Abstract
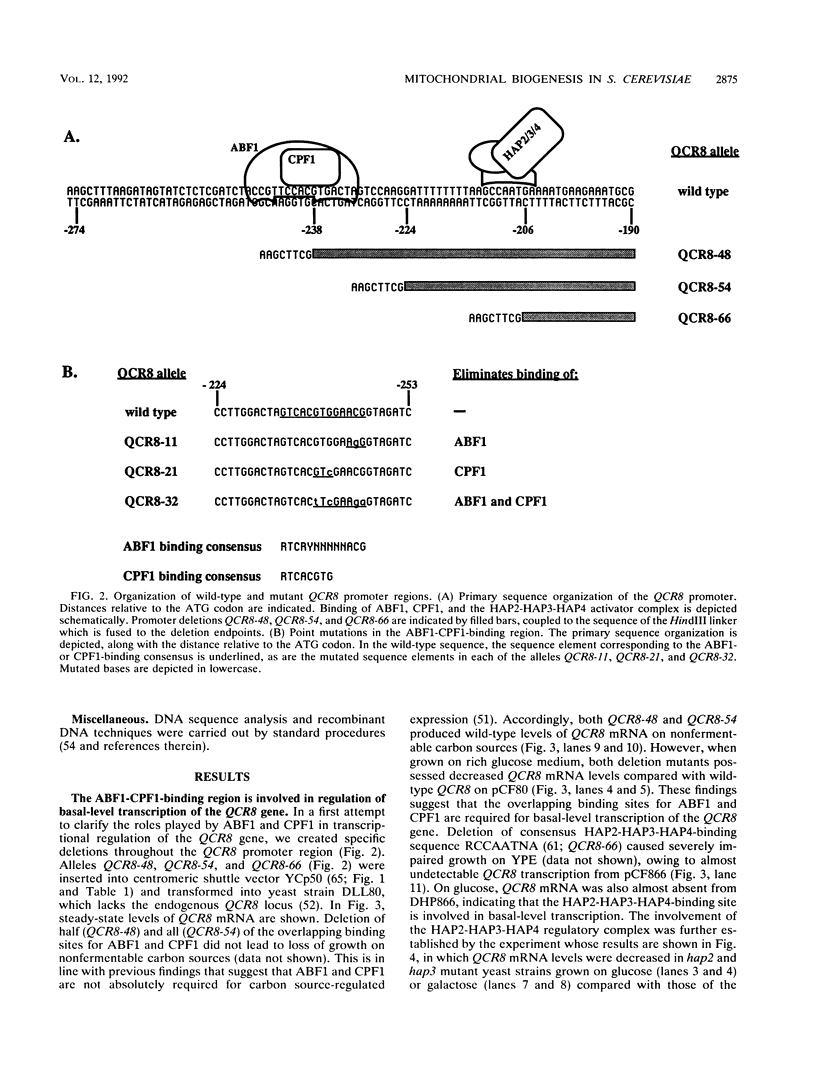
The multifunctional DNA-binding proteins ABF1 and CPF1 bind in a mutually exclusive manner to the promoter region of the QCR8 gene, which encodes 11-kDa subunit VIII of the Saccharomyces cerevisiae mitochondrial ubiquinol-cytochrome c oxidoreductase (QCR). We investigated the roles that the two factors play in transcriptional regulation of this gene. To this end, the overlapping binding sites for ABF1 and CPF1 were mutated and placed in the chromosomal context of the QCR8 promoter. The effects on transcription of the QCR8 gene were analyzed both under steady-state conditions and during nutritional shifts. We found that ABF1 is required for repressed and derepressed transcription levels and for efficient induction of transcription upon escape from catabolite repression, independently of DNA replication. CPF1 acts as a negative regulator, modulating the overall induction response. Alleviation of repression through CPF1 requires passage through the S phase. Implications of these findings for the roles played by ABF1 and CPF1 in global regulation of mitochondrial biogenesis are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Anderson M. S., Yarger J. G., Burck C. L., Poulter C. D. Farnesyl diphosphate synthetase. Molecular cloning, sequence, and expression of an essential gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Nov 15;264(32):19176–19184. [PubMed] [Google Scholar]
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Fitzgerald-Hayes M., O'Brien T. C. Purification of the yeast centromere binding protein CP1 and a mutational analysis of its binding site. J Biol Chem. 1989 Jun 25;264(18):10843–10850. [PubMed] [Google Scholar]
- Baker R. E., Masison D. C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990 Jun;10(6):2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. D., Ljungdahl P. O., Lopez J. L., Trumpower B. L. Isolation and characterization of the nuclear gene encoding the Rieske iron-sulfur protein (RIP1) from Saccharomyces cerevisiae. J Biol Chem. 1987 Jun 25;262(18):8901–8909. [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Kornberg R. D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990 Mar;10(3):887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M. J., Davis R. W. Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol Cell Biol. 1989 Jun;9(6):2544–2550. doi: 10.1128/mcb.9.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Chambers A., Tsang J. S., Stanway C., Kingsman A. J., Kingsman S. M. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol. 1989 Dec;9(12):5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Olesen J., Hahn S., Baldwin A. S., Guarente L., Sharp P. A. A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell. 1988 Apr 8;53(1):25–35. doi: 10.1016/0092-8674(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Creusot F., Verdière J., Gaisne M., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988 Nov 20;204(2):263–276. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- Crivellone M. D., Wu M. A., Tzagoloff A. Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J Biol Chem. 1988 Oct 5;263(28):14323–14333. [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989 Nov 24;246(4933):1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- Dorsman J. C., Doorenbosch M. M., Maurer C. T., de Winde J. H., Mager W. H., Planta R. J., Grivell L. A. An ARS/silencer binding factor also activates two ribosomal protein genes in yeast. Nucleic Acids Res. 1989 Jul 11;17(13):4917–4923. doi: 10.1093/nar/17.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., Gozdzicka-Jozefiak A., van Heeswijk W. C., Grivell L. A. Multi-functional DNA proteins in yeast: the factors GFI and GFII are identical to the ARS-binding factor ABFI and the centromere-binding factor CPF1 respectively. Yeast. 1991 May-Jun;7(4):401–412. doi: 10.1002/yea.320070410. [DOI] [PubMed] [Google Scholar]
- Dorsman J. C., Grivell L. A. Expression of the gene encoding subunit II of yeast QH2: cytochrome c oxidoreductase is regulated by multiple factors. Curr Genet. 1990 Jun;17(6):459–464. doi: 10.1007/BF00313072. [DOI] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Identification of two factors which bind to the upstream sequences of a number of nuclear genes coding for mitochondrial proteins and to genetic elements important for cell division in yeast. Nucleic Acids Res. 1988 Aug 11;16(15):7287–7301. doi: 10.1093/nar/16.15.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Yeast general transcription factor GFI: sequence requirements for binding to DNA and evolutionary conservation. Nucleic Acids Res. 1990 May 11;18(9):2769–2776. doi: 10.1093/nar/18.9.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1989;5:153–180. doi: 10.1146/annurev.cb.05.110189.001101. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Francesconi S. C., Eisenberg S. The multifunctional protein OBF1 is phosphorylated at serine and threonine residues in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4089–4093. doi: 10.1073/pnas.88.10.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A., Pearlman R. E. Transposable element-mediated enhancement of gene expression in Saccharomyces cerevisiae involves sequence-specific binding of a trans-acting factor. Mol Cell Biol. 1988 Jun;8(6):2572–2580. doi: 10.1128/mcb.8.6.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Halfter H., Kavety B., Vandekerckhove J., Kiefer F., Gallwitz D. Sequence, expression and mutational analysis of BAF1, a transcriptional activator and ARS1-binding protein of the yeast Saccharomyces cerevisiae. EMBO J. 1989 Dec 20;8(13):4265–4272. doi: 10.1002/j.1460-2075.1989.tb08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter H., Müller U., Winnacker E. L., Gallwitz D. Isolation and DNA-binding characteristics of a protein involved in transcription activation of two divergently transcribed, essential yeast genes. EMBO J. 1989 Oct;8(10):3029–3037. doi: 10.1002/j.1460-2075.1989.tb08453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe R. J., Harriss J. V., Sharp Z. D., Douglas M. G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983 Nov;25(2-3):333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
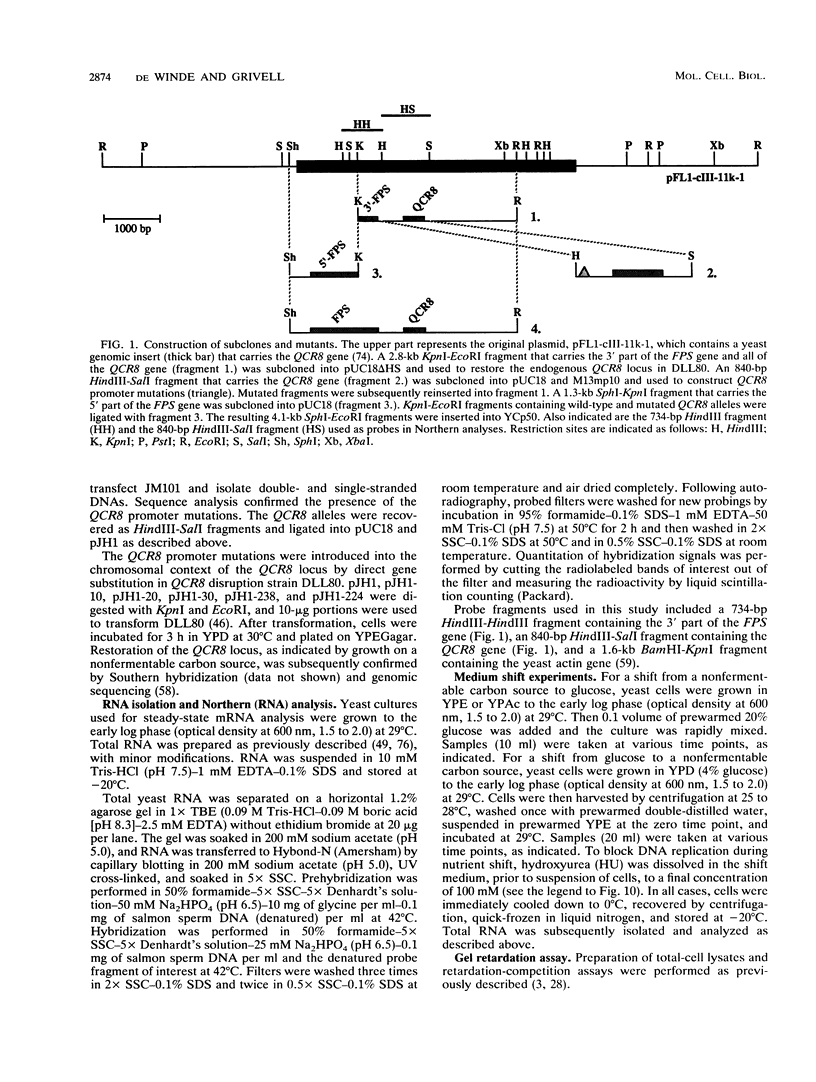
- Kwakman J. H., Konings D., Pel H. J., Grivell L. A. Structure-function relationships in a self-splicing group II intron: a large part of domain II of the mitochondrial intron aI5 is not essential for self-splicing. Nucleic Acids Res. 1989 Jun 12;17(11):4205–4216. doi: 10.1093/nar/17.11.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodi T., Guiard B. Complex transcriptional regulation of the Saccharomyces cerevisiae CYB2 gene encoding cytochrome b2: CYP1(HAP1) activator binds to the CYB2 upstream activation site UAS1-B2. Mol Cell Biol. 1991 Jul;11(7):3762–3772. doi: 10.1128/mcb.11.7.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
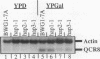
- Maarse A. C., De Haan M., Schoppink P. J., Berden J. A., Grivell L. A. Inactivation of the gene encoding the 11-kDa subunit VIII of the ubiquinol-cytochrome-c oxidoreductase in Saccharomyces cerevisiae. Eur J Biochem. 1988 Feb 15;172(1):179–184. doi: 10.1111/j.1432-1033.1988.tb13870.x. [DOI] [PubMed] [Google Scholar]
- Maarse A. C., Grivell L. A. Nucleotide sequence of the gene encoding the 11-kDa subunit of the ubiquinol-cytochrome-c oxidoreductase in Saccharomyces cerevisiae. Eur J Biochem. 1987 Jun 1;165(2):419–425. doi: 10.1111/j.1432-1033.1987.tb11455.x. [DOI] [PubMed] [Google Scholar]
- Maarse A. C., de Haan M., Bout A., Grivell L. A. Demarcation of a sequence involved in mediating catabolite repression of the gene for the 11 kDa subunit VIII of ubiquinol-cytochrome c oxidoreductase in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 11;16(13):5797–5811. doi: 10.1093/nar/16.13.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Jiang W., Funk M., Rathjen J., Barnes C. A., Hinz T., Hegemann J. H., Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990 Dec;9(12):4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Rathjen J., Jiang W., Barnes C. A., Dowell S. J. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991 Jun 11;19(11):2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C. M., Hinnebusch A. G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen J. T., Guarente L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 1990 Oct;4(10):1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode P. R., Sweder K. S., Oegema K. F., Campbell J. L. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 1989 Dec;3(12A):1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Slater M. L. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973 Jan;113(1):263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Sousa R., Arcangioli B. A point mutation in the CYC1 UAS1 creates a new combination of regulatory elements that activate transcription synergistically. EMBO J. 1989 Jun;8(6):1801–1808. doi: 10.1002/j.1460-2075.1989.tb03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trawick J. D., Rogness C., Poyton R. O. Identification of an upstream activation sequence and other cis-acting elements required for transcription of COX6 from Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5350–5358. doi: 10.1128/mcb.9.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Copy number control by a yeast centromere. Gene. 1983 Aug;23(2):221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- Van Loon A. P., Van Eijk E., Grivell L. A. Biosynthesis of the ubiquinol-cytochrome c reductase complex in yeast. Discoordinate synthesis of the 11-kd subunit in response to increased gene copy number. EMBO J. 1983;2(10):1765–1770. doi: 10.1002/j.1460-2075.1983.tb01655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C., Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975 Jun;122(3):855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S., Marres C. A. The mitochondrial respiratory chain of yeast. Structure and biosynthesis and the role in cellular metabolism. Biochim Biophys Acta. 1987;895(3):205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- van Loon A. P., de Groot R. J., van Eyk E., van der Horst G. T., Grivell L. A. Isolation and characterization of nuclear genes coding for subunits of the yeast ubiquinol-cytochrome c reductase complex. Gene. 1982 Dec;20(3):323–337. doi: 10.1016/0378-1119(82)90201-3. [DOI] [PubMed] [Google Scholar]