Abstract
Clinical trials and animal studies have suggested that lycopene, the red carotenoid found in tomatoes, might be useful for the prevention of prostate cancer in the diet or as a dietary supplement through a variety of chemoprevention mechanisms. Since most mechanism of action studies have used prostate cancer cells or involved men with existing prostate cancer, we investigated the effects of lycopene on protein expression in human primary prostatic epithelial cells. After treatment with lycopene at a physiologically relevant concentration (2 μM) or placebo for 48 h, the primary prostatic epithelial cells were lysed and fractionated using centrifugation into cytosolic/membrane and nuclear fractions. Proteins from lycopene-treated and placebo-treated cells were trypsinized and derivatized for quantitative proteomics using isobaric tags for relative and absolute quantitation (iTRAQ) reagent. Peptides were analyzed using 2-dimensional microcapillary HPLC-tandom mass spectrometry to identify proteins that were significantly up-regulated or down-regulated following lycopene exposure. Proteins that were most affected by lycopene were those involved in antioxidant responses, cytoprotection, apoptosis, growth inhibition, androgen receptor signaling, and the Akt/mTOR cascade. These data are consistent with previous studies suggesting that lycopene can prevent cancer in human prostatic epithelial cells at the stages of cancer initiation, promotion and/or progression.
Keywords: prostate cancer, lycopene, chemoprevention, human primary prostatic epithelial cells, proteomics
Introduction
Prostate cancer is the second leading cause of cancer-related deaths in men in the United States with an average age of 67 years at diagnosis, and 241,740 new cases and 28,170 deaths are anticipated for 2012 (1). Carcinogenesis is a multistep process that are characterized by alterations in pathways involving cellular proliferation, differentiation, apoptosis, and senescence (2). Preneoplastic lesions in prostate tissue represent intermediate stages between normal and malignant epithelium, and since these lesions may appear decades before diagnosis of prostate cancer, the slow progression of prostate cancer might provide an opportunity for intervention and chemoprevention.
Although lacking provitamin A activity, lycopene is the most abundant carotenoid in tomatoes and is the most potent antioxidant among the naturally occurring carotenoids (3). Studies in humans and animal models indicate that blood lycopene levels and intake of tomato-derived products are inversely correlated with prostate cancer risk (4). Clinical trials have shown that dietary tomato sauce or lycopene supplementation in men increases lycopene levels in prostate tissue and serum, reduces DNA oxidation in prostate tissue and lowers serum prostate specific antigen (5, 6).
Postulated mechanisms of cancer chemoprevention by lycopene include inhibition of 5-lipoxygenase (7), regulation of the IGF-1/IGFBP-3 system (8), modulation of immune function (9), up-regulation of phase II cytoprotective enzymes (10), inhibition of growth and induction of differentiation in cancer cells by modulating the expression of cell cycle regulatory proteins (11), prevention of oxidative DNA damage (12), up-regulation of Cx43 and increased gap junction intercellular communication (13), balancing redox signaling (14), and inhibition of inflammation and androgen receptor signaling (15). Evidence for these mechanisms of lycopene action has usually been obtained by studying a single pathway instead of looking at multiple mechanisms and hasinvolved studies of cancer cells or pre-existing cancer instead of primary cells or healthy individuals.
Previously, we used iCAT quantitative proteomics to examine how lycopene alters protein expression in the LNCaP human prostate cancer cell line (16) and found that phase II enzymes involved in protecting cells from oxidative stress and detoxifying electrophilic xenobiotics were up-regulated by lycopene. Here, more sensitive iTRAQ proteomics was used to determine how cellular protein levels are altered upon exposure of primary human prostatic epithelial (PrE) cells to physiologically relevant concentrations of lycopene. The use of healthy prostate epithelial cells instead of prostate cancer cells is ideal for prostate cancer chemoprevention studies since most prostate cancers are of epithelial origin (17, 18).
Materials and Methods
Materials
Prostate epithelial growth medium was purchased from Lonza (Walkersville, MD). Water dispersible beadlets containing 10% (w/w) lycopene and identical placebo beadlets were gifts from DSM (Basel, Germany). Protease inhibitor cocktail was purchased from Sigma-Aldrich (St. Louis, MO). iTRAQ reagents were purchased from AB Sciex (Foster City, CA), and trypsin was purchased from Promega (Madison, WI). The BioRad protein assay kit was purchased from Bio-Rad (Hercules, CA). Monoclonal antibodies for western blots were purchased from Cell Signaling Technology (Danvers, MA) except for Rad23b (hHR23b) which was purchased from Abcam (Cambridge, MA). Buffers, HPLC-grade solvents and all other chemicals and were purchased from Thermo Fisher (San Jose, CA).
Human primary prostatic epithelial (PrE) cell culture
PrE cell cultures were derived from histologically benign areas of the prostate peripheral zone from radical prostatectomy specimens of patients who had received no prior chemical, hormonal or radiation therapy as described previously (19). Briefly, the prostate tissues were minced and digested overnight with collagenase. The digested tissues were plated onto collagen-coated dishes, and PrE cells were cultured in prostate epithelial growth media. PrE cells from three different patients were used for this study (PrE-1, PrE-2, PrE-3).
When the cells reached 70% confluence, they were aliquoted into 25–50 freezer vials and cryopreserved in liquid nitrogen storage. Individual frozen aliquots of PrE cells were thawed for each set of experiments, and cell cultures were incubated in a humidified incubator with 95% air/5% CO2 at 37°C in serum-free PrEGM. Cells were treated with 2 μM lycopene or placebo beadlets for 48 h after reaching 70% confluence. Lycopene beadlets have been shown to be stable in cell culture medium for up to 72 h (20).
Subcellular fractionation
After incubation with lycopene or placebo, the cell culture medium was removed, and cells were washed twice with ice cold PBS buffer. Hypotonic buffer (200 μL) containing 20 mM Tris (pH 7.5), 5 mM MgCl2, 5 mM CaCl2, 1 mM DTT, 1 mM EDTA, and 1% protease inhibitor cocktail were added to the cells which were scraped into buffer and transferred to a new tube. Cells were disrupted by repeated freezing and thawing. The crude nuclear pellet was collected by centrifugation at 1,800 × g for 15 min at 4°C. The supernatant containing the cytosolic and membrane proteins were frozen at −80°C until use. The crude nuclear pellet was resuspended on ice in 0.5 volumes of low salt buffer containing 20 mM Tris (pH 7.5), 5 mM MgCl2, 20 mM KCl, 1 mM DTT, 1 mM EDTA, and 1% protease inhibitor cocktail. While the nuclei were on ice, 0.5 nuclear volumes of high salt buffer containing 20 mM Tris (pH 7.5), 5 mM MgCl2, 1.2 M KCl, 1 mM DTT, 1 mM EDTA, and 1% protease inhibitor cocktail were added slowly to solubilize nuclear proteins. Triton-X100 (1%) was added to the suspension which was sonicated 4 times and centrifuged at 25,000 × g for 30 min at 4°C to pellet nuclear debris. The supernatant, which contained nuclear and nuclear membrane proteins, was stored at −80°C until use. The protein concentration of each cell fraction was determined by using the BioRad protein assay according to manufacturer’s instructions.
Protein labeling by iTRAQ
Proteins from each fraction were digested by using trypsin and labeled with iTRAQ reagents following the manufacturer’s protocol with some modification. Briefly, 100 μg protein from each fraction was precipitated by acetone at −20°C for 2 h. Each protein pellet was dissolved in 0.5 M triethylammonium bicarbonate buffer with 0.1% sodium dodecylsulfate and reduced in 5 mM tris(2-carboxyethyl)phosphine at 60°C for 1 h. The reduced protein was blocked in 10 mM methyl methanethiosulfonate by incubating at room temperature for 20 min and then digested at 37°C overnight by trypsin (Promega, Madison, WI) with shaking. iTRAQ reagent in ethanol was added to each sample (>60% ethanol in the reaction), and the reaction mixture was incubated at room temperature for 2 h. The reaction was stopped by adding an equal volume of water, and the experiment and control samples were mixed together for mass spectrometric analysis.
Two-dimensional microcapillary HPLC-tandem mass spectrometry (μLC-MS/MS)
A PolySulfoethyl A SCX column (5 μm, 200 Å, 4.6 × 100 mm) from PolyLC (Columbia, MD) was used to fractionate digested iTRAQ labeled peptides prior to reversed phase μLC-MS/MS. Mobile phase A consisted of 10 mM potassium phosphate (pH<3) and 25% acetonitrile, and mobile phase B consisted of 10 mM potassium phosphate (pH<3), 1 M KCl and 25% acetonitrile. Labeled peptides were diluted with 25% acetonitrile in water (pH<3) at least 10-fold to reduce the concentration of buffer and iTRAQ reagents, loaded onto the SCX column and eluted as follows: 100% mobile phase A for 5 min, 0% to 10% mobile phase B over 5 min, 10% to 25% mobile phase B over 25 min, 25% to 50% mobile phase B over 10 min, 50% B for 5 min, and then 100% mobile phase A for 20 min. Fractions were collected each minute and combined according to UV 280 nm absorbance. The fractions were evaporated to dryness under a stream of nitrogen and reconstituted in 4% acetonitrile in water containing 0.1% formic acid immediately prior to μLC-MS/MS analysis.
Labeled peptides were analyzed using a Thermo (San Jose, CA) LTQ linear ion trap mass spectrometer equipped with a Dionex (Auburn, CA) μHPLC system. Reverse phase μHPLC was carried out using an Agilent (Santa Clara, CA) Zorbax 300SB C18 column (3.5 μm, 75 μm × 150 mm) and Dionex/LC Packings C18 PepMap precolumn cartridge (5 μm, 0.3 mm × 5 mm). A linear gradient was used from 5% to 55% solvent B over 120 min (solvent A: 95:5:0.1; and solvent B: 5:95:0.1, water/acetonitrile/formic acid, v/v/v) at a flow rate of 250 nL/min. Positive ion nanoelectrospray mass spectra were acquired in data-dependent mode in which each MS scan (m/z 400 to m/z 2000) was followed by 4 MS/MS scans using hybridization of pulsed Q dissociation (PQD) with a normalized collision energy of 31%, activation Q value of 0.6, activation time of 0.4 ms, and collision-induced dissociation with collision energy of 35%, activation Q of 0.25, and activation time of 30 ms. The 4 most abundant peptide ions in each mass spectrum except singly charged ions were dynamically selected to generate tandem mass spectra. Dynamic exclusion was used to prevent repetitive selection of the same ions during the next 60 s.
The iTRAQ experiments were carried out with 3 independent biological replicates. Each of the 3 biological experiments was analyzed 3 times by μLC-MS/MS as technical replicates to gather reliable quantitative information. Collision-induced dissociation and PQD were used to obtain reliable quantitation and sequencing of peptides ions.
Western Blot
PrE cells were used on the second passage at 70% confluency. Lycopene or placebo beadlets were dissolved in cell culture media at a final concentration of 2 μM (or equivalent mass of placebo), and cell media were replaced with the test containing media for 48 h. Protein samples were collected in Cell Lysis Buffer (Cell Signaling Technology). After a 10 min centrifugation at 10,000 × g and 4°C, clarified protein lysate (10 μg) was run on a 10% bis-Tris NuPAGE gel (Life Technologies, Carlsbad, CA) and transferred to a PVDF membrane. Membranes were blocked in either 5% milk or bovine serum albumin depending on antibody manufacturer’s recommendations and incubated with antibodies at the following concentrations: HSP90B1 (Grp94) 1:1000; NDRG1 1:5000; GSTP1 1:5000, and Rad23b (Aka hHR23b) 1:500. Anti-mouse-HRP and anti-rabbit-HRP (Cell Signaling) were used at 1:5000 and the bands visualized with Western Lightning ECL Pro (PerkinElmer, Waltham, MA). Densitometry measurements of the band intensities were carried out based on mean peak height with ImageJ 1.46r (NIH).
Data analysis
The raw data from the MS/MS analyses were extracted automatically and converted to mzXML and MGF formats by using in house software. Two different search engines, MassMatrix (University of Illinois at Chicago, IL) (21, 22) and Mascot (Matrix Science, London, UK) were used to search against the International Protein Index (IPI.human.V3.5, 2008) with decoy option to improve confidence levels of protein quantitation and identification. The mass tolerance for precursor ions and fragment ions was set to 2 and 0.8 Da, respectively. Up to 2 missed trypsin cleavages were allowed. Variable modification was permitted to allow for the detection of methionine oxidation, and fixed modifications included β-methylthiolation of cysteine and the iTRAQ MS/MS tag of peptide N-terminals and lysine. Mascot searching was used to identify the protein hits, and the Mascot search results were further visualized and validated by Scaffold (Proteome Software, Portland, OR). MassMatrix was used to quantify the relative changes in protein expression and the individual protein expression ratio change was normalized by protein global expression ratio as described by Armenta et al. (23). For chemoprevention, a 10% change in protein expression was defined as significant.
Results and Discussion
Proteomics of human primary prostatic epithelial cells
A model of transit amplifying cells of the basal prostatic epithelium, PrE cells have a basal phenotype as evidenced by p63 and CK5 positivity and lack of an androgen receptor as determined using RT-qPCR and have a high rate of proliferation (17, 24). They express basal and transitional cell type markers (25) and cytokeratins 5 and 14, which were confirmed during proteomics measurements (Supplement Table 1). These basal cells are believed to be the cell origin for common prostatic adenocarcinomas, which has been confirmed by Goldstein et al. (18) using human primary prostate basal cells with lentivirus carrying genetic alteration genes in immunodeficient mice.
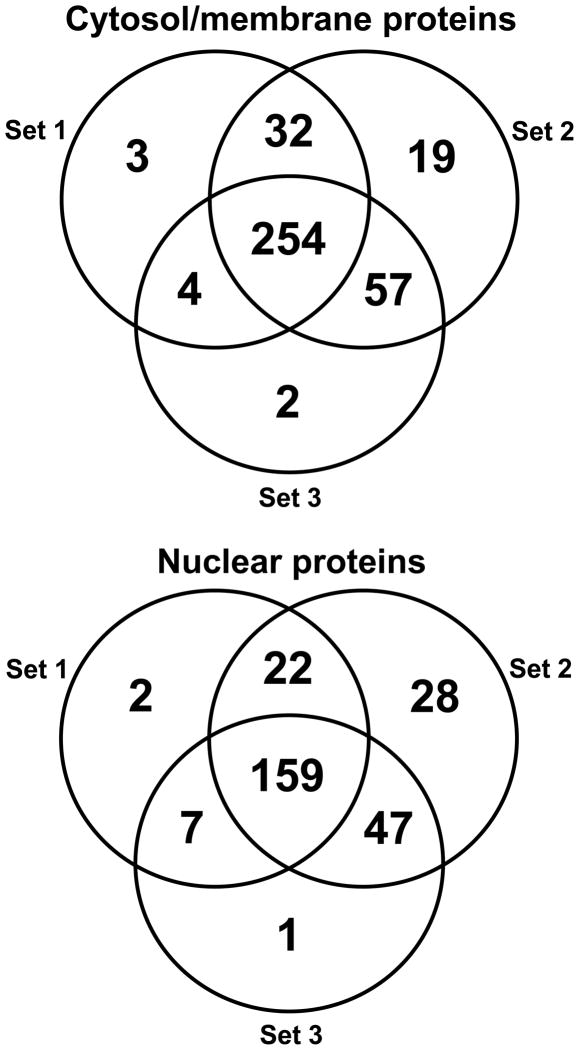
In the cytosolic and membrane fraction of the PrE cells, 371 proteins were identified including 254 proteins shared by all 3 experiments, 93 proteins identified in 2 experiments and 24 proteins identified in just a single experiment with a false discovery rate of 0.0% protein and peptides (Figure 1). All the proteins identified have a probability score of at least 95% (p-value ≤ 0.05) with at least 2 unique peptides, and the peptide probability score was ≥90%. Gene ontology analysis of biological processes of the identified proteins in the cytosolic and membrane fractions indicated that most of these proteins are involved in metabolism, development, response to stimulus, and regulatory processes; the remainder of the identified proteins are involved in immune, growth, multi-cellular, and multi-organism processes. Gene ontology analysis of molecular function indicated that most of the identified proteins have binding and catalytic activities, and that the other proteins have primarily antioxidant, transporter, translation regulator, enzyme regulator, carrier, and motor activities. Therefore, many proteins that were identified in the cytosolic and membrane fraction might be involved in mechanisms of action related to lycopene.
Figure 1.
Venn diagrams of proteins identified during 3 independent biological experiments in the cytosol/membrane fraction and in the nuclear fraction of human primary prostate epithelial (PrE) cells treated with lycopene.
In the nuclear extract fraction, 266 proteins were identified including 159 proteins by all 3 experiments, 76 proteins in 2 experiments and 31 proteins identified in just one experiment. The false discovery rate for protein and peptides was 0.8% and 0.1%, respectively. The gene ontology analysis of biological processes of the proteins identified in nuclear extracts indicated that most of the identified proteins were involved in metabolism, development, cell proliferation, cell growth, and regulatory processes. The other nuclear proteins that were identified were involved in communication, cell killing, immune response, and cell aging processes. Gene ontology analysis of molecular function of the nuclear proteinsindicated that most of the identified proteins have binding, catalytic and structural activities, followed by transporter, translation, transcription, and antioxidant functions.
Differential protein expression induced by lycopene
The cytosolic-membrane fraction and nuclear extract protein expression level changes showed an average change close to 1.0, and this distribution met normalization requirements based on protein global expression ratio (23). The proteins modulated by lycopene treatment compared with placebo were identified and are listed in Table 1 according to their cellular functions which included modulating cell redox homeostasis, apoptosis, cytoprotection, and androgen signaling. Note that these proteins showed the same expression change in at least 2 independent experiments.
Table 1.
Human primary prostate epithelial cellular proteins differentially expressed due to lycopene treatment.
| Accession Number | Protein | Unique peptides | Fold Change (p ≤ 0.05) | Function |
|---|---|---|---|---|
| Cytosol/membrane (non-nuclear) | ||||
| Apoptosis | ||||
| IPI00021263 | Protein kinase C inhibitor protein 1 | 7 | 0.85±0.13 | Anti-apoptosis process |
| IPI00304925 | Heat shock 70 kDa protein 1A/1B | 6 | 0.88±0.10 | Anti-apoptosis process |
| IPI00025512 | Heat shock protein beta-1 | 4 | 0.90±0.04 | Anti-apoptosis process |
| IPI00003815 | Rho GDP-dissociation inhibitor 1 | 3 | 0.89±0.09 | Anti-apoptosis process |
| IPI00550900 | Translationally-controlled tumor protein | 2 | 0.90±0.07 | Anti-apoptosis process |
| IPI00220766 | Lactoylglutathione lyase | 4 | 0.87±0.01 | Anti-apoptosis process |
| IPI00003362 | 78 kDa glucose-regulated protein | 14 | 0.84±0.09 | Anti-apoptosis process |
| IPI00007074 | Tyrosyl-tRNA synthetase | 3 | 1.20±0.09 | Induction of apoptosis |
| IPI00011253 | 40S ribosomal protein S3 | 6 | 1.15±0.07 | Activation of caspase, induction of apoptosis |
| IPI00479186 | Pyruvate kinase isozymes M2 | 19 | 1.15±0.07 | Nuclear translocation induction of cell death |
| IPI00010896 | Chloride intracellular channel protein 1 | 6 | 0.65±0.05 | Suppression of CLIC1 which induces apoptosis and inhibits tumor growth |
| Antioxidation and cytoprotection | ||||
| IPI00386755 | ERO1-like protein alpha | 10 | 0.80±0.06 | Responsible for a significant proportion of reactive oxygen species (ROS) |
| IPI00642936 | Glutathione S-transferase omega 1 | 4 | 1.11±0.05 | Functions in the GSH-ascorbate cycle as part of antioxidant metabolism |
| IPI00009634 | Sulfide:quinone oxidoreductase | 8 | 1.13±0.06 | Catalyzes oxidation of hydrogen sulfide with the help of a quinone |
| IPI00640741 | Peroxiredoxin-1 | 7 | 1.14±0.09 | Reduces hydrogen peroxide and alkyl hydroperoxides |
| IPI00219757 | Glutathione S-transferase P 1 | 10 | 1.17±0.04 | Conjugates GSH with electrophiles |
| IPI00000877 | Hypoxia up-regulated protein 1 | 4 | 1.56±0.19 | Cytoprotective mechanisms triggered by oxygen deprivation |
| IPI00008223 | UV excision repair protein RAD23 homolog B | 3 | 1.49±0.35 | Recognizes helix-distorting lesions in DNA and initiates global genome repair |
| Signal transduction | ||||
| IPI00298547 | Protein DJ1 | 4 | 0.77±0.13 | Positive regulator of androgen receptor-dependent transcription |
| IPI00027230 | Heat shock protein 90 kDa beta 1 | 9 | 0.89±0.08 | AR chaperon protein which stabilize AR in the cytosol |
| IPI00646689 | Thioredoxin domain-containing protein 17 | 3 | 0.87±0.07 | Deficiency enhances TNF-alpha induced apoptosis |
| IPI00293276 | Macrophage migration inhibitory factor | 2 | 0.76±0.04 | Proinflammatory cytokine, positive regulation of ERK1 and ERK2 cascade |
| IPI00022078 | Protein NDRG1 | 4 | 1.42±0.21 | Act as tumor suppressor in many cell types |
| IPI00411765 | Epithelial cell marker protein 1 | 7 | 0.88±0.08 | Regulates protein synthesis and epithelial cell growth by stimulating Akt/mTOR pathway |
| Nucleus | ||||
| IPI00021405 | Prelamin-A/C | 7 | 0.84±0.14 | Disrupts mitosis and induces DNA damage |
| IPI00411765 | Epithelial cell marker protein 1 | 7 | 0.84±0.08 | Regulates protein synthesis and epithelial cell growth by stimulating Akt/mTOR pathway |
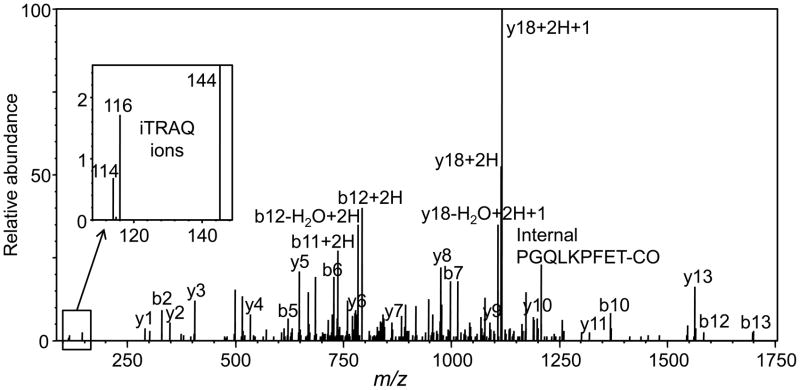
A representative positive ion electrospray PQD and CID tandem mass spectrum of a peptide from glutathione-S-transferase-pi-1 (GSTP1) is shown in Figure 2. The amino acid sequence of this peptide was determined based on the indicated b ions and y ions. Figure 2B shows the low mass region containing the iTRAQ ion of m/z 145 and the iTRAQ reporter ions of m/z 114 and m/z 116 from which the relative amounts of the peptides were determined.
Figure 2.
Positive ion nanoelectrospray PQD and collision-induced dissociation product ion tandem mass spectrum of peptide ALPGQLKPFETLLSQNQGGK from GSTP1 protein which was up-regulated in the cytosolic/membrane fraction. The triply protonated precursor ion was observed at m/z 853.831 corresponding to an uncharged mass of 2558.47 u (ΔM 2.2 ppm). Note the iTRAQ reporter ions of m/z 114, m/z 116 and m/z 144 that were used for the determination of relative protein expressionin PrEcells treated with lycopene or placebo beadlets.
Apoptosis
Lycopene treatment of the PrE cells up-regulated proteins associated with apoptosis induction and down-regulated proteins involved in anti-apoptosis processes (Table 1). The proteins associated with apoptosis induction that were up-regulated were tyrosyl-tRNA synthetase (TyrRS) (up-regulated by 20%), 40S ribosomal protein S3 (RPS3) (up by 15%), and pyruvate kinase isozyme M2 (PKM2) (up by 15%). Wakasugi et al. (26) reported that upregulation of TyrRS is associated with cellular apoptosis. Up-regulation of RPS3 can result in apoptosis through activation of caspase, while up-regulation of PKM2 is important for caspase-independent cell death of tumor cells.
The apoptosis proteins down-regulated by lycopene treatment (Table 1) included chloride intracellular channel protein 1 (CLIC1) (down-regulated by 35%), heat shock 70 kDa protein (HSP70) 1A/1B (down by 12%), heat shock protein beta 1 (HSP27) (down by 10%), Rho GDP-dissociation inhibitor 1 (Rho GDI 1) (down by 11%), translationally-controlled tumor protein (TCTP) (down by 10%), lactoylglutathione lyase (down by 13%), 78 kDa glucose-regulated protein (Grp78) (down by 16%), and protein kinase C inhibitor protein 1 (KCIP1) (down by 15%). Suppression of CLIC1 protein can induce apoptosis, enhance TNFα induced apoptosis and inhibit tumor growth as reported by Suh et al. (27). HSP70 directly inhibits apoptosis by blocking the recruitment of procaspase-9 to the Apaf-1/dATP/cytochrome c apoptosome complex (28). HSP27 protects cancer cells against apoptosis by interacting with the outer mitochondrial membranes and interfering with the activation of cytochrome/c/Apaf-1/dATP complex to inhibit the activation of procaspase-9 (29). Zhang et al. (30) reported that Rho GDI 1 protects cancer cells against drug-induced apoptosis. TCTP is stabilized by anti-apoptotic protein myeloid cell leukemia sequence (31), and its level is down-regulated through activation of the tumor suppressor protein p53 (32). Grp78 expression is associated with the development of castration-resistant prostate cancer (33). KCIP1 is known to bind to MEKK1, which is activated by caspase 3 to block apoptosis (34).
The apoptotic activity of lycopene is consistent with Ivanov et al. (35) and Hwang et al. (36) who reported apoptosis induction in prostate cancer cells by lycopene and Bowen et al. (37) and Kim et al. (38) who found that lycopene increased the apoptotic index in neoplastic and hyperplastic human prostate tissues. These results indicate that lycopene exerts pro-apoptotis effects in PrE cells by down-regulating expression of anti-apoptotic chaperone proteins, activating caspase activity and promoting the caspase independent cell death pathway. However, treatment of PrE cell cultures with lycopene for 48 h did not produce any observable apoptosis.
Cytoprotection and redox homeostasis
GSTP1, which catalyzes the conjugation of exogenous and endogenous electrophilic compounds with glutathione, was up-regulated 17% in PrE cells treated with lycopene (Table 1). Silencing of the gene for GSTP1 by promoter hypermethylation occurs frequently in prostate cancer and has been detected in proliferative inflammatory atrophy lesions of the prostate (39, 40). Glutathione-S-transferase omega 1 (GSTO1), which functions in the glutathione-ascorbate cycle to detoxify hydrogen peroxide, was up-regulated by 11%. Peroxiredoxin 1, an antioxidant enzyme that reduces hydrogen peroxide and alkyl hydroperoxides, was up-regulated by 14%, and sulfide-quinone oxidoreductase, which catalyzes the oxidation of hydrogen sulfide, was up-regulated by 13% in PrE cells treated with lycopene (Table 1). These effects indicate that another function of lycopene is to increase levels of phase II protective enzymes that can prevent cytotoxicity due to xenobiotic electrophiles and carcinogens.
These results are consistent with those of Ben-Dor et al. (10) who reported that lycopene induced the antioxidant response element in MCF-7 human mammary cancer cells and in HepG2 human hepatocellular carcinoma cells. Our results show that lycopene up-regulates phase II enzymes in PrE cells and not just in mammary or liver cells, and our data show a chemoprevention effect in primary prostate epithelial cells instead of cancer cells. Establishing up-regulation of phase II enzymes after lycopene exposure is significant in terms of confirming that this prostate cancer chemoprevention mechanism occurs in human PrE cells.
Hypoxia up-regulated protein 1, which participates in cytoprotective cellular mechanisms triggered by oxygen deprivation, was up-regulated by 56% in PrE cells treated with lycopene (Table 1). The UV excision repair protein RAD23 homolog B, which has a central role in DNA repair, was up-regulated by 49% (Table 1). Both of these effects indicate that lycopene stimulates cytoprotective functions in PrE cells that can help prevent prostate cancer initiation.
Lycopene treatment of PrE cells down-regulated ERO1-like protein alpha by 20% (Table 1). ERO1-like protein alpha is a cytosolic protein responsible for a significant proportion of reactive oxygen species (ROS) in cells. CLIC1, which is usually expressed during oxidative stress, was down-regulated by 35% (Table 1). These results show that lycopene can reduce ROS generation and reduce oxidative stress in PrE cells. These results are consistent with reports that lycopene reduces oxidative DNA damage in CV1-P monkey cell culture (41), in rat prostate tissue (42) and in human prostate tissue (43).
Signal transduction interference
Lycopene has been reported to alter signaling in the prostate including inflammation signaling, growth factor signaling, and steroid hormone signaling (44). Lycopene treatment of PrE cells down-regulated thioredoxin domain-containing protein 17 (TXNDC17) by 13% (Table 1). Deficiency of TXNDC17 enhances tumor necrosis factor alpha (TNFα) induced activation of caspase and subsequent apoptosis (45). Macrophage migration inhibitory factor, which is a proinflammatory cytokine and a positive regulator of the MAPK pathway, was down-regulated by 24% (Table 1). This effect can reduce inflammation, inhibit cell proliferation and slow prostate cancer progression to androgen-independent growth (4, 46).
Protein DJ1, a positive regulator of androgen receptor dependent transcription, was down-regulated by 23% in PrE cells (Table 1), indicating that lycopene exerts anti-androgen activity at the level of gene expression. HSP90, a chaperone protein that helps to stabilize the androgen receptor in the cytosol, was down-regulated by 11% (Table 1). Inhibitors of HSP90 cause androgen receptor degradation and are in clinical trials for prostate cancer therapy (47). These results are consistent with those of Herzog et al. (48) and Siler et al. (15) who showed that lycopene reduced androgen signaling and lowered expression of androgen receptor target genes including prostatic steroid binding protein C1 and C3, cystatin related protein 2 and seminal vesicle secretion protein IV. Down-regulation of androgen signaling by lycopene is an anti-cancer effect that should decrease androgen-dependent prostate growth.
Protein NDGR1, which is a tumor suppressor in many cell types and is necessary for the p53/TP53 mediated caspase activation and apoptosis, was up regulated by 42% in PrE cells treated with lycopene (Table 1). Epithelial cell marker protein 1 (SFN), which regulates protein synthesis and epithelial cell growth by stimulating the AKT/mTOR pathway, was down-regulated by 12% (Table 1). Estimated to be up-regulated 30–50% in most prostate cancers (2), the PI3K/AKT/mTOR pathway is a target for prostate cancer therapy. Reduced AKT signaling by lycopene might prevent the progression of prostate cancer to an androgen-independent state. These observations are consistent with lycopene growth inhibitory effects reported by other groups studying prostate cancer cells and PrE cells (35, 36, 49). Repression of the AKT/mTOR pathway by down-regulation of SFN would contribute to chemoprevention by lycopene by inhibiting prostate cancer progression to an androgen independent state.
Compared with cytosolic and membrane proteins, the expression of relatively few nuclear proteins in PrE cells was found to be altered by lycopene. Down-regulated by 12% in the cytosol, the expression of SFN in the nucleus was reduced 16% in response to lycopene treatment (Table 1). Finally, lycopene down-regulated prelamin-A/C 16% in the nucleus (Table 1). Prelamin-A/C disrupts mitosis and induces DNA damage.
To help confirm the proteomics results, identification and differential expression of 4 proteins in cell cultures established from 3 different patients were investigated using western blot analysis. The presence of RAD23B, NDRG1, GSTP1, and HSP90 B1 was confirmed in all 3 cell lines (Supplement Figure 1), and differential expression of each protein due to lycopene treatment was similar to the proteomics results. For example, HSP90 B1 was downregulated in 2 of 3 cell lines, and RAD23B, NDRG1 and GSTP1 were upregulated in 2 of 3 cell lines.
Conclusions
Prostate cancer initiation, promotion and progression involve multiple molecular pathways, many of which interact with each other. Investigations of changes in the levels of one or just a few proteins due to lycopene exposure cannot detect such pleiotropic effects. Our proteomics approach shows how lycopene exerts multiple protective effects on human PrE cells, while avoiding the use of less relevant models of chemoprevention such as prostate cancer cells or even non-prostate cell lines.
Several proteins that were up-regulated or down-regulated by lycopene indicate reduced oxidative stress in the cell. Up-regulation of phase II enzymes such as GSTP1, GSTO1 and SQR can help prevent cancer initiation by detoxifying potentially carcinogenic electrophiles. Lycopene was found to inhibit proliferation of PrE cells by down-regulating the AKT/mTOR pathway and by up-regulating genes that have growth inhibitory effects. Lycopene up-regulated proteins that can promote apoptosis and down-regulate several proteins involved in anti-apoptosis. Lycopene was also found to alter several signaling pathways, including inhibition of androgen signaling, down-regulation of TNFα signaling and deactivation of the MAPK pathway. All of these lycopene effects on cellular proteins contribute to the prevention of cancer initiation, promotion, and progression and are consistent with many previous studies that have addressed just one or a few of these effects of lycopene.
Supplementary Material
Acknowledgments
Grant Support
This research was supported by the National Cancer Institute (grant R01 CA101052; Richard B. van Breemen).
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.National Cancer Institute. [accessed 10 August 2012];Surveillance Epidemiology and End Results. http://seer.cancer.gov/statfacts/html/prost.html.
- 2.Morgan TM, Koreckij TD, Corey E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr Cancer Drug Targets. 2009;9:237–249. doi: 10.2174/156800909787580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Breemen RB, Sharifi R, Viana M, Pajkovic N, Zhu D, Yuan L, et al. Antioxidant effects of lycopene in African American men with prostate cancer or benign prostate hyperplasia: a randomized, controlled trial. Cancer Prev Res. 2011;4:711–718. doi: 10.1158/1940-6207.CAPR-10-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Mascio P, Kaiser S, Sies H. Lycopene as the most efficient biological carotenoid singlet oxygen quencher. Arch Biochem Biophys. 1989;274:532–538. doi: 10.1016/0003-9861(89)90467-0. [DOI] [PubMed] [Google Scholar]
- 5.Riso P, Visioli F, Grande S, Guarnieri S, Gardana C, Simonetti P, et al. Effect of a tomato-based drink on markers of inflammation, immunomodulation, and oxidative stress. J Agric Food Chem. 2006;54:2563–2566. doi: 10.1021/jf053033c. [DOI] [PubMed] [Google Scholar]
- 6.Haseen F, Cantwell MM, O’Sullivan JM, Murray LJ. Is there a benefit from lycopene supplementation in men with prostate cancer? A systematic review. Prostate Cancer Prostatic Dis. 2009;12:325–332. doi: 10.1038/pcan.2009.38. [DOI] [PubMed] [Google Scholar]
- 7.Hazai E, Bikadi Z, Zsila S, Lockwood SF. Molecular modeling of the non-covalent binding of the dietary tomato carotenoids lycopene and lycophyl, and selected oxidative metabolites with 5-lipoxygenase. Biorg Med Chem. 2006;14:6859–6867. doi: 10.1016/j.bmc.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 8.Karas M, Amir H, Fishman D, Danilenko M, Segal S, Nahum A, et al. Lycopene interferes with cell cycle progression and insulin-like growth factor I signaling in mammary cancer cells. Nutr Cancer. 2000;36:101–111. doi: 10.1207/S15327914NC3601_14. [DOI] [PubMed] [Google Scholar]
- 9.Chew BP, Park JS. Carotenoid action on the immune response. J Nutr. 2004;134:257S–261S. doi: 10.1093/jn/134.1.257S. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Dor A, Steiner M, Gheber L, Danilenko M, Dubi N, Linnewiel K, et al. Carotenoids activate the antioxidant response element transcription system. Mol Cancer Ther. 2005;4:177–186. [PubMed] [Google Scholar]
- 11.Levy J, Bosin E, Feldman B, Giata Y, Miinstera A, Danilenkoa M, et al. Lycopene is a more potent inhibitor of human cancer cell proliferation than either alpha-carotene or beta-carotene. Nutr Cancer. 1995;24:257–266. doi: 10.1080/01635589509514415. [DOI] [PubMed] [Google Scholar]
- 12.Rao AV, Fleshner N, Agarwal S. Serum and tissue lycopene and biomarkers of oxidation in prostate cancer patients: a case-control study. Nutr Cancer. 1999;33:159–164. doi: 10.1207/S15327914NC330207. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L-X, Cooney RV, Bertram JS. Carotenoids enhance gap junctional communication and inhibit lipid peroxidation in C3H/10T1/2 cells: relationship to their cancer chemopreventive action. Carcinogenesis. 1991;12:2109–2114. doi: 10.1093/carcin/12.11.2109. [DOI] [PubMed] [Google Scholar]
- 14.Palozza P, Parrone N, Simone R, Catalano A. Role of lycopene in the control of ROS-mediated cell growth: implications in cancer prevention. Curr Med Chem. 2011;18:1846–1860. doi: 10.2174/092986711795496845. [DOI] [PubMed] [Google Scholar]
- 15.Siler U, Barella L, Spitzer V, Schnorr J, Lein M, Goralczyk R, et al. Lycopene and vitamin E interfere with autocrine/paracrine loops in the Dunning prostate cancer model. FASEB J. 2004;18:1019–1021. doi: 10.1096/fj.03-1116fje. [DOI] [PubMed] [Google Scholar]
- 16.Goo YA, Li Z, Pajkovic N, Shaffer S, Taylor G, Chen J, et al. Systematic investigation of lycopene effects in LNCaP cells by use of novel large-scale proteomic analysis software. Proteomics Clin Appl. 2007;1:513–523. doi: 10.1002/prca.200600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uzgare AR, Xu Y, Isaacs JT. In vitro culturing and characteristics of transit amplifying epithelial cells from human prostate tissue. J Cell Biochem. 2004;91:196–205. doi: 10.1002/jcb.10764. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peehl DM. Growth of prostatic epithelial and stromal cells in vitro. In: Russell PJ, Jackson P, Kingsley EA, editors. Prostate Cancer Methods and Protocols. Totowa: Human Press; 2003. pp. 41–57. [DOI] [PubMed] [Google Scholar]
- 20.Liu A, Pajkovic N, Pang Y, Zhu D, Calamini B, Mesecar AL, van Breemen RB. Absorption and subcellular localization of lycopene in human prostate cancer cells. Mol Cancer Ther. 2006;5:2879–2885. doi: 10.1158/1535-7163.MCT-06-0373. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armenta JM, Hoeschele I, Lazar IM. Differential protein expression analysis using stable isotope labeling and PQD linear ion trap MS technology. J Am Soc Mass Spectrom. 2009;20:1287–1302. doi: 10.1016/j.jasms.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 24.Peehl DM. Are primary cultures realistic models of prostate cancer? J Cell Biochem. 2004;91:185–195. doi: 10.1002/jcb.10691. [DOI] [PubMed] [Google Scholar]
- 25.Peehl DM. Primary cell cultures as models of prostate cancer development. Endocr Relat Cancer. 2005;12:19–47. doi: 10.1677/erc.1.00795. [DOI] [PubMed] [Google Scholar]
- 26.Wakasugi K, Schimmel P. Highly differentiated motifs responsible for two cytokine activities of a split human tRNA synthetase. J Biol Chem. 1999;274:23155–23159. doi: 10.1074/jbc.274.33.23155. [DOI] [PubMed] [Google Scholar]
- 27.Suh KS, Mutoh M, Gerdes M, Crutchley JM, Mutoh T, Edwards LE, et al. Antisense suppression of the chloride intracellular channel family induces apoptosis, enhances tumor necrosis factor α-induced apoptosis, and inhibits tumor growth. Cancer Res. 2005;65:562–71. [PubMed] [Google Scholar]
- 28.Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, et al. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol. 2000;2:469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- 29.Sarto C, Binz PA, Mocarelli P. Heat shock proteins in human cancer. Electrophoresis. 2000;21:1218–1226. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1218::AID-ELPS1218>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Zhang Y, Dagher MC, Shacter E. Rho GDP dissociation inhibitor protects cancer cells against drug-induced apoptosis. Cancer Res. 2005;65:6054–6062. doi: 10.1158/0008-5472.CAN-05-0175. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D, Li F, Weidner D, Mnjoyan ZH, Fujise K. Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and fortilin. J Biol Chem. 2002;277:37430–37438. doi: 10.1074/jbc.M207413200. [DOI] [PubMed] [Google Scholar]
- 32.Tuynder M, Susini L, Prieur S, Besse S, Fiucci G, Amson R, et al. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Sci STKE. 2002;99:14976–14981. doi: 10.1073/pnas.222470799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pootrakul L, Datar RH, Shi SR, Cai J, Hawes D, Groshen SG, et al. Expression of stress response protein Grp78 is associated with the development of castration-resistant prostate cancer. Clin Cancer Res. 2006;12:5987–5993. doi: 10.1158/1078-0432.CCR-06-0133. [DOI] [PubMed] [Google Scholar]
- 34.Fanger GR, Widmann C, Porter AC, Sather S, Johnson GL, Vaillancourt RR. 14-3-3 proteins interact with specific MEK kinases. J Biol Chem. 1998;273:3476–3483. doi: 10.1074/jbc.273.6.3476. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov NI, Cowell SP, Brown P, Rennie PS, Guns ES, Cox ME. Lycopene differentially induces quiescence and apoptosis in androgen-responsive and -independent prostate cancer cell lines. Clin Nutr. 2007;26:252–263. doi: 10.1016/j.clnu.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Hwang ES, Bowen PE. Cell cycle arrest and induction of apoptosis by lycopene in LNCaP human prostate cancer cells. J Med Food. 2004;7:284–289. doi: 10.1089/jmf.2004.7.284. [DOI] [PubMed] [Google Scholar]
- 37.Bowen P, Chen L, Stacewicz-Sapuntzakis M, Duncan C, Sharifi R, Ghosh L, et al. Tomato sauce supplementation and prostate cancer: lycopene accumulation and modulation of biomarkers of carcinogenesis. Exp Biol Med (Maywood) 2002;227:886–893. doi: 10.1177/153537020222701008. [DOI] [PubMed] [Google Scholar]
- 38.Kim HS, Bowen P, Chen L, Duncan C, Ghosh L, Sharifi R, et al. Effects of tomato sauce consumption on apoptotic cell death in prostate benign hyperplasia and carcinoma. Nutr Cancer. 2003;47:40–47. doi: 10.1207/s15327914nc4701_5. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama M, Bennett CJ, Hicks JL, Epstein JI, Platz EA, Nelson WG, et al. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am J Pathol. 2003;163:923–933. doi: 10.1016/s0002-9440(10)63452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee WH, Isaacs WB, Bova GS, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol Biomarkers Prev. 1997;6:443–450. [PubMed] [Google Scholar]
- 41.Matos HR, Di Mascio P, Medeiros MH. Protective effect of lycopene on lipid peroxidation and oxidative DNA damage in cell culture. Arch Biochem Biophys. 2000;383:56–59. doi: 10.1006/abbi.2000.2035. [DOI] [PubMed] [Google Scholar]
- 42.Matos HR, Marques SA, Gomes OF, Silva AA, Heimann JC, Di Mascio P, et al. Lycopene and beta-carotene protect in vivo iron-induced oxidative stress damage in rat prostate. Braz J Med Biol Res. 2006;39:203–210. doi: 10.1590/s0100-879x2006000200006. [DOI] [PubMed] [Google Scholar]
- 43.Chen L, Stacewicz-Sapuntzakis M, Duncan C, Sharifi R, Ghosh L, van Breemen RB, et al. Oxidative DNA damage in prostate cancer patients consuming tomato sauce-based entrees as a whole-food intervention. J Natl Cancer Inst. 2001;93:1872–1879. doi: 10.1093/jnci/93.24.1872. [DOI] [PubMed] [Google Scholar]
- 44.Wertz K. Lycopene effects contributing to prostate health. Nutr Cancer. 2009;61:775–783. doi: 10.1080/01635580903285023. [DOI] [PubMed] [Google Scholar]
- 45.Jeong W, Chang TS, Boja ES, Fales HM, Rhee SG. Roles of TRP14, a thioredoxin-related protein in tumor necrosis factor-{alpha} signaling pathways. Sci STKE. 2004;279:3151–3159. doi: 10.1074/jbc.M307959200. [DOI] [PubMed] [Google Scholar]
- 46.Kim GY, Kim JH, Ahn SC, Lee HJ, Moon DO, Lee CM, et al. Lycopene suppresses the lipopolysaccharide-induced phenotypic and functional maturation of murine dendritic cells through inhibition of mitogen-activated protein kinases and nuclear factor-kappaB. Immunology. 2004;113:203–211. doi: 10.1111/j.1365-2567.2004.01945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taplin ME. Drug insight: role of the androgen receptor in the development and progression of prostate cancer. Nat Clin Pract Oncol. 2007;4:236–244. doi: 10.1038/ncponc0765. [DOI] [PubMed] [Google Scholar]
- 48.Herzog A, Siler U, Spitzer V, Seifert N, Denelavas A, Hunziker PB, et al. Lycopene reduced gene expression of steroid targets and inflammatory markers in normal rat prostate. FASEB J. 2005;19:272–274. doi: 10.1096/fj.04-1905fje. [DOI] [PubMed] [Google Scholar]
- 49.Obermuller-Jevic UC, Olano-Martin E, Corbacho AM, Eiserich JP, van der Vliet A, Valacchi G, et al. Lycopene inhibits the growth of normal human prostate epithelial cells in vitro. J Nutr. 2003;133:3356–3360. doi: 10.1093/jn/133.11.3356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.