that Angel2 arrests cell cycles, thereby functioning as an anti-proliferating gene [103]. Angel2 interacts with p21/Cip1 mRNA, a direct mediator of cell cycle arrest at the G1-to-S transition [104] and up-regulates p21/Cip1 expression by stabilizing its mRNA [103]. Even though it is not clear whether this interaction with p21 mRNA occurs in the nucleus or cytoplasm, this study clearly indicates that Angel2 can not only function as a deadenylase but also as an RNA-binding protein to alter target gene expression. Angel2 is expressed in various tissues and cell lines, with strong expression in liver and skeletal muscle in human [103], suggesting an important role in these tissues.
Highlights.
PARN deadenylase binds to mRNA 5’ caps with functional and biological implications
PARN plays a role in early development, DNA damage and possibly cancer
Nocturin deadenylase shows rhythmic expression under circadian control
Nocturnin is rapidly induced in response to serum shock, LPS and other stimuli
Nocturnin functions in adipogenesis and lipid metabolism.
Acknowledgements
The authors wish to thank Ryan Thummel, Ph.D., Wayne State University, for aid with Figure 1. Research on fundamental aspects of RNA stability are funded in the Wilusz laboratory by NIH award GM072481 and in the Green laboratory by GM076626 and GM090247.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Åström J, Åström A, Virtanen A. In vitro deadenylation of mammalian mRNA by a HeLa cell 3’ exonuclease. EMBO J. 1991;10:3067–3071. doi: 10.1002/j.1460-2075.1991.tb07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]].Åström J, Åström A, Virtanen A. Properties of a HeLa cell 3’ exonuclease specific for degrading poly(A) tails of a mammalian mRNA. J. Biol. Chem. 1992;267:18154–18159. [PubMed] [Google Scholar]
- [3].Körner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3’-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- [4].Körner CG, Wormington CG, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Milone J, Wilusz J, Bellofatto V. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA. 2004;10:448–457. doi: 10.1261/rna.5180304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Opyrchal M, Anderson JR, Sokoloski KJ, Wilusz CJ, Wilusz J. A cell-free mRNA stability assay reveals conservation of the enzymes and mechanisms of mRNA decay between mosquito and mammalian cell lines. Insect Biochem. Mol. Biol. 2005;35:1321–1334. doi: 10.1016/j.ibmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- [7].Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA. 2004;10:1200–1214. doi: 10.1261/rna.7540204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gao M, Fritz DT, Ford LP, Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5’ Cap influences mRNA deadenylation rates in vitro. Mol. Cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dehlin E, Wormington M, Körner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martînez J, Ren YG, Nilsson P, Ehrenberg M, Virtanen A. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J. Biol. Chem. 2001;276:27923–27929. doi: 10.1074/jbc.M102270200. [DOI] [PubMed] [Google Scholar]
- [13].Seal R, Temperley R, Wilusz J, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Serum-deprivation stimulates cap-binding by PARN at the expense of eIF4E, consistent with the observed decrease in mRNA stability. Nucleic Acids Res. 2005;33:376–387. doi: 10.1093/nar/gki169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell. Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- [16].Wu M, Reuter M, Lilie H, Liu Y, Wahle E, Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Monecke T, Schell S, Dickmanns A, Ficner R. Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m(7)G-cap-binding mode. J. Mol. Biol. 2008;382:827–834. doi: 10.1016/j.jmb.2008.07.073. [DOI] [PubMed] [Google Scholar]
- [18].Niedzwiecka A, Lekka M, Nilsson P, Virtanen A. Global architecture of human poly(A)-specific ribonuclease by atomic force microscopy in liquid and dynamic light scattering. Biophys. Chem. 2011;158:141–149. doi: 10.1016/j.bpc.2011.06.010. [DOI] [PubMed] [Google Scholar]
- [19].Ren Y-G, Kirsebom LA, Virtanen A. Coordination of divalent metal ions in the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 2004;279:48702–48706. doi: 10.1074/jbc.M403858200. [DOI] [PubMed] [Google Scholar]
- [20].He G-J, Liu W-F, Yan Y-B. Dissimilar roles of the four conserved acidic residues in the thermal stability of poly(a)-specific ribonuclease. Int. J. Mol. Sci. 2011;12:2901–2916. doi: 10.3390/ijms12052901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ren Y-G, Martínez J, Virtanen A. Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe(2+)-mediated cleavage. J. Biol. Chem. 2002;277:5982–5987. doi: 10.1074/jbc.M111515200. [DOI] [PubMed] [Google Scholar]
- [22].Liu W-F, Zhang A, Cheng Y, Zhou H-M, Yan Y-B. Effect of magnesium ions on the thermal stability of human poly(A)-specific ribonuclease. FEBS Lett. 2007;581:1047–1052. doi: 10.1016/j.febslet.2007.02.008. [DOI] [PubMed] [Google Scholar]
- [23].Berndt H, Harnisch C, Rammelt C, Stöhr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez J-P, Hüttelmaier S, Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE. Nuclear deadenylation/polyadenylation factors regulate 3’ processing in response to DNA damage. EMBO J. 2010;29:1674–1687. doi: 10.1038/emboj.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- [26].Maragozidis P, Karangeli M, Labrou M, Dimoulou G, Papaspyrou K, Salataj E, Pournaras S, Matsouka P, Gourgoulianis KI, Balatsos NA. Alterations of deadenylase expression in acute leukemias: Evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012;128:39–46. doi: 10.1159/000337418. [DOI] [PubMed] [Google Scholar]
- [27].Lingenfelter BM, Dailey RA, Inskeep EK, Vernon MW, Poole DH, Rhinehart JD, Yao J. Changes of maternal transcripts in oocytes from persistent follicles in cattle. Mol. Repro. Dev. 2007;74:265–272. doi: 10.1002/mrd.20568. [DOI] [PubMed] [Google Scholar]
- [28].Lee JE, Lee JY, Trembly J, Wilusz J, Tian B, Wilusz CJ. The PARN deadenylase targets a discrete set of mRNAs for decay and regulates cell motility in mouse myoblasts. PLoS Genet. 2012;8:e1002901. doi: 10.1371/journal.pgen.1002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mittal S, Aslam A, Doidge R, Medica R, Winkler GS. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell. 2011;22:748–758. doi: 10.1091/mbc.E10-11-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grishin NV. The R3H Motif: a domain that binds single-stranded nucleic acids. Trends Biochem. Sci. 1998;23:329–330. doi: 10.1016/s0968-0004(98)01258-4. [DOI] [PubMed] [Google Scholar]
- [31].Wu M, Nilsson P, Henriksson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H. Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease. Structure. 2009;17:276–286. doi: 10.1016/j.str.2008.11.012. [DOI] [PubMed] [Google Scholar]
- [32].Nagata T, Suzuki S, Endo R, Shirouzu M, Terada T, Inoue M, Kigawa T, Kobayashi N, Güntert P, Tanaka A, Hayashizaki Y, Muto Y, Yokoyama S. The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition. Nucleic Acids Res. 2008;36:4754–4767. doi: 10.1093/nar/gkn458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu W-F, Zhang A, He G-J, Yan Y-B. The R3H domain stabilizes poly(A)-specific ribonuclease by stabilizing the RRM domain. Biochem. Biophys. Res. Comm. 2007;360:846–851. doi: 10.1016/j.bbrc.2007.06.139. [DOI] [PubMed] [Google Scholar]
- [34].Balatsos NA, Nilsson P, Mazza C, Cusack S, Virtanen A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC) J. Biol. Chem. 2006;281:4517–4522. doi: 10.1074/jbc.M508590200. [DOI] [PubMed] [Google Scholar]
- [35].Moraes KCM, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol. Cell. Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AH, Dean JLE. MAPKAP kinase 2 blocks tristetraproline-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 2010;285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lauwers A, Twyffels L, Soin R, Wauquier C, Kruys V, Gueydan C. Post-transcriptional regulation of genes encoding anti-microbial peptides in Drosophila. J. Biol. Chem. 2009;284:8973–8983. doi: 10.1074/jbc.M806778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vindry C, Lauwers A, Hutin D, Soin R, Wauquier C, Kruys V, Gueydan C. dTIS11 Protein-dependent polysomal deadenylation is the key step in AU-rich element-mediated mRNA decay in Drosophila cells. J. Biol. Chem. 2012;287:35527–35538. doi: 10.1074/jbc.M112.356188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ota R, Kotani T, Yamashita M. Biochemical characterization of Pumilio1 and Pumilio2 in Xenopus oocytes. J. Biol. Chem. 2011;286:2853–2863. doi: 10.1074/jbc.M110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- [45].Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- [46].Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Utter CJ, Garcia SA, Milone J, Bellofatto V. PolyA-specific ribonuclease (PARN) function in stage-specific mRNA turnover in Trypanosoma brucei. Eukaryot. Cell. 2011;10:1230–1240. doi: 10.1128/EC.05097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ. AtPARN Is an essential poly(A) ribonuclease in Arabidopsis. Gene. 2004;328:95–102. doi: 10.1016/j.gene.2003.11.028. [DOI] [PubMed] [Google Scholar]
- [49].Nishimura N, Yoshida T, Murayama M, Asami T, Shinozaki K, Hirayama T. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol. 2004;45:1485–1499. doi: 10.1093/pcp/pch171. [DOI] [PubMed] [Google Scholar]
- [50].Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005;44:972–984. doi: 10.1111/j.1365-313X.2005.02589.x. [DOI] [PubMed] [Google Scholar]
- [51].Liu W-F, Zhang A, Cheng Y, Zhou H-M, Yan Y-B. Allosteric regulation of human poly(A)-specific ribonuclease by cap and potassium ions. Biochem. Biophys. Res. Comm. 2009;379:341–345. doi: 10.1016/j.bbrc.2008.12.056. [DOI] [PubMed] [Google Scholar]
- [52].Ren Y-G, Martínez J, Kirsebom LA, Virtanen A. Inhibition of Klenow DNA polymerase and poly(A)-specific ribonuclease by aminoglycosides. RNA. 2002;8:1393–1400. doi: 10.1017/s1355838202021015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Balatsos NA, Anastasakis D, Stathopoulos C. Inhibition of human poly(A)-specific ribonuclease (PARN) by purine nucleotides: Kinetic analysis. J. Enzyme Inhib. Med. Chem. 2009;24:516–523. doi: 10.1080/14756360802218763. [DOI] [PubMed] [Google Scholar]
- [54].Manta S, Agelis G, Botic T, Cencic A, Komiotis D. Unsaturated fluoro-ketopyranosyl nucleosides: Synthesis and biological evaluation of 3-fluoro-4-keto-β-D-glucopyranosyl derivatives of N4-benzoyl cytosine and N6-benzoyl adenine. Eur. J. Med. Chem. 2008;43:420–428. doi: 10.1016/j.ejmech.2007.04.001. [DOI] [PubMed] [Google Scholar]
- [55].Paterson J, Uriel C, Egron M-J, Herscovici J, Anonakis K, Alaoui-Jamali MA. Antiproliferative and apoptotic activities of ketonucleosides and keto-C-glycosides against non-small-cell lung cancer cells with intrinsic drug resistance. Antimicrob. Agents Chemother. 1998;42:779–784. doi: 10.1128/aac.42.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Balatsos NA, Vlachakis D, Maraozidis P, Manta S, Anastasakis D, Kyritsis A, Vlassi M, Komiotis D, Stathopoulos C. Competitive inhibition of human poly(A)-specific ribonuclease (PARN) by synthetic fluoro-pyranosyl nucleosides. Biochemistry. 2009;48:6044–6051. doi: 10.1021/bi900236k. [DOI] [PubMed] [Google Scholar]
- [57].Balatsos N, Vlachakis D, Chatzigeorgiou V, Manta S, Komiotis D, Vlassi M, Stathopoulos C. Kinetic and in silico analysis of the slow-binding inhibition of human poly(A)-specific ribonuclease (PARN) by novel nucleoside analogues. Biochimie. 2012;94:214–221. doi: 10.1016/j.biochi.2011.10.011. [DOI] [PubMed] [Google Scholar]
- [58].Dupressoir A, Barbot W, Loireau MP, Heidmann T. Characterization of a mammalian gene related to the yeast CCR4 general transcription factor and revealed by transposon insertion. J. Biol. Chem. 1999;274:31068–31075. doi: 10.1074/jbc.274.43.31068. [DOI] [PubMed] [Google Scholar]
- [59].Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Puech A, Dupressoir A, Loireau MP, Mattei MG, Heidmann T. Characterization of two age-induced intracisternal A-particle-related transcripts in the mouse liver. Transcriptional read-through into an open reading frame with similarities to the yeast ccr4 transcription factor. J. Biol. Chem. 1997;272:5995–6003. doi: 10.1074/jbc.272.9.5995. [DOI] [PubMed] [Google Scholar]
- [61].Draper MP, Liu HY, Nelsbach AH, Mosley SP, Denis CL. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol. Cell. Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wagner E, Clement SL, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol. Cell. Biol. 2007;27:1686–1695. doi: 10.1128/MCB.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- [64].Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- [65].Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Muller WE, Wang X, Grebenjuk VA, Korzhev M, Wiens M, Schlossmacher U, Schroder HC. Nocturnin in the Demosponge Suberites domuncula: a potential circadian clock protein controlling glycogenin synthesis in sponges. Biochem. J. 2012 doi: 10.1042/BJ20120357. Epub Aug 28. [DOI] [PubMed] [Google Scholar]
- [67].Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc. Natl. Acad. Sci. USA. 1996;93:14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Green CB, Besharse JC. Use of a high stringency differential display screen for identification of retinal mRNAs that are regulated by a circadian clock. Brain research. Mol. Brain Res. 1996;37:157–165. doi: 10.1016/0169-328x(95)00307-e. [DOI] [PubMed] [Google Scholar]
- [70].Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev. Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Curran KL, LaRue S, Bronson B, Solis J, Trow A, Sarver N, Zhu H. Circadian genes are expressed during early development in Xenopus laevis. PloS One. 2008;3:e2749. doi: 10.1371/journal.pone.0002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Grönke S, Bickmeyer I, Wunderlich R, Jäckle H, Kühnlein RP. Curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin. Genetics. 2009;183:219–232. doi: 10.1534/genetics.109.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat. Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Barbot W, Wasowicz M, Dupressoir A, Versaux-Botteri C, Heidmann T. A murine gene with circadian expression revealed by transposon insertion: self-sustained rhythmicity in the liver and the photoreceptors. Biochim. Biophys. Acta. 2002;1576:81–91. doi: 10.1016/s0167-4781(02)00296-8. [DOI] [PubMed] [Google Scholar]
- [75].Gilbert MR, Douris N, Tongjai S, Green CB. Nocturnin expression is induced by fasting in the white adipose tissue of restricted fed mice. PloS One. 2011;6:e17051. doi: 10.1371/journal.pone.0017051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, Clemmons DR, Rosen CJ. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc. Natl. Acad. Sci. USA. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu X, Green CB. A novel promoter element, photoreceptor conserved element II, directs photoreceptor-specific expression of nocturnin in Xenopus laevis. J. Biol. Chem. 2001;276:15146–15154. doi: 10.1074/jbc.M009970200. [DOI] [PubMed] [Google Scholar]
- [78].Liu X, Green CB. Circadian regulation of nocturnin transcription by phosphorylated CREB in Xenopus retinal photoreceptor cells. Mol. Cell. Biol. 2002;22:7501–7511. doi: 10.1128/MCB.22.21.7501-7511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li R, Yue J, Zhang Y, Zhou L, Hao W, Yuan J, Qiang B, Ding JM, Peng X, Cao JM. CLOCK/BMAL1 regulates human nocturnin transcription through binding to the E-box of nocturnin promoter. Mol. Cell. Biochem. 2008;317:169–177. doi: 10.1007/s11010-008-9846-x. [DOI] [PubMed] [Google Scholar]
- [80].Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, Kobayashi H, Iitaka C, Umehara T, Horikoshi M, Kudo T, Shimizu Y, Yano M, Monden M, Machida K, Matsuda J, Horie S, Todo T, Ishida N. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- [83].Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PloS One. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lecka-Czernik B, Rosen CJ, Kawai M. Skeletal aging and the adipocyte program: New insights from an “old” molecule. Cell Cycle. 2010;9:3648–3654. doi: 10.4161/cc.9.18.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Niu S, Shingle DL, Garbarino-Pico E, Kojima S, Gilbert M, Green CB. The circadian deadenylase Nocturnin is necessary for stabilization of the iNOS mRNA in mice. PloS One. 2011;6:e26954. doi: 10.1371/journal.pone.0026954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Douris N, Kojima S, Pan X, Lerch-Gaggl AF, Duong SQ, Hussain MM, Green CB. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr. Biol. 2011;21:1347–1355. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Takada I, Kouzmenko AP, Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat. Rev. Rheumatol. 2009;5:442–447. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- [91].Kawai M, Rosen CJ. PPARgamma: a circadian transcription factor in adipogenesis and osteogenesis. Nat. Rev. Endocrinol. 2010;6:629–636. doi: 10.1038/nrendo.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bridges CB, Morgan TH. The third-chromosome group of mutant characters of Drosophila melanogaster. Publs. Carnegie Instn. 1923;327:152–155. [Google Scholar]
- [93].Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol. 2003;15:991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- [94].Kühn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- [95].Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- [96].Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5’ degradation in yeast require the Lsm2-8p complex. Mol. Cell. Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. Nucleophosmin deposition during mRNA 3’ end processing influences poly(A) tail length. EMBO J. 2011;30:3994–4005. doi: 10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Mangus DA, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- [101].Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- [102].Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yi X, Hong M, Gui B, Chen Z, Li L, Xie G, Liang J, Wang X, Shang Y. RNA processing and modification protein, carbon catabolite repression 4 (Ccr4), arrests the cell cycle through p21-dependent and p53-independent pathway. J. Biol. Chem. 2012;287:21045–21057. doi: 10.1074/jbc.M112.355321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Cmielova J, Rezacova M. p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected] J. Cell. Biochem. 2011;112:3502–3506. doi: 10.1002/jcb.23296. [DOI] [PubMed] [Google Scholar]