Abstract
The optimal management of breast cancer (BC) presents challenges due to the heterogeneous molecular classification of the disease. We performed survival analysis on a cohort of 466 patients with primary invasive ductal carcinoma (IDC), the most frequent type of BC, by integrating mRNA, microRNA (miRNA), and DNA methylation next-generation sequencing data from The Cancer Genome Atlas (TCGA). Expression data from eight other BC cohorts were used for validation. The prognostic value of the resulting miRNA/mRNA signature was compared with that of other prognostic BC signatures. Thirty mRNAs and seven miRNAs were associated with overall survival across different clinical and molecular subclasses of a 466-patient IDC cohort from TCGA. The prognostic RNAs included PIK3CA, one of the two most frequently mutated genes in IDC, and miRNAs such as hsa-miR-328, hsa-miR-484, and hsa-miR-874. The area under the curve of the receiver-operator characteristic for the IDC risk predictor in the TCGA cohort was 0.74 at 60 mo of overall survival (P < 0.001). Most relevant for clinical application, the integrated signature had the highest prognostic value in early stage I and II tumors (receiver-operator characteristic area under the curve = 0.77, P value < 0.001). The genes in the RNA risk predictor had an independent prognostic value compared with the clinical covariates, as shown by multivariate analysis. The integrated RNA signature was successfully validated on eight BC cohorts, comprising a total of 2,399 patients, and it had superior performance for risk stratification with respect to other RNA predictors, including the mRNAs used in MammaPrint and Oncotype DX assays.
Keywords: genomics, prognosis, ncRNA, mutation
Breast cancer (BC) can be influenced by a number of environmental factors and is characterized by molecular heterogeneity (1). Comprising about 80% of all breast cancers, invasive ductal carcinomas (IDC) are the most frequent type of BC. Breast tumors of distinct molecular subtypes [luminal A/B, HER2 enriched (HER2E), and basal-like] have dramatically different mRNA profiles (2). Recently, various groups, including The Cancer Genome Atlas (TCGA) network, analyzed and released data for a large number of primary breast cancers characterized by genomic DNA copy-number arrays, DNA methylation, exome sequencing, messenger RNA arrays, and microRNA sequencing (3–8). Somatic mutations in three genes (TP53, PIK3CA, and GATA3) occur frequently across BC, along with subtype-associated gene mutations (4, 8). The earlier description of the four main BC subtypes (9), characterized by different subsets of genetic and epigenetic abnormalities, suggested the hypothesis that much of the clinically observable plasticity and heterogeneity occurred within, and not across, these biological subclasses of breast cancer. In turn, this is thought to impact on the pathways related with outcome (10). Nonetheless, we wanted to investigate whether there are common underlying mechanisms related to overall survival (OS) in the different BC subclasses.
Although much is known about mRNA, microRNA (miRNA), and DNA methylation profiles in BC, no integrated study concerning their prognostic significance has yet been performed on large patient cohorts. The aim of this work was thus to assess the predictive value of such an integrated profile for OS of patients affected by IDC, the most frequent type of BC.
Results
Integrated Molecular Profile and Clinical Parameters in the TCGA IDC Cohort.
Integrated miRNA/mRNA tumor profiles (7,735 mRNAs and 247 miRNAs; integrated expression matrix in Dataset S1) were analyzed in depth for 466 primary IDCs in the TCGA cohort (8). hsa-miR-210, which had been previously associated with the transition from ductal carcinoma in situ to IDC, and with poor prognosis (11, 12), was the most up-regulated miRNA in primary tumors that had distant metastasis (P = 0.02). Before studying the prognostic values of RNA expression and DNA methylation, univariate survival tests were conducted to assess the relationship between clinical parameters and outcome in the TCGA IDC cohort. N stage, M stage, disease stage, T stage, and intrinsic subtype (SI Appendix, Figs. S1–S5) were significantly associated with OS. Estrogen receptor (ER)-positive patients showed a more favorable outcome and patients with triple-negative breast cancer (TNBC) a worse prognosis (SI Appendix, Figs. S6 and S7). Although somatic mutations in IDC were associated with specific intrinsic subtypes (TP53 with Basal-like and HER2-enriched and PIK3CA with Luminal A) as previously reported, they were not associated with OS (SI Appendix, Figs. S8 and S9). The results of this preliminary assessment indicated that the survival data for the TCGA IDC cohort, although containing a majority of censored data, were informative and appropriate for use in further molecular studies.
Association of OS with miRNA/mRNA/methylated DNA in the TCGA IDC Cohort.
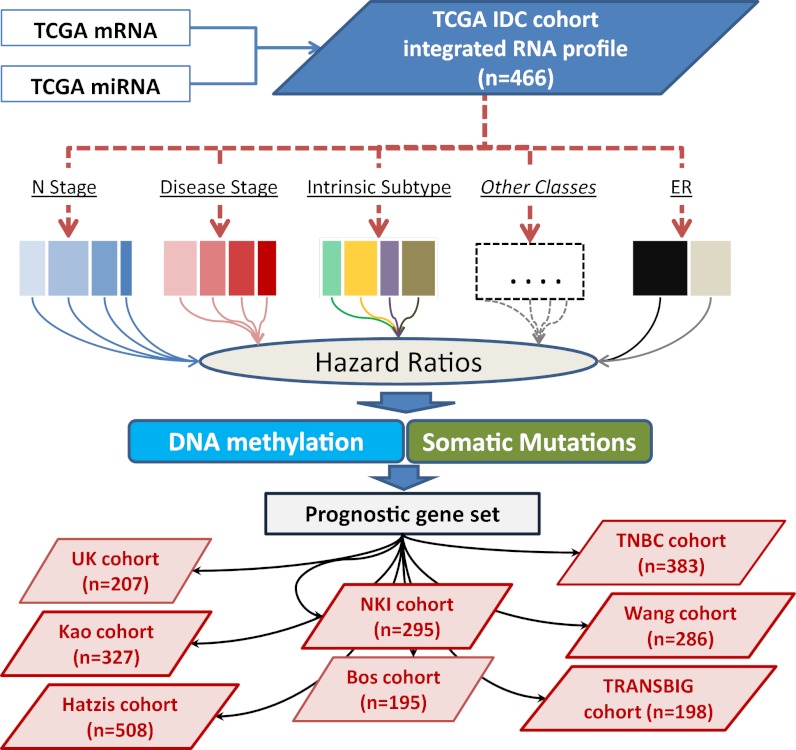
The association of OS with the miRNA, mRNA, and DNA methylation profiles (DNA methylation matrix in Dataset S2) was then studied in detail for the TCGA IDC cohort. The goal of this portion of the study was the identification of a set of common genes, if existing, consistently driving the outcome of the disease across the different clinical or molecular subtypes. The strategy and the underlying rationale are schematically shown in Fig. 1. We conducted univariate survival analyses for OS, using the integrated miRNA/mRNA profile within each of the following independent classes: disease stage, lymph node involvement (N stage), surgical margin, pre- or postmenopause, intrinsic subtype, and somatic mutations (TP53, PIK3CA pathway, GATA3, MAPKs, and remaining less frequently altered genes). The patient subclasses with different clinical or molecular characteristics represented disjoint sets within each class. An mRNA, or an miRNA, was selected only if significant in at least two independent subclasses from the same class (see legend to Fig. 1 for details). The list of genes passing this step, with the respective hazard ratios in each subclass, is shown in the Dataset S3. As an additional step to refine the risk gene set, we retained only mRNAs with known protein mutations in cancer (listed in Dataset S3 according to the Catalogue of Somatic Mutations in Cancer) (13). Because DNA methylation is a key mechanism in transcriptional control (14), we also studied the DNA methylation of coding genes as an additional criterion for association with OS. We first focused on the relation between CpG methylation and mRNA expression using the PIK3CA prognostic gene as a model for subsequent analysis of the candidate prognostic genes. The methylated CpG sites, which correlated with PIK3CA expression, were all located in a 2.2-kb region surrounding PIK3CA’s first exon (SI Appendix, Table S2), a region with strong acetylation of lysine 27 in histone H3 and high-density binding of transcription factors (15). The majority (five of six) of the significant CpG sites in this region had the expected negative correlation between DNA methylation and PIK3CA expression. Based on this finding, we used a majority rule to determine the type of association between a gene’s methylation and OS in the whole TCGA cohort at once. When most of the significant methylation sites for a gene (SI Appendix, Table S3) had hazard ratio (HR) lower than 1, than the correlation between the gene’s methylation and outcome was defined as “negative.” This procedure allowed us to identify the genes that had an association of poor outcome with RNA overexpression and DNA hypo-methylation or vice versa. The DNA methylation test was applied to the coding genes and not to miRNAs because of the limited number of CpG sites assayed in those very small genes. Nevertheless, most miRNAs would have passed the methylation test (Dataset S2).
Fig. 1.
Strategy used to derive and validate prognostic mRNAs and miRNAs in breast cancer. mRNAs and miRNAs were integrated in a single 7982-RNA profile (TCGA IDC cohort, n = 466). Survival analysis was performed within the various subgroups of the following clinical and molecular classes: disease stage, lymph node involvement (N stage), surgical margin, pre- or postmenopause, intrinsic subtype, somatic mutations (TP53, PIK3CA pathway, TP53/PIK3CA double mutants, GATA3, and the remaining less frequently altered genes). The subclasses within a class represented disjoint patient sets, thus enabling immediate validation of the prognostic RNAs within that class. The HRs and Kaplan–Meier curve were generated for every RNA in all independent subclass. RNAs that had significant both HRs and log-rank tests (P < 0.05) in at least two subclasses (within the same clinical or molecular class) were initially selected. Additional criteria, required for the selection of coding genes, were the association of DNA methylation with OS and the presence of somatic mutations in the COSMIC database (www.sanger.ac.uk/genetics/CGP/cosmic/). The association between DNA methylation and OS was carried out on the whole cohort (not on each subclass) using univariate Cox regression (SI Appendix, Tables S2 and S3). The HR was the ratio of hazards for a twofold change in the DNA methylation level. A majority-rule voting procedure was applied to all significant HRs for the CpG sites in the prognostic genes (false discovery rate < 0.001); e.g., the DNA methylation of a gene with the most significant CpG HRs lower than 1 would be defined as negatively correlated to outcome or vice versa. A further step for gene reassessment was then performed in the BC tumor subtype, as detailed in Results. Eight independent validation cohorts (total n = 2,399) were used to evaluate the prognostic miRNA/mRNA signature generated in the TCGA IDC cohort.
The stringent multistep selection that we applied, as shown in Fig. 1, allowed us to (i) identify the common RNAs related to clinical outcome across IDC patients, (ii) validate the prognostic genes in nonoverlapping patient subclasses, (iii) use DNA methylation as an independent molecular parameter to confirm a prognostic role for selected mRNAs, and (iv) identify prognostic genes with bona fide cancer activity (SI Appendix, Table S4). We defined these genes as the common risk integrated gene set. Some known cancer genes (for example, NME3, an isoform of the NM23 family) were associated with outcome only within a single subclass and therefore did not satisfy our selection requirements for common genes.
Integrated IDC Risk Predictor: Common and Subtype-Directed Prognostic Genes.
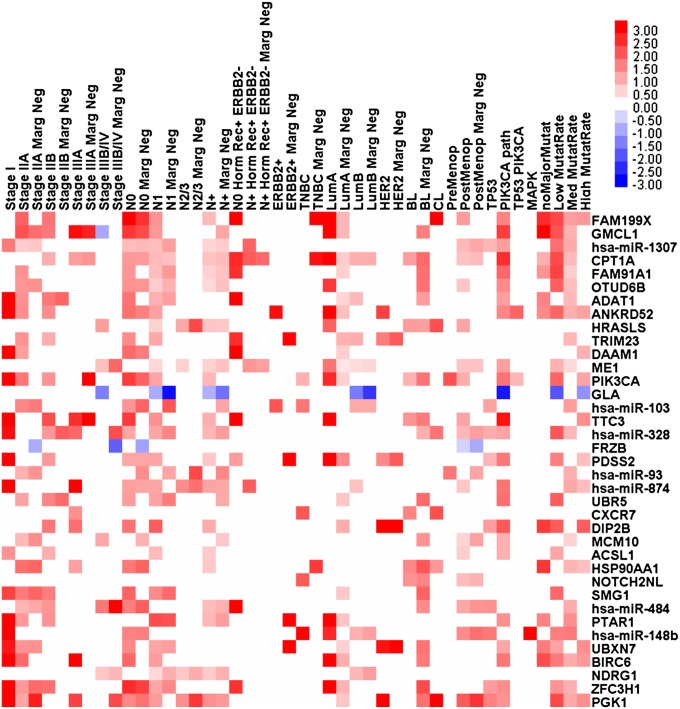
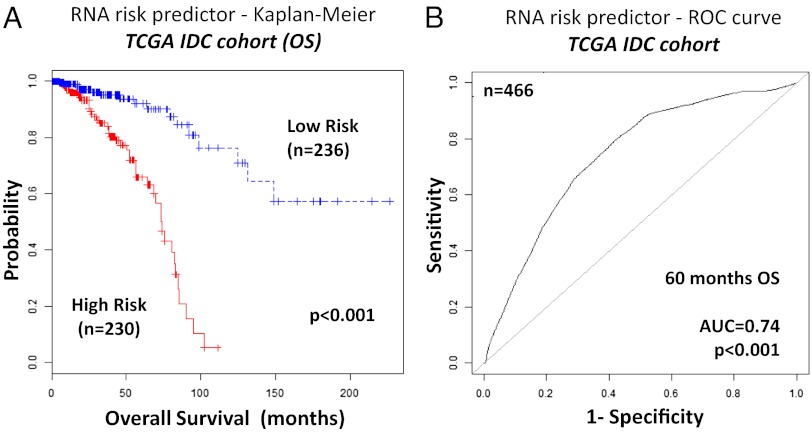
Having determined the common risk genes across different BC subclasses, we wanted to remove any gene that might have divergent prognostic values in the four major BC subtypes, namely Luminal A, Luminal B, Basal-like, and HER2 enriched. We assigned to the HER2-enriched group also tumors not belonging to any of the other three subtypes (i.e., Claudin low and normal breast-like). We used the common prognostic gene set to develop an “RNA model,” using only mRNA and miRNA expression data. Linear risk predictors were constructed using the supervised principal component method (16) to divide the patients in high- and low-risk groups for each of the BC subtypes, and the receiver operating characteristic (ROC) test was used to evaluate their prognostic performance. The risk genes that had divergent association to OS in different subtypes (defined as their weight contribution to the linear risk predictor)—namely FAM208B, C2CD2, CHD9, CHM, DPY19L3, NCOA2, hsa-miR-324, hsa-miR-326, and hsa-miR-365—were removed from the prognostic gene set. Then we added subtype-directed risk genes to the prognostic gene set. We started by reassessing each one of the 195 genes present at step 1 of the selection procedure (Dataset S3). We stopped the subtype-directed gene selection when we obtained the maximal area under curve (AUC) for each one of the four Luminal A, Luminal B, Basal-Like, and HER2E tumor subtypes. The prognostic value for the Luminal B was not as high as for the other three subtypes; therefore, we identified additional markers for this subtype, extending the search to the genes with highest weights in the whole transcriptome. FAM199× and PTAR1 had the largest weights in the predictor for Luminal A tumors; NDRG1, ACSL1, and GLA for the Luminal B tumors; HRASLS, CXCR7, MCM10, and NOTCH2NL for the Basal-like tumors; and PGK1, HSP90AA1, and FRZB for the HER2-enriched group. The prognostic matrix (Fig. 2) visualizes all significant hazard ratios (P < 0.05) for the 30 mRNAs and the seven miRNAs that were finally selected as risk genes. This approach led to the construction of a linear risk predictor, based exclusively on the RNA expression of 37 genes, i.e., the “RNA model,” shown in SI Appendix, Table S5. The cross-validated Kaplan–Meier curves for IDC risk groups obtained from the TCGA cohort (n = 466), using the RNA model, are shown in Fig. 3A. The analysis of AUC for the ROC test was conducted allowing for time-dependent ROC curve estimation with censored data (Fig. 3B). The AUC for the integrated IDC risk predictor was 0.74 at 60 mo of OS (P < 0.001). Because a prognostic biomarker signature in BC is most applicable to early stage disease, we also assessed the risk predictor on stage I and II IDC tumors (n = 348). The integrated miRNA/mRNA signature had an even better performance on early tumors than on the overall cohort (AUC = 0.77, P < 0.001), as shown in SI Appendix, Fig. S10.
Fig. 2.
mRNAs and miRNAs associated with prognosis in different clinical and molecular subclasses of invasive ductal carcinoma (TCGA cohort). The matrix visualizes the significant HRs for the 30 mRNAs and seven miRNAs in the TCGA IDC cohort (listed in SI Appendix, Table S5). The HRs for expression with significant univariate Cox regression (P < 0.05) are displayed on a log2 scale. Red squares indicate HRs >1 and blue squares indicate HRs <1.
Fig. 3.
Kaplan–Meier and ROC curves for the integrated miRNA/mRNA signature (TCGA IDC cohort). (A) The cross-validated Kaplan–Meier curves for IDC risk groups obtained from the TCGA cohort (n = 466), using the integrated signature (“RNA model”). The permutation P value of the log-rank test between risk groups (P < 0.001) was based on 1,000 permutations. (B) The ROC curve had an AUC of 0.74 (P < 0.001). The permutation P value was computed for testing the null hypothesis (AUC = 0.5) using 1,000 permutations.
To evaluate the independent prognostic values of the 37 mRNA/miRNA genes in the integrated RNA predictor, we performed multivariate analysis, including lymph node involvement (N stage), disease stage, T stage, molecular subtype, TP53 mutation status, mutations in the PIK3CA pathway (including AKT1 and PTEN), and ER status (patients were stratified according to age). The final multivariate model contained 10 mRNAs—CPT1A, CXCR7, GLA, HRASLS, NOTCH2NL, PGK1, PIK3CA, TTC3, UBXN7, and ZFC3H1—and two miRNAs—hsa-miR-1307 and hsa-miR-328 (SI Appendix, Table S6). Mutations in the PIK3CA/AKT/PTEN axis, the estrogen receptor status, and N stage were the three remaining clinical or molecular covariates.
Validation of the Integrated miRNA/mRNA Prognostic Signature in Independent BC Cohorts.
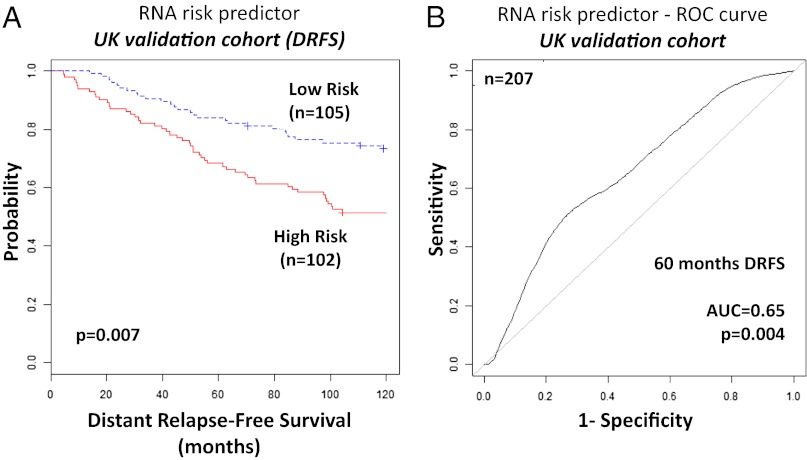
The validation of the prognostic signature was performed on eight independent BC cohorts. At first we used a UK cohort of 207 breast cancer patients because it had both mRNA and miRNA profiles (12). The miRNA/mRNA prognostic gene set was here reassessed for prediction of distant relapse-free survival (DRFS). Nine miRNAs and 11 mRNAs, less than 1/2 of the prognostic genes, were measured in the UK cohort. Both the Kaplan–Meier curve (P = 0.007) and the ROC curve for the prognostic signature (AUC = 0.65, P = 0.004) were significant (Fig. 4). As there were no other available mRNA and miRNA combined expression data from large BC cohorts, we then evaluated the mRNA component of the miRNA/mRNA prognostic signature on the Netherland Cancer Institute (NKI) (17) (n = 295), Hatzis (18) (n = 508), Kao (19) (n = 327), TNBC (20) (n = 383), Bos (21) (n = 195), Wang (22) (n = 286), and TRANSBIG Consortium (20) (n = 198) cohorts. The mRNA component of the prognostic signature was significantly predictive for outcome in all these BC cohorts (SI Appendix, Table S7 and Figs. S11–S17).
Fig. 4.
Kaplan–Meier and ROC curves for the integrated miRNA/mRNA signature in the UK validation cohort. (A) The cross-validated Kaplan–Meier curves for breast cancer risk groups obtained from the validation cohort (n = 207), using the prognostic integrated signature. The permutation P value of the log-rank test between risk groups (P = 0.007) was based on 1,000 permutations. (B) The ROC curve had an AUC of 0.65 (P = 0.004). The permutation P value was computed for testing the null hypothesis (AUC = 0.5) using 1,000 permutations.
Comparison of the Integrated miRNA/mRNA Signature with Other Prognostic BC Signatures.
We compared the prognostic value of the integrated miRNA/mRNA signature to that of different gene sets used for risk stratification of BC: the genes used in the Oncotype DX (23), those used for the Genomic Grade Index (GGI) (24), for MammaPrint (17, 25), the 76-gene (22), the Invasiveness gene (IGS) (26), the 95-gene Japanese (27), and the 10-miRNA signatures (12). We calculated the AUC of the ROC curves for each possible signature/cohort combination (SI Appendix, Table S7). The 10-miRNA signature was predictive of DRFS (12) in the UK dataset, where it was determined (AUC = 0.76, P < 0.001), but not in the TCGA cohort. In the NKI, Hatzis, and TNBC cohorts, all signatures tested were successful with similar performances. The gene signature used in Oncotype DX performed very well in all of the cohorts, with the notable exception of the TCGA cohort, where it was not significant (AUC = 0.58, P = 0.12). The GGI, IGS, and 95-gene signatures had significant AUC in the TCGA cohort. There were only two signatures with significant ROC tests in early stage tumors from the TCGA cohort: the integrated miRNA/mRNA signature (AUC = 0.77, P < 0.001) and the 70-gene NKI signature used in MammaPrint (AUC = 0.66, P = 0.026).
Discussion
Invasive ductal carcinoma is characterized by different molecular subtypes (9) that are thought to impact on the cellular pathways related to clinical outcome (10). miRNAs are modulators of the cellular processes responsible for cancer that are encoded by mRNAs (28), the expression of which in turn is at least partially regulated by DNA methylation. Because of these relations, we performed an integrated survival analysis on a large breast cancer cohort of 466 patients, using genome-wide data for miRNA/mRNA expression and DNA methylation. The resulting integrated prognostic signature, composed of seven miRNA and 30 mRNA genes, was very compact, and it was successfully validated on eight breast cancer cohorts, for a total of 2,399 additional patients.
Some points should be considered. First, as these cohorts were not treatment-naive, the identified RNAs could not only be prognostic but also predictive of response to treatment. Second, the integration of miRNA and mRNA components augmented the prognostic strength of the risk predictor. Third, we used DNA methylation as a criterion to confirm the association between mRNA expression and OS. Fourth, we identified biomarkers that were consistent across nine different and heterogeneous breast cancer cohorts.
Among the few known cancer genes in the prognostic signature, PIK3CA was one of the most prominent. PIK3CA is an example of oncogene addiction (29), including when it is not mutated (30), and thus could be considered as a primary target for therapy. Both PIK3CA expression and the somatic mutations in its pathway (PIK3CA/AKT1/PTEN axis) were retained in the final multivariate model, proving to be important and independent cofactors in prognosis. The prognostic value of the integrated signature was the highest in early stage I and II breast cancers, making this a potentially valuable biomarker signature in the clinical practice.
Methods
Patient Characteristics and Integrated Profiles in the TCGA IDC Cohort.
The miRNA/mRNA tumor profiles (for a total of 19,262 mRNAs and 581 miRNAs) were studied in 466 primary IDCs from female patients with no pretreatment (TCGA IDC cohort) (8). Only patients with fully characterized (mRNA and miRNA profiles) tumors and with at least 1 mo of OS were included in the study. Extended demographics for these patients, characterized by the TCGA consortium (8), are provided in SI Appendix, Table S1. Raw RNA, methylated DNA (meDNA), somatic mutations, and clinical data were obtained from the TCGA data portal. Detailed methods for the integration of miRNA, mRNA, and meDNA data are reported in SI Appendix.
Survival Analysis.
Clinical covariates for the IDC tumors and patients are summarized in SI Appendix, Table S1. The association between continuous RNA expression and OS was carried out using univariate Cox regression. The hazard ratio was the ratio of hazards for a twofold change in the gene expression level. It was equal to exp(b) where b was the Cox regression coefficient. To compute the Kaplan–Meier distribution, the group with gene overexpression was assigned to samples with expression larger than median expression. The test of equality for survival distributions was performed using the log-rank method (Mantel–Cox), except when explicitly stated. For the multivariate analysis, the Cox proportional hazard model was applied, and a backward stepwise selection procedure (Wald) was used to identify genes or covariates with independent prognostic value. All reported P values were two-sided. All analyses were performed using SPSS (version 21) or R/BioConductor (version 2.10).
Definition of Risk Predictor and ROC Curve.
The gene weights for the linear RNA risk predictor were computed using the supervised principal component method (16). The Kaplan–Meier survival curves for the cases predicted to have low or high risks (median cut) were generated using 10-fold cross-validation (31). The statistical significance of the cross-validated Kaplan–Meier curves was determined by repeating the process 1,000 times on random permutations of the survival data. The P value tested the null hypothesis that there was no association between expression data and survival. The ability of the models to predict outcome was assessed by comparing the AUC of the respective ROC curves. Because in all of the survival analyses fewer events occurred after 60 mo (SI Appendix, Figs. S1–S3), we compared the ability of models to predict outcome at, and around, this time point. The ROC curve plots the true-positive vs. false-positive predictions; thus, higher AUC indicates better model performance (with AUC = 0.5 indicating random performance). RNA risk scores and groups (high or low risk defined above) were based on weightings in the linear risk predictor.
Independent Cohorts for the Validation of the miRNA/mRNA Prognostic Signature.
To validate the prognostic signature obtained from the TCGA IDC cohort, we used genome-wide expression data from eight series of primary breast cancer patients for a total of 2,399 patients. In the UK cohort (12) (n = 207), 74% of the patients had IDC, whereas the remaining breast cancers were mostly lobular (12%) or mixed (7%). The clinical endpoints for the UK cohort toward DRFS were distant metastasis detection or death, or the date of last assessment without any such event (censored observation). The expression of miRNAs [Gene Expression Omnibus (GEO) dataset GSE22216] was measured using Illumina miRNA v.1 bead-chip and that of mRNAs (GSE22219) using Illumina Human RefSeq-8 bead-chip. The assays measured 24,332 mRNAs and 488 miRNAs. Quantile normalization was used for both arrays (12) and for the integrated profile. Validation of the mRNA prognostic component was performed on seven additional breast cancer profiles. The NKI cohort was composed of a series of 295 consecutive patients with primary BC. All patients had stage I or II BC and were younger than 53 y old; 151 had lymph-node–negative disease and 144 had lymph-node–positive disease (17). The cancerBreastNKI package from Bioconductor was used to retrieve gene expression data and clinical covariates for the NKI cohort. The Wang cohort (GEO dataset GSE2034) was composed of 180 lymph-node–negative relapse-free patients and 106 lymph-node–negative patients that developed a distant metastasis (22). The Hatzis cohort (GSE25066) included 310 newly diagnosed patients from a prospective multicenter study conducted at the M. D. Anderson Cancer Center and 198 validation patients (99% clinical stage II–III) who received sequential taxane and anthracycline chemotherapy (18). The Kao cohort (GSE20685) was used to identify molecular and clinical subtypes of BC through gene expression profiles of 327 samples (19). The Bos cohort was used to study brain metastasis, one of the most feared complications of BC (GSE29271; n = 195) (21) The TNBC cohort (GSE31519) was assembled from 383 German patients to characterize triple-negative breast cancer (20). The TRANSBIG cohort (GSE7390) was composed of 198 Belgian patients with lymph-node–negative disease (32). DRFS was the clinical endpoint for all of the validation cohorts, with the exceptions of the NKI, Kao, and TRANSBIG cohorts, where OS was used. The clinical data for the validation cohorts are listed in Dataset S4. The eight validation cohorts were also used for the comparison of the miRNA/mRNA integrated signature to other prognostic BC signatures.
Supplementary Material
Acknowledgments
We are indebted to the TCGA consortium for making the mRNA, miRNA, meDNA, and clinical data available. The mutation data were obtained from the Sanger Institute Catalogue of Somatic Mutations in Cancer website (www.sanger.ac.uk/genetics/CGP/cosmic/). We thank Kay Huebner and Kati Maharry for critical reading. S.V. is supported by Associazione Italiana per la Ricerca sul Cancro and Ministero dell’Istruzione, dell’Università e della Ricerca Progetti di Ricerca di Interesse Nazionale (PRIN) grants. C.M.C. is supported by a National Cancer Institute-Early Detection Research Network UO1 grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304977110/-/DCSupplemental.
References
- 1.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens PJ, et al. Oslo Breast Cancer Consortium (OSBREAC) The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koboldt DC, et al. Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 11.Madhavan D, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012;18(21):5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 12.Buffa FM, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71(17):5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 13.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Dunham I, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2(4):E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 18.Hatzis C, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rody A, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13(5):R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 23.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 24.Sotiriou C, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 25.Glas AM, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356(3):217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 27.Naoi Y, et al. Development of 95-gene classifier as a powerful predictor of recurrences in node-negative and ER-positive breast cancer patients. Breast Cancer Res Treat. 2011;128(3):633–641. doi: 10.1007/s10549-010-1145-z. [DOI] [PubMed] [Google Scholar]
- 28.Volinia S, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci USA. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3(8):448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 30.Nijhawan D, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150(4):842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian J, Simon R. An evaluation of resampling methods for assessment of survival risk prediction in high-dimensional settings. Stat Med. 2011;30(6):642–653. doi: 10.1002/sim.4106. [DOI] [PubMed] [Google Scholar]
- 32.Desmedt C, et al. TRANSBIG Consortium Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.