Abstract
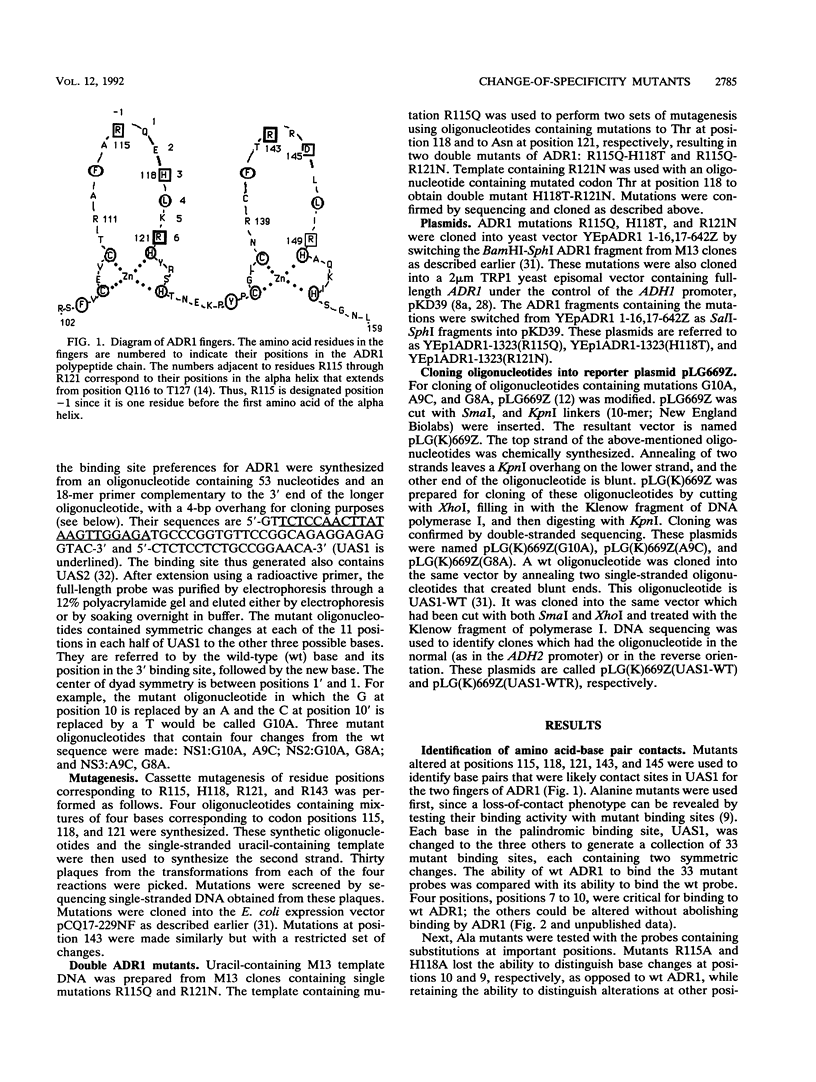
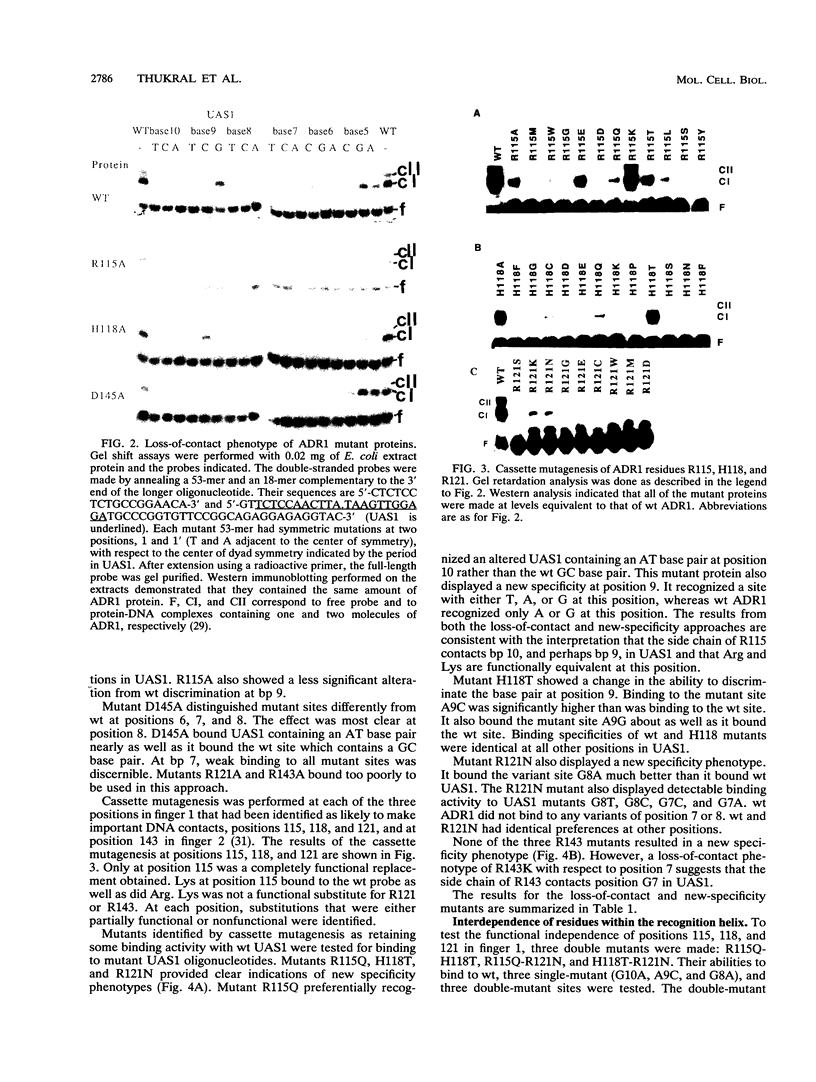
ADR1 is a yeast transcription factor that contains two zinc fingers of the Cys-2-His-2 (C2H2) class. Mutations that change the specificity of DNA binding of ADR1 to its target site, upstream activation sequence 1 (UAS1), have been identified at three positions in the first zinc finger. Mutations Arg-115 to Gln, His-118 to Thr, and Arg-121 to Asn led to new specificities of DNA binding at adjacent positions 10, 9, and 8 (3'-GAG-5') in UAS1. Arg-115 is at the finger tip, and His-118 and Arg-121 are at positions 3 and 6, respectively, in the alpha helix of finger 1. One double mutant displayed the binding specificity expected from the properties of its constituent new-specificity mutations. Mutations in the second finger that allowed its binding site to be identified through loss-of-contact phenotypes were made. These mutations imply a tail-to-tail orientation of the two ADR1 monomers on their adjacent binding sites. Finger 1 is aligned on UAS1 in an amino-to-carboxyl-terminal orientation along the guanine-rich strand in a 3'-to-5' direction. One of the ADR1 mutants was functional in vivo with both its cognate binding site and wild-type UAS1, but the other two mutants were defective in transactivation despite their ability to bind with high affinity to their cognate binding sites.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988 Jan;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Zinc finger domains: hypotheses and current knowledge. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- Blumberg H., Eisen A., Sledziewski A., Bader D., Young E. T. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. 1987 Jul 30-Aug 5Nature. 328(6129):443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- Blumberg H., Hartshorne T. A., Young E. T. Regulation of expression and activity of the yeast transcription factor ADR1. Mol Cell Biol. 1988 May;8(5):1868–1876. doi: 10.1128/mcb.8.5.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. S., Argos P. Fingers and helices. Nature. 1986 Nov 20;324(6094):215–215. doi: 10.1038/324215a0. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989 Feb 10;56(3):409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Gallo C. Constitutive RNA synthesis for the yeast activator ADR1 and identification of the ADR1-5c mutation: implications in posttranslational control of ADR1. Mol Cell Biol. 1986 Nov;6(11):4026–4030. doi: 10.1128/mcb.6.11.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H. Identification of amino acid-base pair contacts by genetic methods. Methods Enzymol. 1991;208:620–640. doi: 10.1016/0076-6879(91)08032-d. [DOI] [PubMed] [Google Scholar]
- Eisen A., Taylor W. E., Blumberg H., Young E. T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988 Oct;8(10):4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T. J., Postma J. P., Brown R. S., Argos P. A model for the tertiary structure of the 28 residue DNA-binding motif ('zinc finger') common to many eukaryotic transcriptional regulatory proteins. Protein Eng. 1988 Sep;2(3):209–218. doi: 10.1093/protein/2.3.209. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevit R. E., Herriott J. R., Horvath S. J. Solution structure of a zinc finger domain of yeast ADR1. Proteins. 1990;7(3):215–226. doi: 10.1002/prot.340070303. [DOI] [PubMed] [Google Scholar]
- Klevit R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991 Sep 20;253(5026):1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Párraga G., Horvath S. J., Eisen A., Taylor W. E., Hood L., Young E. T., Klevit R. E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988 Sep 16;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- Párraga G., Horvath S., Hood L., Young E. T., Klevit R. E. Spectroscopic studies of wild-type and mutant "zinc finger" peptides: determinants of domain folding and structure. Proc Natl Acad Sci U S A. 1990 Jan;87(1):137–141. doi: 10.1073/pnas.87.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J., Yu J., Cox D., Chan R. V., Smith M., Young E. ADR1-mediated regulation of ADH2 requires an inverted repeat sequence. Mol Cell Biol. 1986 Jun;6(6):1894–1902. doi: 10.1128/mcb.6.6.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Adam G., Rapatz W., Spevak W., Ruis H. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol. 1991 Feb;11(2):699–704. doi: 10.1128/mcb.11.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Taylor W. E., Young E. T. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4098–4102. doi: 10.1073/pnas.87.11.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Eisen A., Young E. T. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991 Mar;11(3):1566–1577. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: residues that contact DNA and that transactivate. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Tavianini M. A., Blumberg H., Young E. T. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol Cell Biol. 1989 Jun;9(6):2360–2369. doi: 10.1128/mcb.9.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Donoviel M. S., Young E. T. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jan;9(1):34–42. doi: 10.1128/mcb.9.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]






