Abstract
The calcitonin/calcitonin gene-related peptide (CGRP) primary transcript is alternatively spliced in thyroid C cells and neurons, resulting in the tissue-specific production of calcitonin and CGRP mRNAs. Analyses of mutated calcitonin/CGRP transcription units in permanently transfected cell lines have indicated that alternative splicing is regulated by a differential capacity to utilize the calcitonin-specific splice acceptor. The analysis of an extensive series of mutations suggests that tissue-specific regulation of calcitonin mRNA production does not depend on the presence of a single, unique cis-active element but instead appears to be a consequence of suboptimal constitutive splicing signals. While only those mutations that altered constitutive splicing signals affected splice choices, the action of multiple regulatory sequences cannot be formally excluded. Further, we have identified a 13-nucleotide purine-rich element from a constitutive exon that, when placed in exon 4, entirely switches splice site usage in CGRP-producing cells. These data suggest that specific exon recruitment sequences, in combination with other constitutive elements, serve an important function in exon recognition. These results are consistent with the hypothesis that tissue-specific alternative splicing of the calcitonin/CGRP primary transcript is mediated by cell-specific differences in components of the constitutive splicing machinery.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adema G. J., Baas P. D. A novel calcitonin-encoding mRNA is produced by alternative processing of calcitonin/calcitonin gene-related peptide-I pre-mRNA. J Biol Chem. 1992 Apr 15;267(11):7943–7948. [PubMed] [Google Scholar]
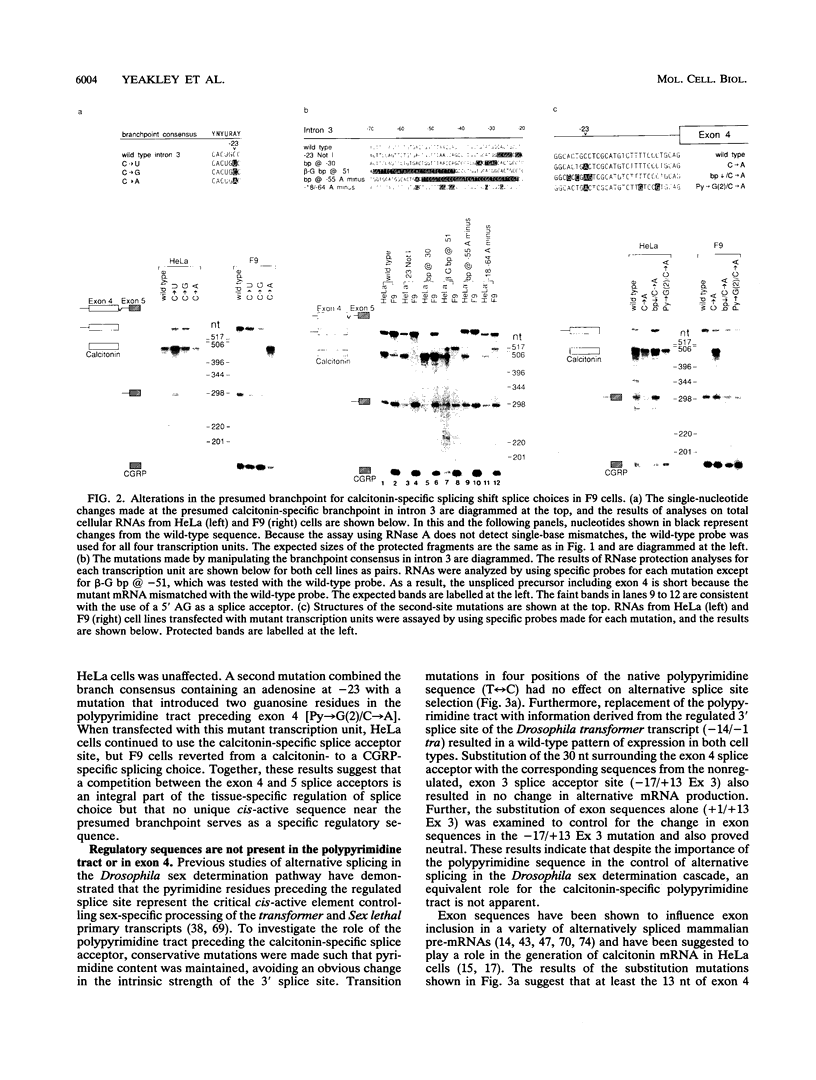
- Adema G. J., Bovenberg R. A., Jansz H. S., Baas P. D. Unusual branch point selection involved in splicing of the alternatively processed Calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 1988 Oct 25;16(20):9513–9526. doi: 10.1093/nar/16.20.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema G. J., van Hulst K. L., Baas P. D. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 1990 Sep 25;18(18):5365–5373. doi: 10.1093/nar/18.18.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara S. G., Arriza J. L., Leff S. E., Swanson L. W., Evans R. M., Rosenfeld M. G. Expression in brain of a messenger RNA encoding a novel neuropeptide homologous to calcitonin gene-related peptide. Science. 1985 Sep 13;229(4718):1094–1097. doi: 10.1126/science.2994212. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Evans R. M., Rosenfeld M. G. Calcitonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol Cell Biol. 1984 Oct;4(10):2151–2160. doi: 10.1128/mcb.4.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Belote J. M., McKeown M., Boggs R. T., Ohkawa R., Sosnowski B. A. Molecular genetics of transformer, a genetic switch controlling sexual differentiation in Drosophila. Dev Genet. 1989;10(3):143–154. doi: 10.1002/dvg.1020100304. [DOI] [PubMed] [Google Scholar]
- Black D. L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991 Mar;5(3):389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- Boden S. D., Kaplan F. S. Calcium homeostasis. Orthop Clin North Am. 1990 Jan;21(1):31–42. [PubMed] [Google Scholar]
- Bovenberg R. A., Adema G. J., Jansz H. S., Baas P. D. Model for tissue specific Calcitonin/CGRP-I RNA processing from in vitro experiments. Nucleic Acids Res. 1988 Aug 25;16(16):7867–7883. doi: 10.1093/nar/16.16.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenberg R. A., Moen T. C., Jansz H. S., Baas P. D. In vitro splicing analysis of mini-gene constructs of the alternatively processed human calcitonin/CGRP-I pre-mRNA. Biochim Biophys Acta. 1989 Jul 7;1008(2):223–233. doi: 10.1016/0167-4781(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Cooper T. A. In vitro splicing of cardiac troponin T precursors. Exon mutations disrupt splicing of the upstream intron. J Biol Chem. 1992 Mar 15;267(8):5330–5338. [PubMed] [Google Scholar]
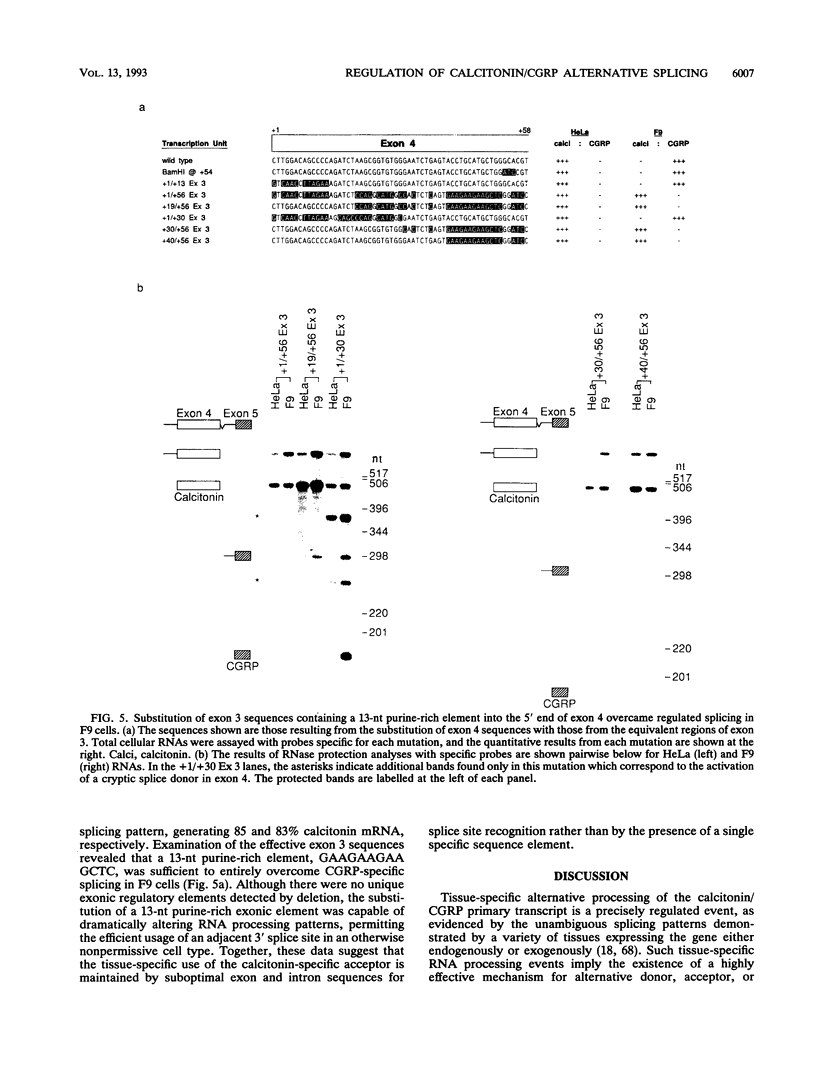
- Cote G. J., Nguyen I. N., Berget S. M., Gagel R. F. Calcitonin exon sequences influence alternative RNA processing. Mol Endocrinol. 1990 Nov;4(11):1744–1749. doi: 10.1210/mend-4-11-1744. [DOI] [PubMed] [Google Scholar]
- Cote G. J., Nguyen I. N., Lips C. J., Berget S. M., Gagel R. F. Validation of an in vitro RNA processing system for CT/CGRP precursor mRNA. Nucleic Acids Res. 1991 Jul 11;19(13):3601–3606. doi: 10.1093/nar/19.13.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote G. J., Stolow D. T., Peleg S., Berget S. M., Gagel R. F. Identification of exon sequences and an exon binding protein involved in alternative RNA splicing of calcitonin/CGRP. Nucleic Acids Res. 1992 May 11;20(9):2361–2366. doi: 10.1093/nar/20.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenshaw E. B., 3rd, Russo A. F., Swanson L. W., Rosenfeld M. G. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987 May 8;49(3):389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- Denis-Donini S. Expression of dopaminergic phenotypes in the mouse olfactory bulb induced by the calcitonin gene-related peptide. Nature. 1989 Jun 29;339(6227):701–703. doi: 10.1038/339701a0. [DOI] [PubMed] [Google Scholar]
- Dietz H. C., Valle D., Francomano C. A., Kendzior R. J., Jr, Pyeritz R. E., Cutting G. R. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993 Jan 29;259(5095):680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol. 1992 May;12(5):2108–2114. doi: 10.1128/mcb.12.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991 Dec;11(12):6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoulin F. L., Raivich G., Haas C. A., Lazar P., Reddington M., Streit W. J., Kreutzberg G. W. Calcitonin gene-related peptide and peripheral nerve regeneration. Ann N Y Acad Sci. 1992 Jun 30;657:351–360. doi: 10.1111/j.1749-6632.1992.tb22782.x. [DOI] [PubMed] [Google Scholar]
- Emeson R. B., Hedjran F., Yeakley J. M., Guise J. W., Rosenfeld M. G. Alternative production of calcitonin and CGRP mRNA is regulated at the calcitonin-specific splice acceptor. Nature. 1989 Sep 7;341(6237):76–80. doi: 10.1038/341076a0. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., O'Brien J. P., Hurwitz J. Alterations in the polypyrimidine sequence affect the in vitro splicing reactions catalyzed by HeLa cell-free preparations. J Biol Chem. 1989 Sep 5;264(25):14631–14637. [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Mayeda A., Maniatis T., Krainer A. R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5' and 3' splice site selection. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H., Zuo P., Manley J. L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991 Jul 26;66(2):373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Graham I. R., Hamshere M., Eperon I. C. Alternative splicing of a human alpha-tropomyosin muscle-specific exon: identification of determining sequences. Mol Cell Biol. 1992 Sep;12(9):3872–3882. doi: 10.1128/mcb.12.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hedley M. L., Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991 May 17;65(4):579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991 May 10;252(5007):833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Hökfelt T., Arvidsson U., Ceccatelli S., Cortés R., Cullheim S., Dagerlind A., Johnson H., Orazzo C., Piehl F., Pieribone V. Calcitonin gene-related peptide in the brain, spinal cord, and some peripheral systems. Ann N Y Acad Sci. 1992 Jun 30;657:119–134. doi: 10.1111/j.1749-6632.1992.tb22762.x. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Higuchi I., Sakamoto H., Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Sakamoto H., Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990 Mar 29;344(6265):461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Mayeda A., Kozak D., Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991 Jul 26;66(2):383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Libri D., Balvay L., Fiszman M. Y. In vivo splicing of the beta tropomyosin pre-mRNA: a role for branch point and donor site competition. Mol Cell Biol. 1992 Jul;12(7):3204–3215. doi: 10.1128/mcb.12.7.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991 Jan 4;251(4989):33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox W., Ryner L., Baker B. S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992 Sep 25;267(27):19023–19026. [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Zahler A. M., Krainer A. R., Roth M. B. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown M. Sex differentiation: the role of alternative splicing. Curr Opin Genet Dev. 1992 Apr;2(2):299–303. doi: 10.1016/s0959-437x(05)80288-6. [DOI] [PubMed] [Google Scholar]
- Mulle C., Benoit P., Pinset C., Roa M., Changeux J. P. Calcitonin gene-related peptide enhances the rate of desensitization of the nicotinic acetylcholine receptor in cultured mouse muscle cells. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5728–5732. doi: 10.1073/pnas.85.15.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen M. P., Smith C. W., Patton J. G., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991 Apr;5(4):642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- Nadal-Ginard B., Smith C. W., Patton J. G., Breitbart R. E. Alternative splicing is an efficient mechanism for the generation of protein diversity: contractile protein genes as a model system. Adv Enzyme Regul. 1991;31:261–286. doi: 10.1016/0065-2571(91)90017-g. [DOI] [PubMed] [Google Scholar]
- Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992 Jun;6(6):1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Niwa M., MacDonald C. C., Berget S. M. Are vertebrate exons scanned during splice-site selection? Nature. 1992 Nov 19;360(6401):277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- Peterson M. L. Balanced efficiencies of splicing and cleavage-polyadenylation are required for mu-s and mu-m mRNA regulation. Gene Expr. 1992;2(4):319–327. [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Rio D. C. RNA binding proteins, splice site selection, and alternative pre-mRNA splicing. Gene Expr. 1992;2(1):1–5. [PMC free article] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos B. A., Yoon M. J., Frelinger A. L., Pensky A. E., Birnbaum R. S., Lambert P. W. Tumor growth and calcitonin during serial transplantation of rat medullary thyroid carcinoma. Endocrinology. 1979 Jul;105(1):27–32. doi: 10.1210/endo-105-1-27. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Amara S. G., Evans R. M. Alternative RNA processing: determining neuronal phenotype. Science. 1984 Sep 21;225(4668):1315–1320. doi: 10.1126/science.6089345. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Amara S. G., Roos B. A., Ong E. S., Evans R. M. Altered expression of the calcitonin gene associated with RNA polymorphism. Nature. 1981 Mar 5;290(5801):63–65. doi: 10.1038/290063a0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Mermod J. J., Amara S. G., Swanson L. W., Sawchenko P. E., Rivier J., Vale W. W., Evans R. M. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983 Jul 14;304(5922):129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- Ryner L. C., Baker B. S. Regulation of doublesex pre-mRNA processing occurs by 3'-splice site activation. Genes Dev. 1991 Nov;5(11):2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- Sosnowski B. A., Belote J. M., McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989 Aug 11;58(3):449–459. doi: 10.1016/0092-8674(89)90426-1. [DOI] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992 Apr 10;256(5054):237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- Uchida S., Yamamoto H., Iio S., Matsumoto N., Wang X. B., Yonehara N., Imai Y., Inoki R., Yoshida H. Release of calcitonin gene-related peptide-like immunoreactive substance from neuromuscular junction by nerve excitation and its action on striated muscle. J Neurochem. 1990 Mar;54(3):1000–1003. doi: 10.1111/j.1471-4159.1990.tb02349.x. [DOI] [PubMed] [Google Scholar]
- Watakabe A., Inoue K., Sakamoto H., Shimura Y. A secondary structure at the 3' splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res. 1989 Oct 25;17(20):8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Tanaka K., Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993 Mar;7(3):407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Lane W. S., Stolk J. A., Roth M. B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992 May;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Neugebauer K. M., Lane W. S., Roth M. B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993 Apr 9;260(5105):219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zaidi M., Moonga B. S., Bevis P. J., Alam A. S., Legon S., Wimalawansa S., MacIntyre I., Breimer L. H. Expression and function of the calcitonin gene products. Vitam Horm. 1991;46:87–164. doi: 10.1016/s0083-6729(08)60683-4. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]