Abstract
miR-21 can act as an oncogene. MSH2 has been reported that it involved in the DNA mismatch repair (MMR) system and overexpression of MSH2 can induce cell apoptosis. We predicted that MSH2-3′-untranslated region (3′-UTR) was targeted by miR-21 using microRNA analysis softwares. To further explore the roles of miR-21 and MSH2 in A549 cells, we constructed pcDNA-GFP-msh-UTR vector (including MSH2-3′-UTR) to transfect A549 cells with miR-21, GFP positive cells were estimated under a fluorescence microscopy and by flow cytometry. We found miR-21 could obviously downregulate the expression of MSH2, which was further proved by western blotting. Moreover, we treated A549 cells with cisplatin and found that cisplatin could inhibit A549 cell growth in vitro and in vivo. We also found that cisplatin could downregulate miR-21 expression, while increase MSH2 expression in A549 cells. Our results demonstrated that cisplatin could upregulate the expression of MSH2 through downregulating miR-21 to inhibit A549 cell proliferation, which provides new gene targets for drug design or cancer therary.
Keywords: microRNA, Lung cancer, MSH2, Cisplatin, Gene expression
1. Introduction
Lung cancer is the leading cause of cancer-related death in the world, with metastasis being the main reason for the mortality [1]. In China, lung cancer is the most common cancer and the incidence of lung cancer continues to increase [2]. According to statistics in 2012, lung cancer is of the first importance reason for cancer deaths in the United States [3]. 85 % of all lung cancers are non-small-cell lung cancer [4], of which adenocarcinoma is one kind of common histology [5]. The diagnosis of non-small-cell lung cancer is often at an advanced stage and the prognosis is poor, although it can be detected in its early stages [4]. Over the past 30 years, the overall 5-year survival from lung cancer remains dismal at around 16%, showing little improvement [3,6].
The defections of DNA repair genes MSH2, RECQL4 and RAD51L1 are typically linked to various cancers [7]. Defection of DNA repair capacity, the susceptibility and risk of lung cancer was increased [8]. MSH2, binding directly to mismatched nucleotides to provide a target for DNA repair, is part of the mismatch repair (MMR) system involved in DNA damage recognition and repair [9,10]. MMR is responsible for the correction of DNA replication errors and therefore essential for maintaining genomic stability or preventing tumor formation [11,12]. Human mutS homolog 2 (hMSH2) and human mutL homolog 1 (hMLH1) are the core MMR proteins, which can form heterodimers with protein homologs hMSH3 or hMSH6 and hMLH3 or hPMS2, respectively [13]. The primarily study about mismatch repair (c-Abl and p73) dependent DNA damage-induced apoptotic response was reported in 1999 [14]. To explore whether MMR machinery was related with apoptotic process, microinjection studies had been used to overexpress MSH2. As a result, overexpression of hMSH2 and hMLH1 induced apoptosis within 24 h [15]. It was reported that more than 95 % of patients with Lynch syndrome were deficiency of MMR [11].
microRNAs (miRNAs) are non-coding RNAs of 20–22–nucleotides (nt) that bind to the 3′-UTRs of cognate mRNAs, negatively regulating the target mRNAs [16,17]. miRNAs has been found that they can modulate growth and death of cells [18]. Dependent upon the nature of their target gene(s), miRNAs may function as tumor suppressors or as oncogenes by downregulating target mRNAs. miR-16, miR-17 and miR-34a-c had been verified to act as tumor suppressors by downregulating the expression of BCL-2 [19]. miR-150 which can downregulate the expression of p53 was confirmed as an oncogene [19,20]. miR-21 has been reported to function as an oncogene and modulate tumorigenesis through regulation of target genes to promote of cell growth, invasion, metastasis, and evasion of apoptosis [21,22].
Cisplatin was an effective anticancer drug for lung cancer therapy [23]. In our previous study, we found that the expression of miR-98 was decreased after treatment with cisplatin, while TP53 expression was enhanced, which indicated that cisplatin involved in the miR-98 regulating TP53 pathway to inhibit A549 cell growth [24]. In this study, to further explore whether cisplatin could affect the expression of miR-21 and MSH2, A549 cells were treated with cisplatin, then MSH2 expression was detected by western blotting. We found that cisplatin might upregulate the expression of MSH2 through downregulating miR-21 to inhibit A549 cell proliferation.
2. Material and methods
2.1. Construction of pcDNA-GFP-msh-UTR vector
Firstly, MSH2-3′-UTR (NM_000251, AK299667) was amplified by PCR from human genomic DNA. The forward primer was: 5′-AGTAATGGAATGAAGGTA-3′; Reverse was 5′-ATAAAATTCAGCA-CATCA -3′. PCR conditions was 30 cycles of denaturation at 94 °C for 45 s, annealing at 47 °C for 45 s and elongation at 72 °C for 60 s, performed in a PCR machine (eppendorf, Germany). The MSH2-3′-UTR production (241 bp) was cloned into T-vector (Takara, Japan) to construct T-MSH2 vector. Then the MSH2-3′-UTR was cut from T-MSH2 and cloned into pcDNA-GFP vector (our previous study [24]) by Kpn I/Hind III to form pcDNA-GFP-msh-UTR vector.
2.2. Cell culture and miRNA transfection
A549 cells (obtained from Shanghai Institute of Cell Biology, China) were maintained in F12 medium (Gibco, USA) supplemented with 10 % calf serum (Hyclone, USA), 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C with 5 % CO2.
For transfection, 1 × 106 cells were treated with 0.5 μg miRNA and 0.5 μg pcDNA-GFP-msh-UTR in 2.5 μL of lipofectamine 2000 (Invitrogen, USA), according to the manufacturer’s instruction. All transfections were carried out in triplicate.
2.3. GFP assays
72 h after transfection, the expression of GFP in A549 cells was observed under one fluorescence microscope. The cells were trypsinized and gently washed with serum-containing medium. The cells was collected and centrifugated at 400 g for 5 min. At last, GFP positive cells were analyzed by flow cytometry (FACS).
2.4. Cisplatin suppressing A549 cell growth
Cells (1 × 104) in each of the 96-well flatbottom microtiter plates were treated with various of concentrations (0 μg/mL, 3 μg/mL, 6 μg/mL, and 9 μg/mL) of cisplatin (synthesized from QiLu Pharmaceutical Co., Ltd, China) for 72 h. The cells in each well were exposed to 10 μL MTT (5 mg/mL) at 4 h before the end of incubation, the supernatant was removed and 100 μL dimethyl sulfoxide (DMSO, signa) was added to determine the OD value at 570 nm using an enzyme-linked immunosorbent assay reader (ELX800, USA). The growth inhibition rate was dectected according to our previous study [20], the growth inhibition rate = (ODcontrol−ODsample)/ODcontrol × 100 (%).
Flow cytometry (FACS) was used to further detect cells apoptosis induced by cisplatin. The specific method was as follows: Cells (8 × 104) in each of 12-well flatbottom microtiter plates were exposed to various of concentrations (0 μg/mL, 3 μg/mL, 6 μg/mL, and 9 μg/mL) of cisplatin for 48 h. Then the cells were dyed with Annexin V-FITC/PI, according to the manufacturer’s instruction (KeyGEN Biotech., China). Annexin V-FITC/PI positive cells were analyzed by FACS.
2.5. A549 lung adenocarcinoma cell xenografts
After trypsinized, 5 × 106 cells in 100 μL PBS were subcutaneously injected into the lower back of 6–8-week old BALB/C-nu mice (nude mice, HFK Bio-Technology, China). When tumor volume (V = length × width2/2, length > width) is ~150 mm3, 3 mg/kg of cisplatin was injected into the body of the nude mice every four days for four times. During treated with cisplatin, tumor volume was measured by a caliper everyday. Four days after the last treatment, nude mice were sacrificed and tumors were collected for further analysis. The control mice were treated with physiological saline in the same amount. All animal experiments were approved by the Committee on the Ethics of Animal Experiments of Binzhou Medical University.
2.6. Reverse transcription-polymerase chain reaction (RT-PCR) and real-time PCR
miRNAs were isolated by mirVana miRNA Kit (Ambion) and added ploy (A) using poly (A) polymerase (Ambion). The cDNA was synthesized by RT primer 5′-AACATGTACAGTCCATG-GATGd(T)30N(A,G,C or T)-3′. The forward primer used to amplify miR-21 was 5′-AGCTTATCAGACTGATGTTGACTG-3′, and the reverse was 5′-ACATGTACAGTCCATGGATG-3′. Then, the Quantitect SYBR-Green kit (Qiagen) was used to assess the expression of miR-21 in one RG3000 system (Corbett Research) as follows: denaturing at 95 °C for 3 min; 40 cycles of 95 °C for 30 s, 60 °C annealing for 20 s and extension at 72 °C for 20 s. At the each extension step of 72 °C, fluorescence was detected at 585 nm.
2.7. Western blotting
After A549 cells or mice were treated with cisplatin, the samples were lysed with lysis buffer (Cell lysis buffer for Western of Beyotime, China). 30 μg protein of each sample was loaded respectively to each lane of polyacrylamide gel, then the protein was transferred to PVDF membranes. After transferring, the membranes were blocked with 5 % non-fat milk in TBST (50 mmol/L Tris-HCl [pH 7.6], 150 mmol/L NaCl, 0.1 % Tween-20) for 2 h at room temperature. The membranes were incubated with MSH2 antibody (1:400, Boster) in TBST at 4 °C overnight. The membranes were washed with TBST for three times. The HRP-labeled goat anti-mouse IgG (1:6000, Beijing Zhong Shan-Golden Bridge Technology Co., Ltd., China) was used to incubate the membrane for 1 h at room temperature. At last, the membrane was put in ECL reagent (Boster Immunoleader, China) and detected. Actin was used as the control.
2.8. Statistics
SAS software was used to analyze the significance of all results. The Student’s t-test was used for inter-group comparison. A P value less than 0.05 was considered significant.
3. Results
3.1. Construction of pcDNA-GFP-msh-UTR vector
The relationship between MSH2-3′-UTR and miR-21 was predicted using microRNA analysis software online (http://www.microrna.org/microrna/getMirnaForm.do, or http://www.targetscan.org/index.html), which showed that MSH2-3′-UTR was targeted by miR-21 (Fig. 1A). Then, MSH2-3′-UTR was amplified by PCR and pcDNA-GFP-msh-UTR vector was constructed, which was identified by restriction endonuclease (Hind III and EcoR I) digestion (Fig. 1B) and an automatic DNA sequencer (data now shown).
Fig. 1.
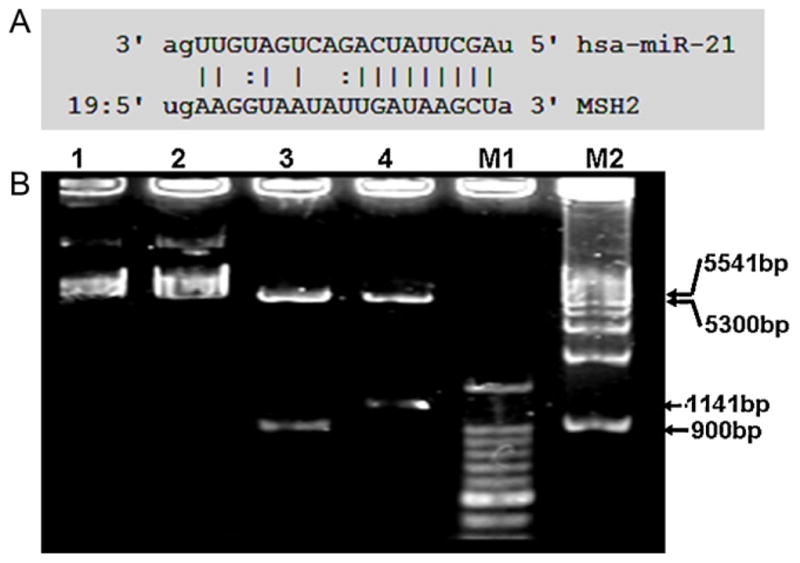
The miR-21 targeting site on MSH2-3′UTR and construction of pcDNA-GFP-msh-UTR vector. A. showing the targeting site of miR-21 on MSH2-3′-UTR. B. Lane 1 and 2, pcDNA-GFP and pcDNA-GFP-msh-UTR plasmids, respectively. Lane 3, pcDNA-GFP plasmid was digested by Hind III and EcoR I, producing 5541 bp and 900 bp. Lane 4, pcDNA-GFP-msh-UTR plasmid was digested by Hind III and EcoR I, producing 5300 bp and 1141 bp. M1, 1 kb Marker. M2, 100 bp Marker.
3.2. 3′-UTR of MSH2 was regulated by miR-21
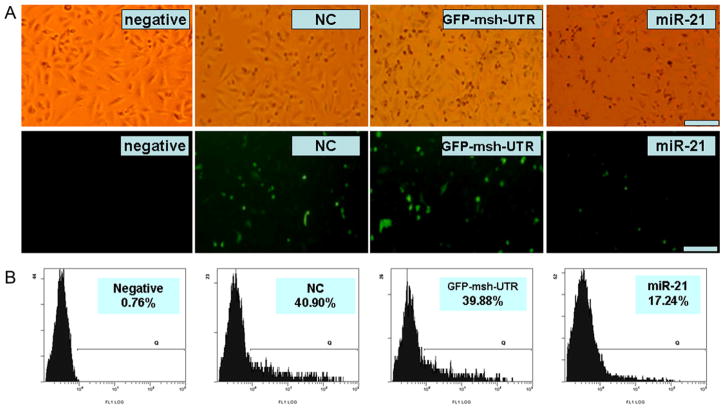
To determine the regulation role of miR-21 to MSH2 3′-UTR, A549 cells were co-transfected with pcDNA-GFP-msh-UTR vector and miR-21. The GFP positive cells were observed using fluorescence microscopy and FACS. Our results showed that the intensity of fluorescence and the number of GFP positive cells in miR-21-treated cultures were obviously decreased compared with control cultures (Fig. 2A), and the percentage of positive GFP cells transfected with miR-21 was also much lower than that in controls by FACS analysis (Fig. 2B). The above results indicated that miR-21 could regulate GFP expression by targeting 3′-UTR of MSH2.
Fig. 2.
Analyzing GFP expression by microscope and FACS. A. Upper, natural light. Lower, the fluorescence. Scale bar = 100 μM. Under microscope, the number of GFP positive cells and intensity of GFP fluorescence in miR-21-treated cultures were found to be decreased obviously compared with control cultures. B. FACS results showed that the ratio of GFP positive cells in miR-21-treated cultures was much lower than that of controls.
3.3. miR-21 negatively regulating MSH2 expression in A549 cells
To further study whether MSH2 expression is regulated by miR-21, MSH2 expression in A549 cells was detected by western blotting after miR-21 treatment. Our results showed that MSH2 expression was obviously decreased in A549 cells overexpressed miR-21 than that in controls (Fig. 3A and B), which demonstrated that miR-21 could negatively regulate MSH2 expression.
Fig. 3.
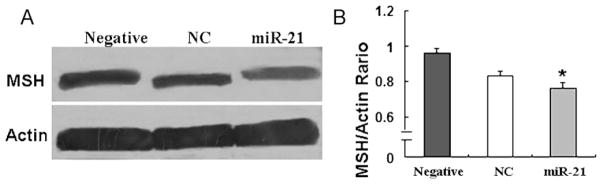
MSH2 protein expression detected by western blotting. A. The results showed that MSH2 expression was decreased in the miR-21-treated cells than those in the control cells. B. Relative values for MSH2 vs Actin of Fig. 3A are indicated to the right of the gel. *P < 0.05.
3.4. Cisplatin inhibiting A549 cell growth by downregulation of miR-21
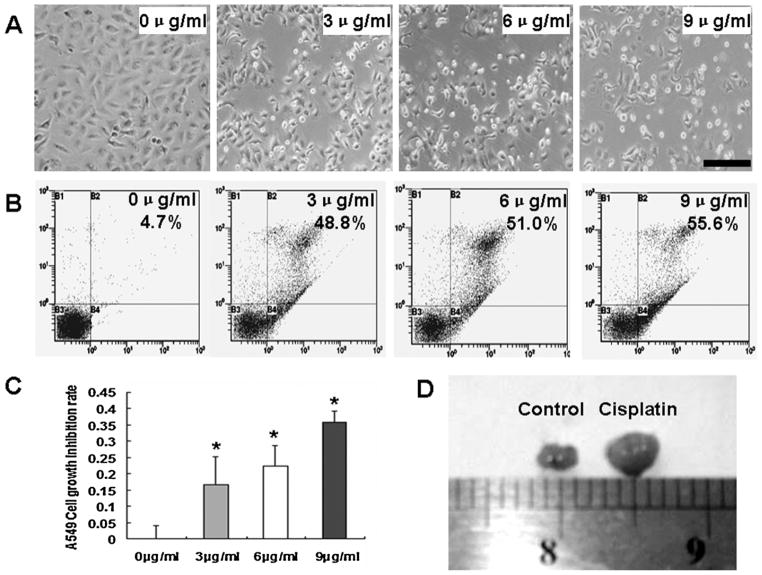
It was reported that cisplatin is one kind of anticancer drugs for lung cancer therapy and could inhibit A549 cell growth [23,24]. To further investigate the anticancer mechanism of cisplatin, A549 cells were treated with different concentrations of cisplatin for 36 h. Our results showed that cisplatin could effectively inhibit A549 cell growth (Fig. 4A), and more apoptotic A549 cells were found after cisplatin treatment comparing to control cultures (Fig. 4B). The MTT assay further proved that cisplatin could effectively inhibit A549 cell growth, and the inhibition rate was enhanced as the concentration increasing (Fig. 4C). In vivo study, we also found that the volume and weight of A549 lung cancer xenografts in nude mice treated with cisplatin were much smaller and lesser than those of controls with physiological saline treatment (n = 3, Fig. 4D, Table 1).
Fig. 4.
Cisplatin inhibiting A549 cells growth. A. This figure showed that cisplatin could inhibit A549 cell proliferation under one microscope. Fewer alive cells were found with the increasing concentration of cisplatin. Scale bar = 100 μM. B. FACS analysis showed that the percentage of apoptotic cells was increased with the higher concentration of cisplatin. C. MTT assay showed that the inhibiting rate of A459 cell growth was increased with the higher concentration of cisplatin. D. The volumes of A549 cancer xenografts were detected after cisplatin treatment, which showed that the tumor volume became smaller with cisplatin treatment than saline treatment.
Table 1.
Growth inhibition of A549 lung cancer xenografts in nude mice by cisplatin.
| Treatment groups (n = 3) | Average (g) |
|---|---|
| Control | 0.182333 ± 0.052351 |
| Cisplatin | 0.093875 ± 0.028734a |
The control mice were treated with physiological saline.
P <0.05 vs control group.
To study the effect of cisplatin on miR-21 expression, real-time PCR analysis showed that miR-21 expression was decreased in A549 cells with cisplatin treatment than that in control cultures (Fig. 5A). We also found that miR-21 expression was inhibited in A549 lung cancer xenografts in nude mice treated with cisplatin than that treated with physiological saline (n = 3, Fig. 5B), which indicated that cisplatin could suppress A549 cell growth in vitro and in vivo by decreasing miR-21 expression.
Fig. 5.
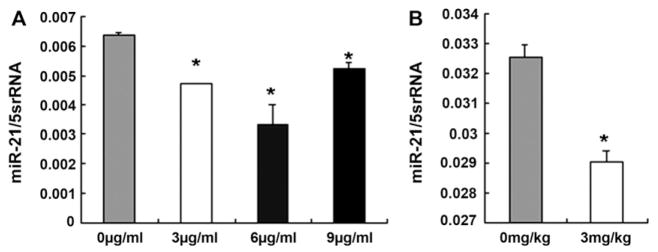
miR-21 expression was decreased by real-time PCR. A. miR-21 expression was decreased obviously in A549 cells treated with 3–9 μg/mL cisplatin (*P < 0.05). B. miR-21 expression was decreased significantly in A549 lung cancer xenografts after the mice treated with cisplatin than that in control mice (*P < 0.01).
3.5. Cisplatin inhibiting A549 cell growth by increasing MSH2 through miR-21
Considering miR-21 could regulating MSH2 expression, the expression of MSH2 in A549 cells and A549 cancer xenografts was further studied after cisplatin treatment. Different to the effect of cisplatin on miR-21 expression, the MSH2 expression was found to be increased both in A549 cells and in A549 cancer xenografts (Fig. 6A–D), which demonstrated that cisplatin could inhibit A549 cell growth by upregulating MSH2 expression.
Fig. 6.
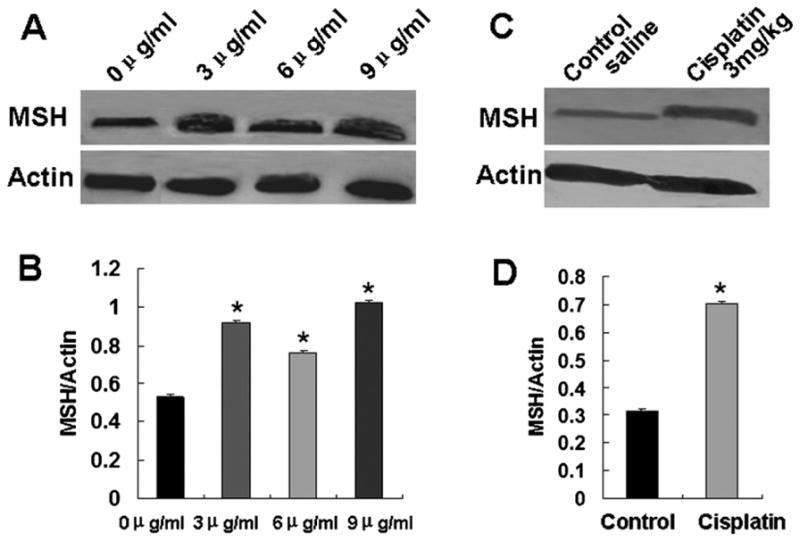
MSH2 expression was detected by western blotting after cisplatin treatment. A. MSH2 expression was increased in A549 cells as increasing cisplatin concentration. B. Relative values for MSH2 vs Actin of Fig. 6A. *P < 0.05. C. MSH2 expression was increased in A549 cancer xenografts in nude mice treated with cisplatin than that in mice treated with saline. D. Relative values for MSH2 vs Actin of Fig. 6C. *P < 0.01.
4. Discussion
miRNAs are involved in cell processes, including proliferation, apoptosis, and stress responses [25]. miRNAs can negatively regulate target mRNAs by binding with their 3′-UTRs [26]. In this study, we predicted MSH2 3′-UTRs was targeted by miR-21 and further confirmed miR-21 could negatively regulate MSH2 expression in A549 cells. We also found that cisplatin could inhibit A549 cell proliferation in vitro and in vivo. After cisplatin treatment, miR-21 expression was found to be decreased, while MSH2 was upregulated in A549 cells and A549 cancer xenografts compared with their controls. Our results demonstrated that cisplatin could inhibit A549 cell growth by upregulating the expression of MSH2 through suppressing miR-21.
miR-21 has been reported commonly overexpressed in a number of human solid tumors including colon cancer, breast cancer, lung cancer, etc. High miR-21 expression is associated with poor survival and poor therapeutic outcome [27,28], which indicated miR-21 can act as oncogene. miR-21 can be used as a tumor marker and its inhibition may prove to be useful in controlling cancers with upregulated miR-21 [29]. It is reported that miR-21 can regulate a variety of cellular pathways through regulation their multiple target genes [30]. It was shown that aberrant expression of miR-21 can contribute to HCC growth, migration and invasion by modulating PTEN expression, which involved in mediating phenotypic characteristics of cancer cells, such as cell growth, migration, and invasion [31]. miR-21 overexpression can also reduce PDCD4 protein to function as a transformation suppressor in gastric cancers [32] and downregulate Tropomyosin 1 (TPM1, a tumor suppressor gene), which further inhibit tumor growth [33]. Similarly, in this study, we also found that miR-21 could act as an oncogene by suppressing MSH2 expression in A549 cells.
MSH2 protein involves in initiation of MMR by recognizing the mismatched bases from replication errors and insertion-deletion loops that lead to frame shift mutations [34]. Germline mutations in MSH2, MSH6, and PMS2 of MMR genes can cause autosomal dominant hereditary disease Lynch syndrome [35], and germline mutations had been found in MSH2 in HNPCC [36]. Our results showed that cisplatin could increase MSH2 expression in apoptotic A549 cells, indicating that MSH2 might act as a suppressing role in cancer growth. The suppressing role of MSH2 was also supported by Lin DP’s study, which demonstrated that MSH2 could mediate cell apoptosis which was important to tumor suppression, and deficiency of mismatch repair caused by missense mutations remained sensitivity to chemotherapeutic agents [37].
Cisplatin is an effective chemotherapeutic agent that elicits its antineoplastic activity by binding to DNA and disrupting template functions [38]. Some studies have showed that cisplatin can regulate the expression of miRNA to induce tumor cell apoptosis. We also find that cisplatin could effectively induce A549 cell apoptosis, and the growth inhibition rate was enhanced as the concentration increasing. In previous study, we have found that cisplatin could upregulate the expression of miR-16, miR-34a-c, miR-17-5p and miR-125 in apoptotic K562 cells, while inhibit miR-106 and miR-150 expression [39]. To further study the effect of cisplatin to miR-21 expression, we detected that miR-21 expression was decreased in A549 cells or A549 cancer xenografts with cisplatin treatment than those in controls. It has been also confirmed that regression of miR-21 can reduce MCF-7 xenografts tumor growth, relative to downregulation of bcl-2 expression [21]. Differently, in our study, we found that miR-21 could regulate MSH2 expression. Following the decreased expression of miR-21 by cisplatin, we detected that MSH2 was high expressed in A549 cells or xenografts with cisplatin treatment compared with controls, which indicated that cisplatin could inhibit A549 cell proliferation by overexpression of MSH through miR-21.
In summary, we found that miR-21 was the upstream factor to inhibit MSH2 expression. Cisplatin could inhibit A549 cell proliferation in vitro and in vivo by suppressing miR-21 expression to further increase MSH2. Our results provide new gene targets for drug design and lung cancer therapy.
Acknowledgments
This study was supported by the NCET-10-0919, “Taishan scholar” position (No.tshw20110515) and National Natural Science Foundation (No. 30801324, 81141114, 81200601), the Shandong Science and Technology Committee (ZR2009CQ033, ZR2012HQ035) and the Foundation of ShanDong Educational Committee of China (No. J10LC60, J11LC01).
Abbreviations
- miRNAs
microRNAs
- MMR
mismatch repair
- 3′-UTR
3′-untranslated region
- hMSH2
Human mutS homolog 2
- hMLH1
human mutL homolog 1
- nt
nucleotides
- FACS
Flow cytometry
- RT-PCR
Reverse transcription-polymerase chain reaction
- TPM1
Tropomyosin 1
Footnotes
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
References
- 1.Chen J, Ye L, Xie F, Yang Y, Zhang L, Jiang WG. Expression of bone morphogenetic protein 7 in lung cancer and its biological impact on lung cancer cells. Anticancer Res. 2010;30:1113–20. [PubMed] [Google Scholar]
- 2.Zhao P, Dai M, Chen W, Li N. Cancer trends in China. Jpn J Clin Oncol. 2010;40:281–5. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer Statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Heymach JV, Lippman SM. Lung Cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selvaggi G, Scagliotti GV. Histologic subtype in NSCLC: does it matter? Oncology (Williston Park) 2009;23:1133–40. [PubMed] [Google Scholar]
- 6.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 7.Wibom C, Sjöström S, Henriksson R, Brännström T, Broholm H, Rydén P, et al. DNA repair gene variants are associated with glioblastoma survival. Acta Oncol. 2012;51:325–32. doi: 10.3109/0284186X.2011.616284. [DOI] [PubMed] [Google Scholar]
- 8.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12:689–98. [PubMed] [Google Scholar]
- 9.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: Revising the mutator hypothesis. Cancer Res. 2001;61:7369–74. [PubMed] [Google Scholar]
- 10.Fishel R, Ewel A, Lescoe MK. Purified human MSH2 protein binds to DNA containing mismatched nucleotides. Cancer Res. 1994;54:5539–42. [PubMed] [Google Scholar]
- 11.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 12.Jun SH, Kim TG, Ban C. DNA mismatch repair system classical and fresh roles. FEBS J. 2006;273:1609–19. doi: 10.1111/j.1742-4658.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 13.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–26. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 14.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–9. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Richards B, Wilson T, Lloyd M, Cranston A, Thorburn A, et al. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–7. [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–21. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 18.Ambros V. MicroRNA pathways in flies mini review and worms: growth, death, fat, stress and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 19.Wang PY, Li YJ, Zhang S, Li ZL, Yue Z, Xie SY, et al. Regulating A549 cells growth by ASO inhibiting miRNAexpression. Mol Cell Biochem. 2010;339:163–71. doi: 10.1007/s11010-009-0380-2. [DOI] [PubMed] [Google Scholar]
- 20.Li YJ, Zhang YX, Wang PY, Chi YL, Zhang C, Xie SY, et al. Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol Rep. 2012;27:129–34. doi: 10.3892/or.2011.1466. [DOI] [PubMed] [Google Scholar]
- 21.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 22.Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer. 2011;30:371–80. doi: 10.5732/cjc.011.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan T, Li R, Todd NW, Qiu Q, Fang HB, Wang H, et al. Upregulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–6. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S, Zhang C, Li Y, Wang P, Yue Z, Xie S. miR-98 regulates cisplatin-induced A549 cell death by inhibiting TP53 pathway. Biomed Pharmacother. 2011;65:436–42. doi: 10.1016/j.biopha.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 27.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci Ucehsp sp025/Scehsp sp025/A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaman MS, Shahryari V, Deng G, Thamminana S, Saini S, Majid S, et al. Upregulation of microRNA-21 correlates with lower kidney cancer survival. PLoS One. 2012;7:e31060. doi: 10.1371/journal.pone.0031060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusenda B, Mraz M, Mayer J, Pospisilova S. MicroRNA biogenesis, functionality and cancer relevance. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:205–15. doi: 10.5507/bp.2006.029. [DOI] [PubMed] [Google Scholar]
- 31.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepato-cellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Z, Yoon JH, Nam SW, Lee JY, Park WS. PDCD4 expression inversely correlated with miR-21 levels in gastric cancers. J Cancer Res Clin Oncol. 2012;138:611–9. doi: 10.1007/s00432-011-1140-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 34.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci Ucehsp sp025/Scehsp sp025/A. 1996;93:13629–34. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartley AN, Luthra R, Saraiya DS, Urbauer DL, Broaddus RR. Identification of cancer patients with Lynch syndrome: clinically significant discordances and problems in tissue based mismatch repair testing. Cancer Prev Res (Phila) 2012;5:320–7. doi: 10.1158/1940-6207.CAPR-11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–74. [PubMed] [Google Scholar]
- 37.Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, et al. An MSH2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–22. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 38.Dedoussis GV, Mouzaki A, Theodoropoulou M, Menounos P, Kyrtsonis MC, Karameris A, et al. Endogenous interleukin 6 conveys resistance to cis-dia-mminedichloroplatinum-mediated apoptosis of the K562 human leukemic cell line. Exp Cell Res. 1999;249:269–78. doi: 10.1006/excr.1999.4442. [DOI] [PubMed] [Google Scholar]
- 39.Xie SY, Li YJ, Wang PY, Jiao F, Zhang S, Zhang WJ. miRNA-regulated expression of oncogenes and tumor suppressor genes in the cisplatin-inhibited growth of K562 cells. Oncol Rep. 2010;23:1693–700. doi: 10.3892/or_00000813. [DOI] [PubMed] [Google Scholar]