Abstract
To rationalize the disentangling action of type II topoisomerases, an improved wormlike DNA model was used to delineate the degree of unknotting and decatenating achievable by selective segment passage at specific juxtaposition geometries and to determine how these activities were affected by DNA circle size and solution ionic strength. We found that segment passage at hooked geometries can reduce knot populations as dramatically as seen in experiments. Selective segment passage also provided theoretical underpinning for an intriguing empirical scaling relation between unknotting and decatenating potentials.
Closed DNA circles can be unknotted, knotted, or linked (catenated). Such topological entanglements of DNA molecules have important impact on biological processes [1–3]. Unlike “knots” in proteins that are resolvable without breaking their linear chains [4], knots in DNA circles cannot be disentangled by continuous deformation [5]. In the living cell, DNA topology is regulated by enzymes called topoisomerases, which function by transporting either a single-stranded or a double-stranded DNA (dsDNA) segment through another. We focus here on the unknotting and decatenating actions of type-II topoisomerases (topo IIs), which have the ability to pass one dsDNA through another [6,7]. Topo II disentangling is often compared against a baseline referred to as “topological equilibrium” in which the distribution of topological states is the same as if the chain segments of DNA circles were free to pass through each other. Rybenkov et al. demonstrated that topo IIs can reduce knot and catenane populations to steady-state values that are tens of times less than those at topological equilibrium [8]. This observation implies that topo IIs do not pass dsDNA segments randomly but, instead, selectively act on entangled molecules. How does a topo II recognize the topology of a DNA molecule that is much larger than itself?
In this paper, we evaluate a promising answer [9] to this puzzle. Several hypotheses have been proposed to meet the conceptual challenge [8–12], almost all of them envision the topo II actively deforming or probing the DNA to ascertain its topology (reviewed in Ref. [3]), like Maxwell’s demon [13]. In contrast, the hooked juxtaposition hypothesis [9] relies on pre-existing DNA conformational statistics. It stipulates that a simple rule of allowing segment passage only when a DNA juxtaposition is hooked (when the two segments curve toward each other, see top inset in Fig. 1) should account for many capabilities of topo II.
FIG. 1.
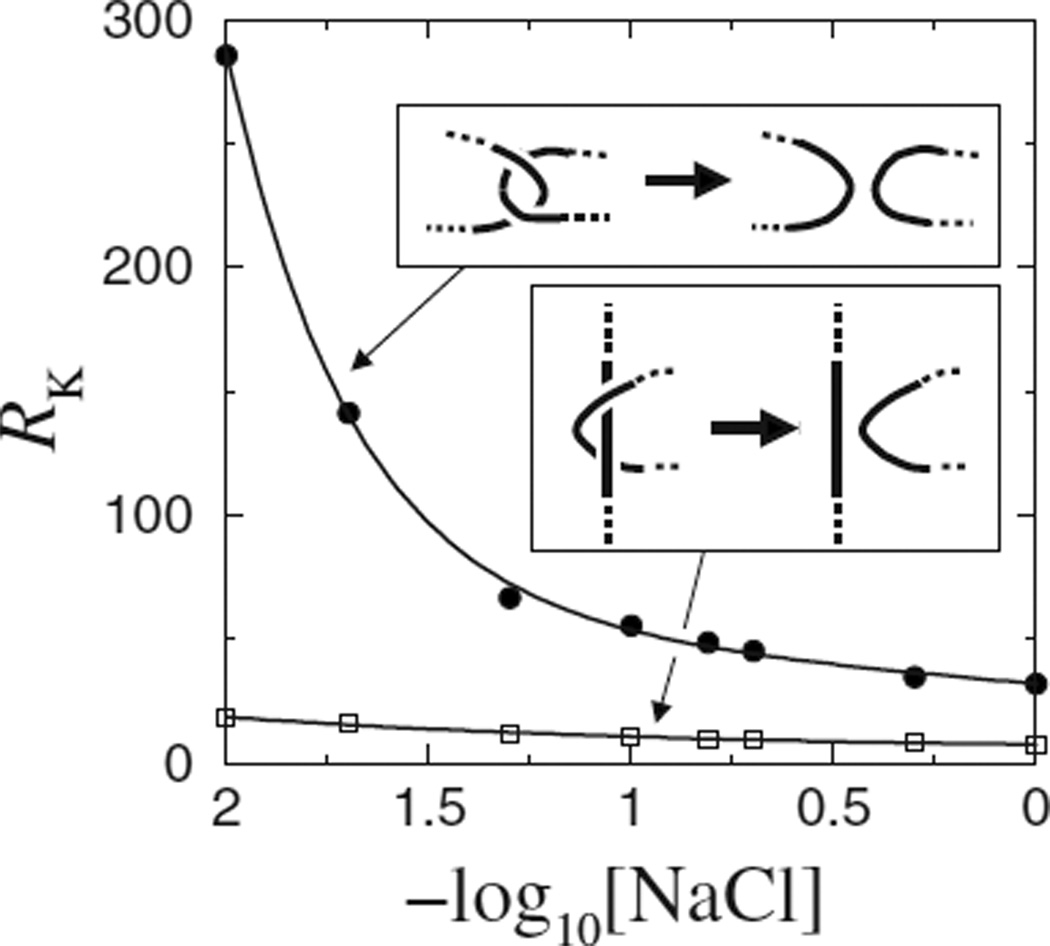
Modeled knot reduction factor (RK) for 3.5 kb DNA circles varies with [NaCl] for selective segment passages (insets) at the hooked (dots) or the half-hooked (squares) juxtaposition. Throughout this paper, each data point was simulated using 6 × 108–3 × 109 attempted chain moves; curves through data points are merely guides for the eyes.
The hooked juxtaposition hypothesis is amenable to direct computational evaluation using a “juxtaposition-centric” algorithm that enumerates or samples conformations subject to the constraint of any given preformed two-segment juxtaposition of interest. Applications of this method to self-avoiding walks on the simple-cubic lattice have shown that the hypothesis can, in principle, lead to very significant disentangling [14,15]. An independent study using freely jointed chains reached a similar conclusion [16]. However, the biophysical viability of the hypothesis remains to be demonstrated using geometrically more realistic DNA models [17,18]. To do so here, we begin by comparing the effects of segment passages at the four specific juxtaposition geometries in Table I. Besides the hooked, the other juxtapositions are of interest because (i) the straight juxtaposition is useful as a baseline, (ii) the effect of the free juxtaposition is expected to be essentially opposite to that of the hooked juxtaposition [9,14,15], and (iii) the effect of the half-hooked juxtaposition is essentially equivalent to that predicted by an active bending model of topo II action [11] (see discussion in Ref. [15]).
TABLE I.
Knotting and unknotting as a function of juxtaposition geometry. 𝒥(j)’s are fractions of segment-passage events via a given juxtaposition geometry j that resulted in various transitions among unknotted (U) and knotted (K) states, computed for 3.5 kb DNA circles under ionic strength [NaCl] = 0.154 M in our model. RK is the knot reduction factor.
| Juxtaposition (j) |
|
|
|
|
RK | |||||
| hooked | 0.858 | 0.0028 | 0.1370 | 0.0023 | 48.9 | |||||
| half-hooked | 0.914 | 0.0077 | 0.0761 | 0.0025 | 9.8 | |||||
| straight | 0.954 | 0.0240 | 0.0198 | 0.0021 | 0.82 | |||||
| free | 0.894 | 0.1014 | 0.0027 | 0.0022 | 0.027 | |||||
The lengths and shapes of our model juxtapositions were inspired by a recent x-ray crystal structure for a topo II–DNA complex showing that the interacting DNA segment has length ~10 nm and is bent into an ~150° arc [19]. Although this configuration might not be exactly that of DNA–topo II interaction in vivo or in solution, it is a good starting point for our analysis. Accordingly, the four juxtapositions in Table I were constructed as follows: each straight or curved segment has length l = 10 nm; each curved segment is a circular arc with subtending angle α = 150°; the midpoints of the two segments of a juxtaposition are separated by d =5 nm; and the two segments are perpendicular with crossing angle θ =90° (i.e., the scalar product of the segments’ midpoint tangents is zero). This set of parameters was used for Table I and Figs. 1 and 2 but later we investigate the consequences of varying these parameters.
FIG. 2.
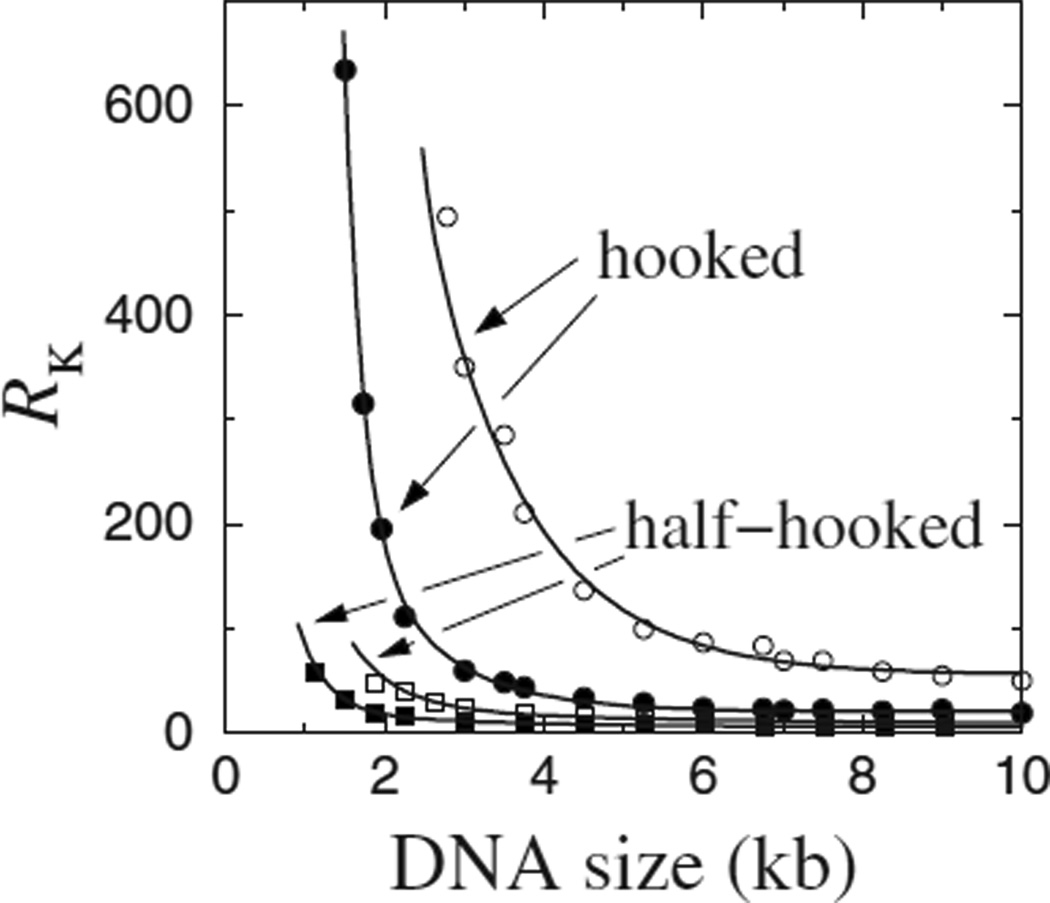
Knot reduction factor, RK, varies with DNA circle size. Model results are shown for selective segment passages at the hooked and half-hooked juxtaposition geometries for [NaCl]=0.01 (open symbols) and 0.154 (filled symbols).
Here we use a wormlike DNA model [17] with a potential that includes stretching, bending, torsion, excluded volume terms, and an electrostatic interaction that depends on salt concentration [20,21]. In parallel with our lattice studies [14,15], preformed juxtapositions were imposed as constraints in Monte Carlo simulations. Following Ref. [17], we used the generalized Madras-Orlitsky-Shepp (MOS) moves to sample conformations across different topologies [22] and the length-changing T± moves to sample conformations with the preformed juxtaposition situated at different locations of the model DNA circle. The topological state of each conformation was determined by computing its HOMFLY polynomial [23]. Mimicking topo II action, virtual segment passages [14,15], which change the crossing sign of the selected juxtaposition (see insets in Fig. 1), were performed on the conformations to ascertain the transition probabilities between various topological states.
In Table I, denotes the probability that a segment passage at juxtaposition j converts an unknot to a knot; 𝒥(j)’s for other transitions are similarly defined [15]. We measure unknotting power by a knot-reduction factor
| (1) |
where (PU) and (PK) are, respectively, the unknot and knot populations and the subscripts refer to topological equilibrium (eq) and the steady state (st) maintained by segment passage through the selected juxtaposition. Because knot populations are generally small [(PU)st, (PU)eq ≈ 1], RK ≈ (PK)eq/(PK)st. A simple master-equation derivation [15] showed that .
Table I provides results under ~ physiologically relevant solution conditions ([NaCl]=0.154 M) for circles with equilibrium knot probability (PK)eq < 0.4%. If the circle contains a hooked juxtaposition, the knot probability is much higher at , but an overwhelming majority of these knots are converted to unknots upon segment passage, resulting in ≈ 50 times reduction in knot population in the steady state . Remarkably, the topological information embodied in a small juxtaposition with two 10 nm segments is sufficient to effect such a significant knot reduction in relatively much larger DNA circles with chain length ≈ 1100 nm. Selective segment passages at the half-hooked juxtaposition also reduced knot populations, but much less effectively (RK=9.8). The effect of selective segment passage at the straight juxtaposition was approximately neutral (RK ≈ 1), as expected from its symmetric geometry. Finally, Table I shows that selective segment passages at the free juxtaposition promote rather than reduce knotting (RK ≪ 1). Because we are primarily interested in unknotting, we will focus mainly on the hooked and half-hooked juxtapositions below.
The effective diameter of DNA and its knotting probability are affected by salt [24]. Recent studies of topo II action using lattice [14,15] and freely jointed [16] chain models correspond, respectively, to solution conditions with low and high salts [17]. The present simulations allow changes in salt concentration to be modeled by varying the electrostatic term [17,20,21] to explore how salt may affect the outcome of topo II action. For the hooked juxtaposition, the knot reduction factor (Fig. 1) shows a surprising salt sensitivity: RK increases from ≈ 32 for [NaCl]=1 M to ≈ 150 and 280, respectively, for [NaCl]=0.02 and 0.01 M. In contrast, RK for the half-hooked juxtaposition is generally low, ranging from 7.5 to 19.0 with [NaCl] varying between 0.01 and 1.0 M (Fig. 1). This clear difference in predicted variation of RK with salt for the two juxtapositions may offer a test of the hooked juxtaposition hypothesis [9] and the active bending model [11] by future experiment.
Figure 2 shows knot reduction as a function of DNA circle size. For the hooked juxtaposition, RK increases sharply when the DNA circle size decreases below ~4 kb. When DNA circle size increases, RK first decreases then stabilizes for large circle sizes. This behavior, now obtained using a wormlike chain model with real DNA-like parameters, is consistent with lattice model results [15] and the original ansatz [9]. The robust trend supports the hooked juxtaposition hypothesis because it is in line with an experiment [8] showing that the RK achieved by a particular topo II for a 10 kb DNA is 50 whereas that for a 7 kb DNA is 90. In Fig. 2, RK≈ 22.0 for 10 kb DNA under physiological ionic strength ([NaCl]=0.154 M). This result is almost identical to that obtained using a 32-bead freely jointed chain model for the same DNA circle size [16]. It has the same order of magnitude as but is smaller than the experimental RK=50 in Rybenkov et al. [8]. At lower salt, RK is larger (≈ 57 for 10 kb DNA) and decays more slowly with increasing DNA circle size. In comparison, the RK values for the half-hooked juxtaposition in Fig. 2—as in the lattice model [15]—do not show much increase with decreasing DNA circle size.
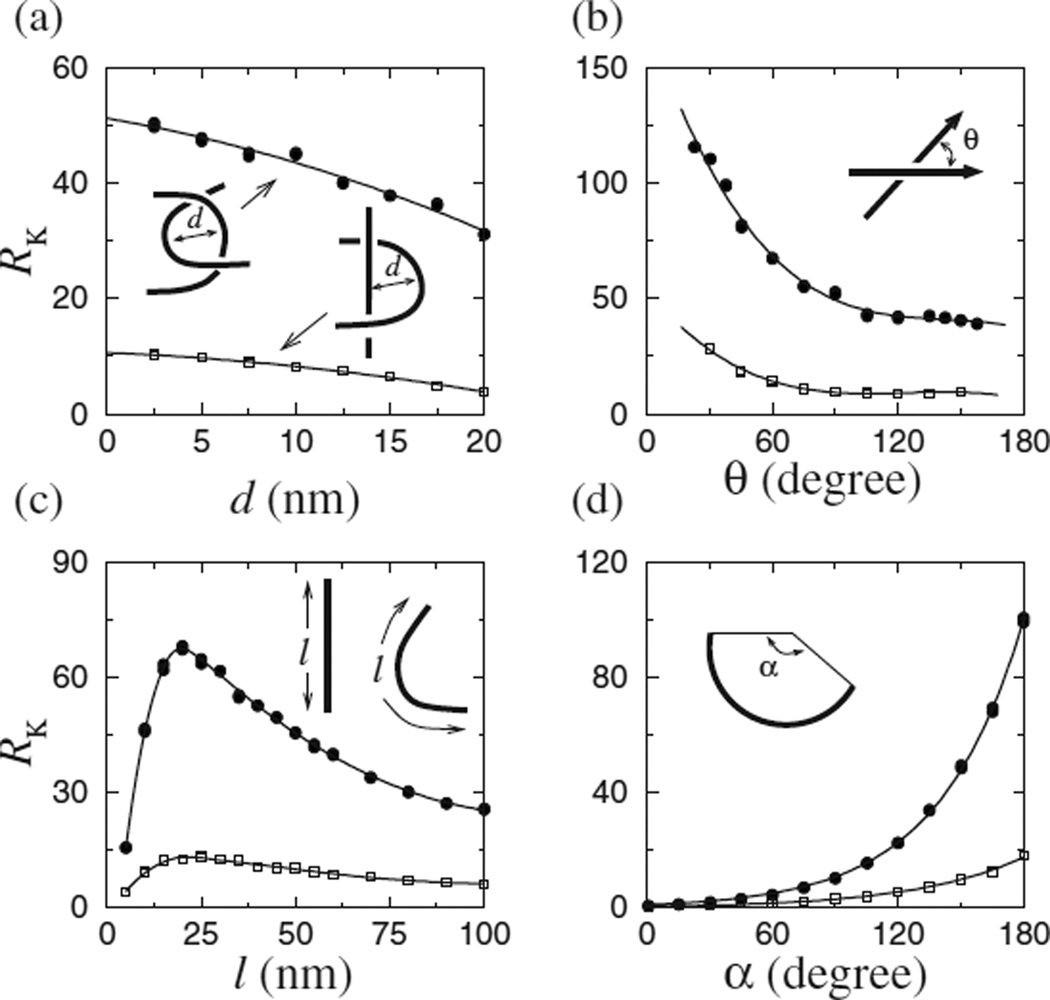
RK varied tremendously for the different juxtapositions with fixed geometries (Table I). We next varied the geometrical parameters in Fig. 3 to study a spectrum of juxtapositions based on the hooked and half-hooked geometries. RK is affected, to different degrees, by all four parameters. RK increases with decreasing segment-segment separation d, but only mildly. Even for the hooked juxtaposition that shows more variation, RK only increases from ≈ 32 to 50 for a decrease of d from 20 to 2.5 nm [Fig. 3(a)]. The crossing angle θ has a larger effect [Fig. 3(b)]. Because of the directionality of the segments (arrows), RK is not symmetric with respect to θ=90°, showing an increase with decreasing θ. Interestingly, the structural data suggest that topo II does not act on juxtapositions with a 90° angle and this may rationalize the enzyme’s apparent chirality-sensing ability [25]. This question deserves future effort.
FIG. 3.
Effects of juxtaposition geometry on knot reduction. RK for model 3.5 kb DNA circles, [NaCl]=0.154 M, was computed for variations of the hooked (dots) and half-hooked (squares) juxtapositions by changing the following parameters: (a) d is the midpoint separation between two equal-length segments of the juxtaposition, (b) θ is their crossing angle, (c) l is segment length, and (d) α is the arc angle subtended by the circular segment. Solid curves in (d) are single exponential fits. Except the parameter being varied in each panel, d=5 nm, α =150°, θ =90°, and l=10 nm.
Figure 3(c) shows that for the hooked juxtaposition, RK increases sharply with segment length for l≲20 nm, which corresponds to a physically plausible regime given the size of topo II. For larger l, RK decreases gradually. Among the geometric parameters examined, the arc angle α shows the most dramatic effect on RK. Figure 3(d) shows an exponential increase in RK with respect to α, which is consistent with the lattice results [15] showing a correlation between log RK with the hookedness parameter H defined in Ref. [14] because H∝α. Taken together, Fig. 3 demonstrates that even subtle changes in the geometry of the selected juxtaposition can lead to large variations in RK. These results may provide insight into the wide range of different RK’s between 5 and 90 that have been observed among topo IIs from different organisms [8].
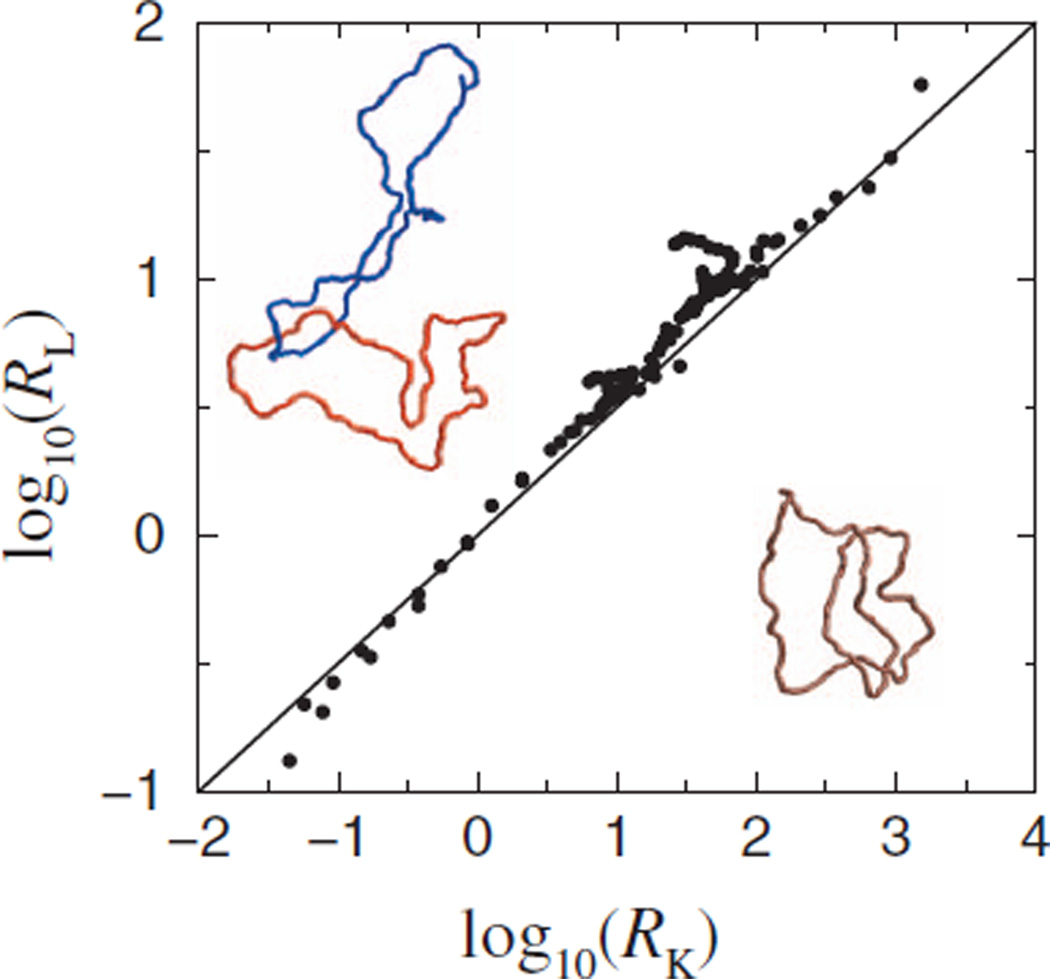
Although the disentangling powers of topo IIs from different organisms differ, experiments on six topo IIs suggest that their unknotting and decatenating potentials follow an approximate scaling law RK≈ (RL)1.6, where RL is the link (catenane) reduction factor [Fig. 3A of Ref. [8]]. Motivated by this finding, we examined (Fig. 4) the relationship between RK and RL in our model for the juxtaposition geometries studied in Figs. 1–3 plus others with α < 0 (e.g., the free juxtaposition) in which a circular segment curved away from the other segment. RL in our model is defined in the same way as RK in Eq. [1] except “U” now stands for unlinked and “K” (knotted) is replaced by “L” (linked); the corresponding 𝒥(j)’s were determined using two-circle juxtaposition-centric simulations [14]. The slope (=2) of the fitted line in Fig. 4 shows that our model data follow the power law RK=(RL)2 to a good approximation. The same scaling was deduced from our lattice model [15]. The robustness of this behavior is suggestive of deeper mathematics yet to be discovered. To date, our results offer the only physical rationalization for the empirical scaling, despite a small ≈ 0.4 difference in the exponents, the resolution of which will require more extensive experimental data and perhaps further theoretical analyses.
FIG. 4.
(Color) Scatter plot reveals an approximate scaling law between models RL and RK computed for selective segment passages via a variety of juxtaposition geometries and for circles of various sizes (see text). Each data point is for a given juxtaposition and a given circle size. Also shown are two catenated circles (top left) and a knotted circle (bottom right) as example conformations in the wormlike DNA chain model.
In summary, using an improved wormlike model [17], we have shown that selective segment passage at juxtapositions with a hooklike geometry can lead to significant knot and link reduction. Because the wormlike model is reasonably realistic for DNA, our results demonstrate that the hooked juxtaposition hypothesis is a biophysically viable mechanism to account for topo II action. Many of the present results are very similar to those predicted by lattice [14,15] as well as freely jointed [16] chain models, pointing to underlying general principles governing the relationship between local conformational preference and global topology. Building on these advances, much biology and much physics remain to be learnt from topo II-mediated changes in DNA topology.
Acknowledgments
Z.L. is supported by the Ministry of Science and Technology (Grant No. 2009CB918500) and the National Natural Science Foundation (Grant No. 20973016) of China. L.Z. is supported by the National Institutes of Health of the U.S.A (Grant No. RO1 AI054830). H.S.C. is supported by the Natural Sciences and Engineering Research Council of Canada (Grant No. 216901).
References
- 1.Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 2.Deibler RW, Mann JK, Sumners DWL, Zechiedrich L. BMC Mol. Biol. 2007;8:44. doi: 10.1186/1471-2199-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu ZR, Deibler RW, Chan HS, Zechiedrich L. Nucleic Acids Res. 2009;37:661. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virnau P, Mirny LA, Kardar M. PLOS Comput. Biol. 2006;2:e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wallin S, Zeldovich KB, Shakhnovich EI. J. Mol Biol. 2007;368:884. doi: 10.1016/j.jmb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sułkowska JI, Sułkowski P, Szymczak P, Cieplak M. Phys. Rev. Lett. 2008;100 doi: 10.1103/PhysRevLett.100.058106. 058106. [DOI] [PubMed] [Google Scholar]; Sułkowska JI, Sułkowski P, Onuchic J. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3119. doi: 10.1073/pnas.0811147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katritch V, Bednar J, Michoud D, Scharein RG, Dubochet J, Stasiak A. Nature (London) 1996;384:142. doi: 10.1038/384122a0. [DOI] [PubMed] [Google Scholar]; Flammini A, Maritan A, Stasiak A. Biophys. J. 2004;87:2968. doi: 10.1529/biophysj.104.045864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JC. Nat. Rev. Mol. Cell Biol. 2002;3:430. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Schoeffler AJ, Berger JM. Q. Rev. Biophys. 2008;41:41. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 8.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Science. 1997;277:690. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 9.Buck GR, Zechiedrich EL. J. Mol. Biol. 2004;340:933. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 10.Yan J, Magnasco MO, Marko JF. Nature (London) 1999;401:932. doi: 10.1038/44872. Phys. Rev. E 63, 031909 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3045. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trigueros S, Salceda J, Bermudez I, Fernández X, Roca J. J. Mol. Biol. 2004;335:723. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Pulleyblank DE. Science. 1997;277:648. doi: 10.1126/science.277.5326.648. [DOI] [PubMed] [Google Scholar]
- 14.Liu ZR, Zechiedrich EL, Chan HS. Biophys. J. 2006;90:2344. doi: 10.1529/biophysj.105.076778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu ZR, Mann JK, Zechiedrich EL, Chan HS. J. Mol. Biol. 2006;361:268. doi: 10.1016/j.jmb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Burnier Y, Weber C, Flammini A, Stasiak A. Nucleic Acids Res. 2007;35:5223. doi: 10.1093/nar/gkm532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZR, Chan HS. J. Chem. Phys. 2008;128:145104. doi: 10.1063/1.2899022. 131, 049902 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Micheletti C, Marenduzzo D, Orlandini E, Sumners DW. Biophys. J. 2008;95:3591. doi: 10.1529/biophysj.108.137653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong KC, Berger JM. Nature (London) 2007;450:1201. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 20.Stigter D, Dill KA. J. Phys. Chem. 1993;97:12995. [Google Scholar]
- 21.Vologodskii A, Cozzarelli N. Biopolymers. 1995;35:289. doi: 10.1002/bip.360350304. [DOI] [PubMed] [Google Scholar]
- 22.Our ensembles include circles with all supercoiling states with different linking numbers Lk, as in Ref. [17].
- 23.Freyd P, Yetter D, Hoste J, Lickorish WBR, Millett K, Ocneanu A. Bull. Am. Math. Soc. 1985;12:239. [Google Scholar]
- 24.Rybenkov VV, Cozzarelli NR, Vologodskii AV. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5307. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. J. Mol. Biol. 2005;351:545. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]