Abstract
The high selectivity of protein farnesyltransferase was used to regioselectively append farnesyl analogues bearing bioorthogonal alkyne and azide functional groups to recombinant Schistosoma japonicum glutathione S-transferase (GSTase) and the active modified protein was covalently attached to glass surfaces. The cysteine residue in a C-terminal CVIA sequence appended to N-terminally His6-tagged glutathione S-transferase (His6-GSTase-CVIA) was posttranslationally modified by incubation of purified protein or cell-free homogenates from E. coli M15/pQE-His6-GSTase-CVIA with yeast protein farnesyltransferase (PFTase) and analogues of farnesyl diphosphate (FPP) containing ω-azide and alkyne moieties. The modified proteins were added to wells on silicone-matted glass slides whose surfaces were modified with PEG units containing complementary ω–alkyne and azide moieties and covalently attached to the surface by a Cu(I)-catalyzed Huisgen [3+2] cycloaddition. The wells were washed and assayed for GSTase activity by monitoring the increase in A340 upon addition of 1-chloro-2,4-dinitrobenzene (CDNB) and reduced glutathione (GT). GSTase activity was substantially higher in the wells spotted with alkyne (His6-GSTase-CVIA-PE) or azide (His6-GSTase-CVIA-AZ) modified glutathione-S-transferase than in control wells spotted with farnesyl-modified enzyme (His6-GSTase-CVIA-F).
INTRODUCTION
Since their inception, protein chips have proven to be useful in high throughput analysis for drug discovery, diagnosis, and in enhancing our understanding of key biological interactions.1–4 In contrast to non-selective approaches for anchoring proteins on surfaces in random orientations, the function of protein chips is enhanced when proteins are immobilized regioselectively. For example, the binding capacity of RNAse A for ribonuclease inhibitor proteins was four-fold higher when the protein was linked to a gold surface regioselectively through a cysteine residue at position 19, relative to protein immobilized in random orientations by amide linkages to surface lysines.5 Various approaches have been designed to achieve regioselectivity by selective modification of innate or engineered functional groups.6–12 Despite these developments, immobilization of enzymes that retain catalytic function remains a challenge.
Protein farnesyltransferase (PFTase) is a eukaryotic enzyme that catalyzes the attachment of a C15 isoprenoid (farnesyl) moiety to the cysteine residue in a C-terminal CaaX recognition motif, where C is cysteine, a is usually an amino acid with a small aliphatic side chain, and X is alanine, serine, phenylalanine, methionine, or glutamine.13 Although recognition of a CaaX sequence by PFTase can be context-dependent, protein prenylation is one of nature’s strategies for docking soluble proteins to membranes and the reaction is general for any soluble protein or peptide bearing this recognition sequence.14, 15 Proteins targeted to membranes by posttranslational prenylation is a prominent feature in cellular signal transduction networks.16
Protein prenylation strategies with natural and modified farnesyl or geranylgeranyl diphosphate substrates have proved to be highly valuable in studying various biological signaling events.17–22 We previously reported that PFTase accepts a broad range of farnesyl diphosphate (FPP) analogues as substrates, and we used yeast PFTase to chemo- and regioselectively append short hydrocarbon moieties containing bioorthogonal ω-terminal azide and alkyne groups to the cysteine residue.23,24 We and others have utilized this approach to site specifically modify proteins for various applications.21, 25–28 In our application, the modified proteins were immobilized site specifically to activated glass slides by either a Cu (I)-catalyzed Huisgen [3+2] cycloaddition or a Staudinger ligation (Scheme 1).24 The conditions for these immobilization steps are sufficiently mild to preserve the tertiary fold of green fluorescent protein (GFP) and the covalent attachments are robust enough that bound GFP can be detected with antibodies after treatment of the slides with detergent at high temperatures (~80 °C).
Scheme 1.
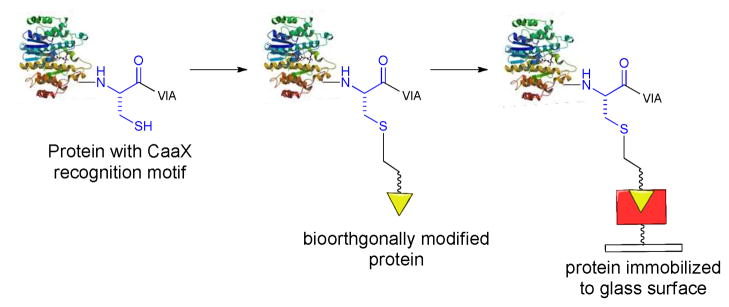
Protein immobilization by post-translational modification of cysteine of the CaaX motif with bioorthogonal functionality followed by ligation to glass surface.
Previously, prenylation of CaaX-containing GSTase was used as a test protein to compare the catalytic activity of yeast and mammalian farnesylytransferase29 and in our preliminary work to immobilize proteins on a glass surface.24 The enzyme was selected as a demanding model for this study because its activity depends on its tertiary fold and maintenance of its homodimeric quaternary structure.30 In addition, the catalytic site is located at the interface of the two monomeric units in GSTases, and we were especially interested to see if the enzyme was active after C-terminal bioorthogonal modification and subsequent covalent attachment to the surface.31 We now report the results of experiments with purified GSTase and enzyme from cell-free homogenates, which demonstrate that the regioselectively immobilized protein retains its catalytic activity.
EXPERIMENTAL PROCEDURES
Protein immobilization
A solution of His6-GSTase-CVIA-PE or His6-GSTase-CVIA-F (3 μL, 10 μM) was spotted into individual wells. Click reagent (7 μL of a solution prepared by mixing 40 μL of 100 mM CuSO4, 8.0 μL of 500 mM TCEP and 400 μL of 10 mM TBTA in 3.6 mL of 3:1 H2O:glycerol) was added to each well. The slide was kept humid at 4 °C in a cold room with gentle swirling for 6 h. Each well was washed with 10 μL of phosphate buffered saline containing Tween20® (PBST) followed by addition of 4 μL of Block IT™ solution and gentle shaking for 4 h at 4 °C. Each well was washed three times for 15 min with 10 μL of PBST before 4 μL of anti-GSTase Alexa Fluor (40 μg/mL in Tris-buffered saline TBS) was added. The slide was incubated overnight at 4 °C. The silicone mat was carefully removed; the slide was washed with several portions of PBST over 2h; and the slide was scanned with a phosphorimager. After scanning, the slide was immersed in stripping solution (125 mM glycine, 500 mM sodium chloride, 2.5% Tween20® pH=2.0) for 6 h at 80 °C. The slide was scanned again after rinsing with PBST. The slide was blocked, washed, incubated with anti-GST (40 μg/mL) overnight at 4 °C and imaged. Spots were quantified using ImageQuant® software.
General procedure for assay of immobilized GSTase
ArrayIT™ glass slides derivatized with L-AZ or L-PE were immersed in 2 mL of 3:1 H2O:glycerol for 1 h at 4 °C with constant swirling under N2. The slides were matted in a 96-well microarray format using a commercially available hardware sandwich and gasket system (ArrayIT, MMH4x24). A stock solution of His6-GSTase-CVIA-PE or His6-GSTase-CVIA-AZ (986 μM) was serially diluted with a buffer consisting of 100 mM sodium phosphate and 1 mM dithiothreitol (DTT), at pH 6.5 to give solutions containing 33, 67, 133, and 267 μM protein. Thirty μL portions of these samples were spotted into wells on the slide. A 70 μL portion of click reagent cocktail was added to each well and the slide was incubated at 4 °C for 4 h with gentle swirling. The wells were rinsed once with PBS (200 μL per well) and with PBST (200 μL per well) overnight. The wells were emptied and replenished with 100 μL of a solution containing 250 μM 1-chloro-2,4-dinitrobenzene (CDNB), 1 mM glutathione in 100 mM sodium phosphate buffer, pH 6.5. The plates were gently shaken at 21 °C for 30 min. A 75 μL portion was removed from each well and the absorbance at 340 nm was measured in order to calculate the activity of the immobilized enzyme.
RESULTS AND DISCUSSION
Cloning, expression and prenylation of recombinant glutathione S-transferase
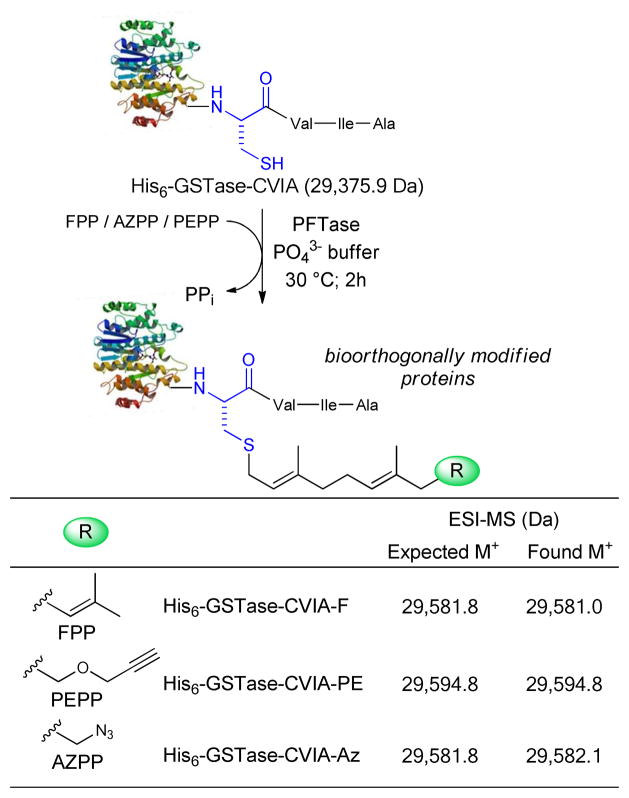
pQE -His6-GSTase-CVIA was constructed by inserting the gene for Schistosoma japonicum GSTase into pQE-30Xa to encode GSTase with an N-terminal histidine tag (His6) and a C-terminal CVIA PFTase CaaX recognition motif. IPTG induction of expression strain E. coli M15-pQE-His6-GSTase-CVIA gave recombinant His6-GSTase-CVIA. The enzyme was purified to near homogeneity by chromatography on a Ni2+-NTA matrix (see Figure S1 in Supporting Information for SDS PAGE analysis). The gel showed higher molecular weight bands for GSTase multimers that persisted even after dialysis under reducing conditions in the presence of 1 mM β-mercaptoethanol (β-ME) or 1 mM dithiothreitol (DTT). ESI-MS also showed a peak consistent with truncated GSTase that co-eluted with the full-length protein. Purified His6-GSTase-CVIA was farnesylated by incubation with FPP and yeast PFTase as shown in Scheme 2. The progress of the reaction was followed by ESI-MS by observing the disappearance of the ESI-MS signal at m/z 29,376 (unmodified His6-GSTase-CVIA) and the concomitant increase of a new signal at m/z 29,581. The 205 Da difference between the two signals confirmed the addition of a C15 farnesyl unit to His6-GSTase-CVIA. After 3 h at 30 °C, HPLC analysis indicated that the unmodified protein was consumed and farnesylated His6-GSTase-CVIA-F was the sole product.
Scheme 2.
Protein farnesyltransferase (PFTase) catalyzed prenylation of His6-GSTase-CVIA leading to site-specific modifications with FPP, AZPP and PEPP. Incubations were in 50 mM sodium phosphate, pH 7.0; containing 10 mM MgCl2; 10 mM ZnCl2, 5 mM β-ME with 35 mM FPP, AZPP or PEPP, 35 mM purified His6-GSTase-CVIA and 12 nM yeast PFTase. The mixture was incubated at 30 °C for 3 h.
His6-GSTase-CVIA was prenylated with the alkyne analogue PEPP or the azide analogue AZPP under identical incubation conditions. We previously found that these compounds are excellent substrates for PFTase with catalytic efficiencies (V/K) that are comparable to that of FPP, where (V/K)relFPP = 1.0, (V/K)relAZPP = 2.1, and (V/K)relPEPP = 0.71, and the rates for prenylation of the CaaX cysteine are similar, where kcatFPP = 1.31 s−1, kcatAZPP = 1.10 s−1, and kcatPEPP = 1.06 s−1.24 Thus, the alkyne and azide moieties can be placed in either the protein or the complementary linker.
Prenylation of His6-GSTase-CVIA modified with PEPP yielded a protein with an ESI-MS signal at m/z 29,595, confirming the attachment of the alkyne-containing prenyl chain. Similarly, prenylation with AZPP gave a protein with an ESI-MS signal at m/z 29,582 for attachment of the azide-containing prenyl chain. These values differ from the mass of unmodified His6-GSTase-CVIA by 219 and 206 Da respectively.
Bioorthogonal modifications of glass surfaces
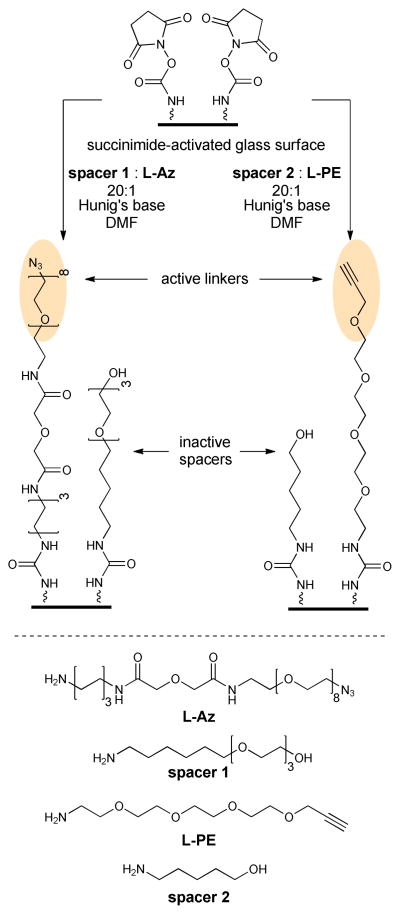
Glass slides were coated with azide (L-Az) or alkyne (L-PE) linkers and hydroxyl-terminated spacers (see Scheme 3).32, 33 Commercially available amine-terminated slides were activated by treatment with a mixtutre of disuccinimidyl carbonate and Hunig’s base in DMF for 1 h and rinsed with DMF.
Scheme 3.
Glass surface modification with bioorthogonal azide and alkyne functionalities.
The activated slides were treated with a 20:1 mixture of spacer and linker as illustrated in Scheme 3. The slides were shaken for 12 h with a mixture of L-Az and ω-aminohexylPEG-3 ether (spacer 1) or L-PE and 5-amino-1-pentanol (spacer 2) in DMF containing Hunig’s base in a sealed chamber. Ethanolamine was added to cap any remaining active carbamate sites. Contact angle measurements confirmed an increase in the hydrophobicity of the glass surface upon attachment of the linkers and spacers. Values for the contact angle increased from 0 ° for the free amine-coated slides to ~ 50 ° after treatment with L-Az or L-PE, confirming an increase in the hydrophobicity of the glass surface. The slides were stored in a mixture of water:glycerol at 4 °C until used.
Immobilization of purified His6-GSTase-CVIA-PE
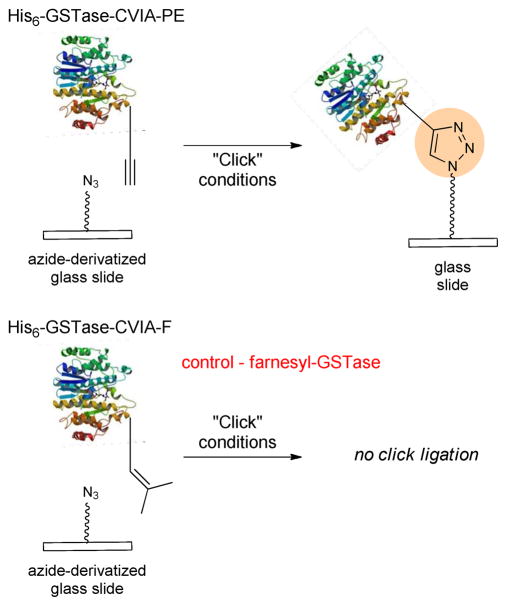
Azide-coated glass slides were matted with a 24-well microplate format. Increasing concentrations of His6-GSTase-CVIA-PE in pH 6.5 phosphate buffer, containing DTT were added to the wells, followed by a mixture of Cu(I) salt and TBTA (Figure 1). We compared the activity of covalently immobilized GSTase to a non-specifically bound control using farnesylated GSTase, which lacks the complementary azide or alkyne required for the Huisgen cycloaddition.
Figure 1.
Immobilization of His6-GSTase-CVIA-PE immobilization on an azide-coated glass surface using bioorthogonal Cu(I)-catalyzed Huisgen [3+2] cycloaddition
Modification, immobilization, and activity of purified His6-GSTase-CVIA
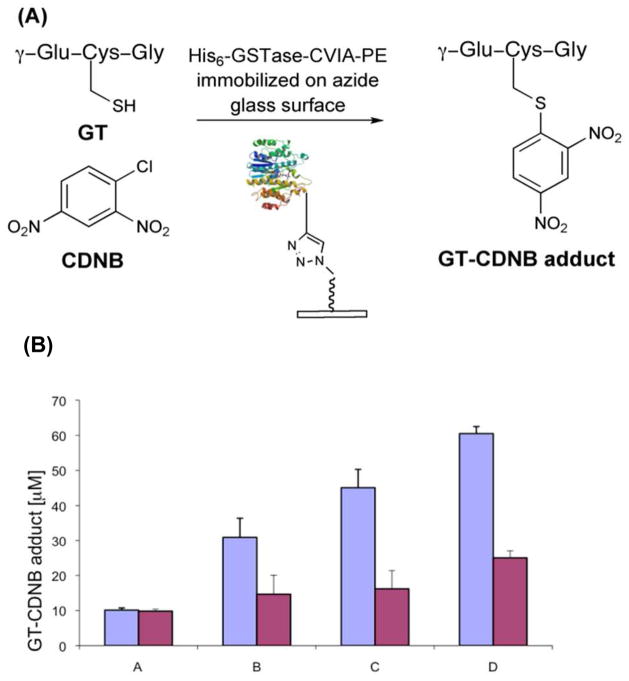
GSTase assay
The UV assay for GSTase activity is based on a 10-fold change in absorbance at 340 nm upon substitution of the chlorine in chloro-2,4-dinitrobenzene (CDNB) by glutathione.34, 35 The difference in the relative rates of enzymatic and non-enzymatic S-alkylation reactions was sufficiently large to permit the catalytic activity of immobilized GSTase to be evaluated (see SI). His6-GSTase-CVIA-F, His6-GSTase-CVIA-PE, or His6-GSTase-CVIA was added to a solution of CDNB and reduced glutathione (GT) in phosphate buffer, pH 6.5, at 21 °C. The rate of product formation was determined from the change in absorbance with time in the linear portion of the progress curve (See Figure S3) to give kcat = 2.1 min−1 for His6-GSTase-CVIA-PE (see Figure S3). His6-GSTase-CVIA-F and His6-GSTase-CVIA-PE retained ~66% of their catalytic activity after modification relative to the unmodified enzyme.
His6-GSTase-CVIA-PE was immobilized on azide-coated glass slides as shown in Figure 1. The activity of immobilized His6-GSTase-CVIA-PE was measured using saturating concentrations of GT and CDNB. Each assay was run in triplicate along with enzyme-free controls to determine the non-enzymatic background for the assay (Figure 2A). Results for wells containing His6-GSTase-CVIA-PE and His6-GSTase-CVIA-F are shown in Figure 2B. Activity, which increased with increasing amounts of enzyme, was detected in wells containing His6-GSTase-CVIA-PE and the His6-GSTase-CVIA-F control. Predictably, this activity increased with increasing concentration of His6-GSTase-CVIA-PE or His6-GSTase-CVIA-F. Similar levels of activity were observed for GSTase-PE and GSTase-F in those wells containing ≤ 5.0 picomole of enzyme/mm2 (Figure 2B, entry A). We attribute this observation to a basal level of non-specifically bound enzyme that was not removed by washing with PBST buffer. In wells containing higher concentrations of GSTase (B, C and D in Figure 2B), the activity of His6-GSTase-CVIA-PE was 2–3-fold higher than His6-GSTase-CVIA-F. Based on the estimated density of immobilized His6-GSTase-CVIA-PE (see below), kcat = 1.4 min−1 for the conjugation of glutathione with CDNB, as compared to kcat = 2.1 min−1 for unmodified GSTase in solution. Thus, qualitatively, the activity of immobilized His6-GSTase-CVIA-PE is similar to what is observed for GSTase in solution. The ~30% decrease in activity is within our ability to accurately estimate the amount of protein covalently bound to the slide. In subsequent experiments described below, we observed that the non-specific adsorption of His6-GSTase-CVIA-F was dramatically reduced when the protein was immobilized from cell-free homogenates.
Figure 2.
(A) Enzyme activity assay for immobilized His6-GSTase-CVIA-PE on azide-derivatized glass surface. (B) Plot of GSTase activity as a function of the concentration of enzyme immobilized. His6GSTase-CVIA-PE -
 ; and His6GSTase-CVIA-F -
; and His6GSTase-CVIA-F -
 . Estimated GSTase concentration on glass surface (pmole/mm2): A: 5.0; B: 10.0 ; C: 20; D: 40. Activity determined by measuring A340 nm in 100 mM NaH2PO4 buffer, pH = 6.5, containing 250 μM CDNB and 1 mM GT after incubation for 30 min at 21 °C.
. Estimated GSTase concentration on glass surface (pmole/mm2): A: 5.0; B: 10.0 ; C: 20; D: 40. Activity determined by measuring A340 nm in 100 mM NaH2PO4 buffer, pH = 6.5, containing 250 μM CDNB and 1 mM GT after incubation for 30 min at 21 °C.
While the precise amount of immobilized GSTase is not known, qualitatively, we observe that the protein is immobilized in an active conformation. An upper limit for the concentration of GSTase on the surface of the slide was estimated from the density of amino groups on the glass surface (8.3 × 10−12 moles of amino groups per mm2) and a spacer:linker ratio of 20:1 to give a surface concentration of 0.42 picomole/mm2 for immobilized GSTase monomer. The surface area for each well is 59 mm2, which corresponds to 250 picomole of immobilized GSTase in each well when the surface is saturated. The estimated maximal amounts of immobilized GSTase in our experiments vary from ~5 – 250 picomole/well.
Prenylation, immobilization and activity of His6-GSTase-CVIA-PE from cell-free homogenates
We also modified His6-GSTase-CVIA in clarified cell-free homogenates with PEPP using conditions identical to those described for the purified protein. ESI-MS gave signal at 29,594 Da, indicative or formation of His6-GSTase-CVIA-PE (See SI, Table 1). No signals were detected for unmodified His6-GSTase-CVIA. An ESI spectrum was also obtained for the modified protein after it was purified by Ni2+-NTA chromatography. Similar reactions with AZPP and FPP gave His6-GSTase-CVIA-Az and His6-GSTase-CVIA-F, respectively, as indicated by ESI-MS analysis.
Cell lysate containing His6-GST-CVIAase-PE or His6-GSTase-CVIA-F (control) were spotted in individual wells on an azide-derivatized glass surface, followed by addition of the Cu(I)-TBTA-TCEP to initiate cycloaddition. The wells were rinsed with PBST. Additional control samples included His6-GST-CVIAase-PE and His6-GSTase-CVIA-F in wells where the Cu(I)-catalyst was not added. Individual wells were assayed for GSTase activity and the results are shown in Figure 3.
Figure 3.
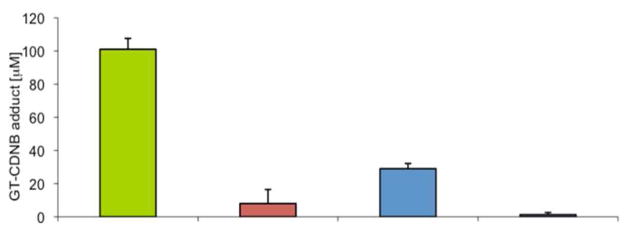
GSTase activity for covalently immobilized His6GSTase-CVIA-PE and His6GSTase-CVIA-F from cell-free homogenates of E. coli M15-pQE(His6-GSTase-CVIA). Activity was determined by measuring A340 in 100 mM NaH2PO4 buffer, pH = 6.5, containing 250 mM CDNB and 1 mM GT after incubation for 60 min. at 21 °C. His6GSTase-CVIA-PE with Cu(I) -
 ; His6GSTase-CVIA-F with Cu(I) -
; His6GSTase-CVIA-F with Cu(I) -
 ; His6GSTase-CVIA-PE -
; His6GSTase-CVIA-PE -
 ; His6GSTase-CVIA-F -
; His6GSTase-CVIA-F -
 .
.
The highest levels of activity were seen in wells where His6-GSTase-CVIA-PE was treated with Cu(I) (column 1). Residual activity was also seen in wells where His6-GSTase-CVIA-F was treated similarly, indicative of a basal level of non-specific, non-covalent adsorption. However, the activity for wells containing His6-GSTase-CVIA-PE in the absence of Cu(I) was significantly higher. This observation might reflect incomplete washing of the well before the assay or a basal level of uncatalyzed cycloaddition.
The difference in the relative amounts of GSTase activity in wells containing covalently immobilized protein and the controls for samples spotted from solutions of purified protein and cell free homogenates is noticeable. We surmise that proteins in the cell free homogenate compete favorable with GSTase for reactive sites on the glass surface responsible for the non-specific immobilization.
Detection of immobilized GSTase with fluorescent antibodies
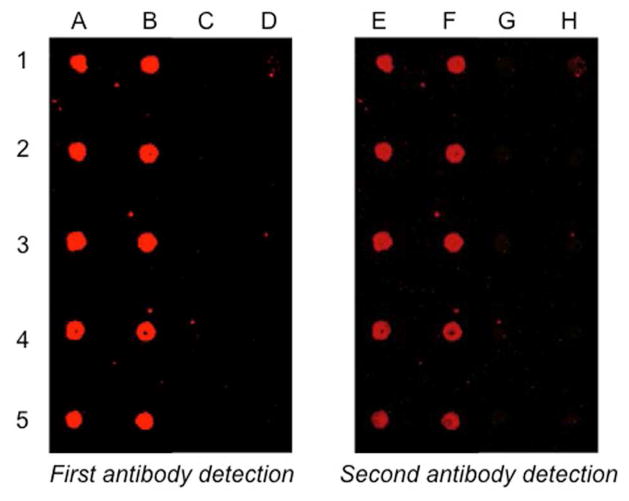
A slide with wells containing covalently immobilized His6-GSTase-CVIA-PE (+Cu(I)) and adsorbed His6-GSTase-CVIA-PE (−Cu(I)), both from cell free homogenates, was treated with a solution of AlexaFluor anti-GST, washed with PBST, and visualized by phosphorimaging. As seen in Figure 4, strong fluorescence was detected for wells containing His6-GSTase-CVIA-PE (columns A and B), while only very faint spots were detected for wells containing His6-GSTase-CVIA-F (columns C and D). All adsorbed proteins were then stripped from the slide by treatment with 25 mM glycine buffer, pH 2.0, containing 500 mM sodium chloride for 2h at 80 °C. Treatment with a second round of antibodies produced the same patterns of fluorescence. These results suggest that a low level of covalent modification occurs without Cu(I), which is substantially increased with addition of the catalyst.
Figure 4.
His6-GSTase-CVIA-PE immobilization from cell-free homogenate of E. coli M15-pQE(His6-GSTase-CVIA) strain without Cu(I). Wells loaded with 10 μM His6-GSTase-CVIA-PE, columns A and B. Wells loaded with 10 μM His6-GSTase-CVIA-F, columns C and D. Wells were treated with 40 μg/mL of AlexaFluor conjugated anti-GSTase antibody prior to imaging.
Immobilization and analysis of His6-GSTase-CVIA-AZ from a cell-free homogenate
We also immobilized GSTase regioselectively by reversing the locations of alkyne and azide moieties. Purified His6-GSTase-CVIA-AZ and enzyme in a cell-free homogenate was spotted on slides coated with L-PE. Figure 5 shows enzymatic activity assay after immobilization of His6-GSTase-CVIA-AZ. The activity in wells containing covalently immobilized His6-GSTase-CVIA-AZ was approximately 9-fold higher than in those containing adsorbed His6-GSTase-CVIA-F.
Figure 5.
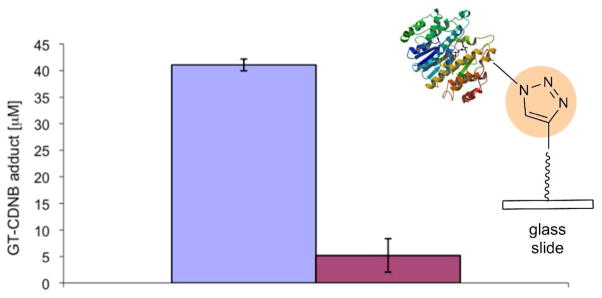
Schematic representation of azide-derivatized GSTase immobilized on alkyne slides. GSTase activity for immobilized His6GSTase-CVIA-Az and His6GSTase-CVIA-F from cell-free homogenate of E. coli M15-pQE(His6-GSTase-CVIA) strain is plotted as a function of product formation. Activity determined by measuring A340 in 100 mM NaH2PO4 buffer, pH = 6.5, containing 250 μM CDNB and 1 mM GT after incubation for 60 minutes at 21°C. His6GSTase-CVIA-Az -
 and His6GSTase-CVIA-F -
and His6GSTase-CVIA-F -

CONCLUSIONS
Bioorthogonal reactions are widely employed in a variety of applications in cellular imaging.36 Among these, the Huisgen [3+2] cycloaddition has proved to be particularly useful because molecules bearing azide or alkyne functional groups can be readily synthesized and are easy to handle, and the reaction is highly selective and efficient.37 The PFTase catalyzed posttranslational alkylation of the cysteine residue in a C-terminal CaaX recognition motif by analogues of FPP provides an opportunity for placing alkyne and azide moieties near the C-terminus of a soluble protein using procedures that are mild and regioselective. The modified proteins can be attached to imaging agents, other proteins, surfaces, etc. by the Huisgen [3+2] cycloaddition under biocompatible conditions that preserve the native fold of the protein.
Purified GSTase-CVIA-AZ or GSTase-CVIE-PE, or more conveniently the proteins from cell-free lysates, were covalently attached to glass slides through their respective isoprenoid appendages. The modification and immobilization reactions were sufficiently mild for the enzyme to retaining its catalytic activity and presumable its native tertiary and quaternary structure. The immobilization procedure should be applicable for immobilization of a wide variety of soluble proteins.
Supplementary Material
Acknowledgments
This work was supported by the NIH Grant GM 21328. The authors would like to thank Chad Nelson at the core Mass Spectral facility at the U. of Utah for analysis of protein samples by ESI-MS; Dr. Joel Harris laboratory for help with contact angle measurements; Dr. Robert Schackman for CaaX peptides used for optimizations of the prenylation reaction; and Dr. James Muller, director of the MS facility. We also thank Drs. Cecile Gauchet, Joel Walker, Steve Rothman for helpful discussions.
Footnotes
Supporting Information contains general experimental details in addition to specific synthesis and protein biochemical protocols followed in this study. General experimental information is provided in S3-S4. Details of the cloning and expression studies of His6-GSTase-CVIA are described in S5-S6. SDS-PAGE analysis of purified GSTase is described in S6. Prenylation studies of GSTase are described in S7-S8. Chemical synthesis of L-Az and L-PE and their spectral data (NMR, HRMS), followed by glass surface activation are provided in pages S9-S15. Protein immobilization protocol and assay procedures are provided in pages S17-S19 of Supporting Information. Specific activity calculations are provided in pages S19-S20 of Supporting Information. SDS-PAGE analysis of GSTase after prenylation from cell-free homogenates is described in S21-S22. Copies of ESI-MS data for bioorthogonally-prenylated GSTase and copies of 1H, 13C NMR data for small molecule linkers are provided in pages S23-S33 of Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 4.Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Curr Opin Chem Biol. 2009;13:398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luk YY, Tingey ML, Dickson KA, Raines RT, Abbott NL. Imaging the binding ability of proteins immobilized on surfaces with different orientations by using liquid crystals. J Am Chem Soc. 2004;126:9024–9032. doi: 10.1021/ja0398565. [DOI] [PubMed] [Google Scholar]
- 6.Soellner MB, Dickson KA, Nilsson BL, Raines RT. Site-specific protein immobilization by Staudinger ligation. J Am Chem Soc. 2003;125:11790–11791. doi: 10.1021/ja036712h. [DOI] [PubMed] [Google Scholar]
- 7.Duckworth BP, Xu JH, Taton TA, Guo A, Distefano MD. Site-specific, covalent attachment of proteins to a solid surface. Bioconjugate Chem. 2006;17:967–974. doi: 10.1021/bc060125e. [DOI] [PubMed] [Google Scholar]
- 8.Duckworth BP, Zhang ZY, Hosokawa A, Distefano MD. Selective labeling of proteins by using protein farnesyltransferase. Chem Bio Chem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 9.Wu P, Shui WQ, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc Natl Acad Sci U S A. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin J, Liu F, Li XH, Walsh CT. Labeling proteins with small molecules by site-specific posttranslational modification. J Am Chem Soc. 2004;126:7754–7755. doi: 10.1021/ja047749k. [DOI] [PubMed] [Google Scholar]
- 11.Foong YM, Fu J, Yao SQ, Uttamchandani M. Current advances in peptide and small molecule microarray technologies. Curr Opin Chem Biol. 2012:16. doi: 10.1016/j.cbpa.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Dirks AJ, Cornelissen J, van Delft FL, van Hest JCM, Nolte RJM, Rowan AE, Rutjes F. From (bio)molecules to biohybrid materials with the click chemistry approach. QSAR Comb Sci. 2007;26:1200–1210. [Google Scholar]
- 13.Lane KT, Beese LS. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res. 2006;47:681–699. doi: 10.1194/jlr.R600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Hougland JL, Lamphear CL, Scott SA, Gibbs RA, Fierke CA. Context-Dependent Substrate Recognition by Protein Farnesyltransferase. Biochemistry. 2009;48:1691–1701. doi: 10.1021/bi801710g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triola G, Waldmann H, Hedberg C. Chemical Biology of Lipidated Proteins. ACS Chem Biol. 2012;7:87–99. doi: 10.1021/cb200460u. [DOI] [PubMed] [Google Scholar]
- 16.Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci U S A. 2007;104:12294–12299. doi: 10.1073/pnas.0701817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelb MH, Brunsveld L, Hrycyna CA, Michaelis S, Tamanoi F, Van Voorhis WC, Waldmann H. Therapeutic intervention based on protein prenylation and associated modifications. Nat Chem Biol. 2006;2:518–528. doi: 10.1038/nchembio818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyro K, Manandhar SP, Mullen D, Schmidt WK, Distefano MD. Photoaffinity labeling of Ras converting enzyme 1 (Rce1p) using a benzophenone-containing peptide substrate. Bioorg Med Chem. 2010;18:5675–5684. doi: 10.1016/j.bmc.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan LN, Hart C, Guo L, Nyberg T, Davies BSJ, Fong LG, Young SG, Agnew BJ, Tamanoi F. A novel approach to tag and identify geranylgeranylated proteins. Electrophoresis. 2009;30 doi: 10.1002/elps.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen UTT, Guo Z, Delon C, Wu YW, Deraeve C, Fraenzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, Wolters D, Alexandrov K. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 22.Krzysiak AJ, Rawat DS, Scott SA, Pais JE, Handley M, Harrison ML, Fierke CA, Gibbs RA. Combinatorial modulation of protein prenylation. ACS Chem Biol. 2007;2:385–389. doi: 10.1021/cb700062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labadie GR, Viswanathan R, Poulter CD. Farnesyl diphosphate analogues with omega-bioorthogonal azide and alkyne functional groups for protein farnesyl transferase-catalyzed Ligation reactions. J Org Chem. 2007;72:9291–9297. doi: 10.1021/jo7017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauchet C, Labadie GR, Poulter CD. Regio- and chemoselective covalent immobilization of proteins through unnatural amino acids. J Am Chem Soc. 2006;128:9274–9275. doi: 10.1021/ja061131o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeGraw AJ, Palsuledesai C, Ochocki JD, Dozier JK, Lenevich S, Rashidian M, Distefano MD. Evaluation of Alkyne-Modified Isoprenoids as Chemical Reporters of Protein Prenylation. Chem Biol -Drug Des. 2010;76:460–471. doi: 10.1111/j.1747-0285.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinrich D, Lin PC, Jonkheijm P, Nguyen UTT, Schroder H, Niemeyer CM, Alexandrov K, Goody R, Waldmann H. Oriented Immobilization of Farnesylated Proteins by the Thiol-Ene Reaction. Angew Chem, Int Ed Engl. 2010;49:1252–1257. doi: 10.1002/anie.200906190. [DOI] [PubMed] [Google Scholar]
- 27.Jonkheijm P, Weinrich D, Schroder H, Niemeyer CM, Waldmann H. Chemical Strategies for Generating Protein Biochips. Angew Chem, Int Ed Engl. 2008;47:9618–9647. doi: 10.1002/anie.200801711. [DOI] [PubMed] [Google Scholar]
- 28.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng JK, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao YM. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez R, Goodman LE, Tripathy SK, Orourke E, Manne V, Tamanoi F. Purified yeast protein prenyltransferase is structurally and functionally similar to its mammalian counterpart. Biochemical J. 1993;289:25–31. doi: 10.1042/bj2890025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allardyce CS, McDonagh PD, Lian LY, Wolf CR, Roberts GCK. The role of tyrosine-9 and the C-terminal helix in the catalytic mechanism of Alpha-class glutathione S-transferases. Biochem J. 1999;343:525–531. [PMC free article] [PubMed] [Google Scholar]
- 31.Lim K, Ho JX, Keeling K, Gilliland GL, Ji XH, Ruker F, Carter DC. 3-Dimensional Structure Of Schistosoma-Japonicum Glutathione-S-Transferase Fused With A 6-Amino Acid Conserved Neutralizing Epitope Of GP41 From HIV. Protein Sci. 1994;3:2233–2244. doi: 10.1002/pro.5560031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun XL, Stabler CL, Cazalis CS, Chaikof EL. Carbohydrate and protein immobilization onto solid surfaces by sequential Diels-Alder and azide-alkyne cycloadditions. Bioconjugate Chem. 2006;17:52–57. doi: 10.1021/bc0502311. [DOI] [PubMed] [Google Scholar]
- 33.Bertozzi CR, Bednarski MD. The Synthesis of Heterobifunctional Linkers For The Conjugation Of Ligands To Molecular Probes. J Org Chem. 1991;56:4326–4329. [Google Scholar]
- 34.Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 35.Ji X, Armstrong RN, Gilliland GL. Snapshots Along the Reaction Coordinate of an SNAr Reaction Catalyzed by Glutathione Transferase. Biochemistry. 1993;32:12949–12954. doi: 10.1021/bi00211a001. [DOI] [PubMed] [Google Scholar]
- 36.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A Comparative Study of Bioorthogonal Reactions with Azides. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 37.Best MD. Click Chemistry and Bioorthogonal Reactions: Unprecedented Selectivity in the Labeling of Biological Molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.