Abstract
Background
Up to 25% of patients discontinue adjuvant aromatase inhibitor (AI) therapy due to intolerable symptoms. Predictors of which patients will be unable to tolerate these medications have not been defined. We hypothesized that inherited variants in candidate genes are associated with treatment discontinuation because of AI-associated toxicity.
Methods
We prospectively evaluated reasons for treatment discontinuation in women with hormone receptor-positive breast cancer initiating adjuvant AI through a multicenter, prospective, randomized clinical trial of exemestane versus letrozole. Using multiple genetic models, we evaluated potential associations between discontinuation of AI therapy because of toxicity and 138 variants in 24 candidate genes, selected a priori, primarily with roles in estrogen metabolism and signaling. To account for multiple comparisons, statistical significance was defined as p<0.00036.
Results
Of the 467 enrolled patients with available germline DNA, 152 (33%) discontinued AI therapy because of toxicity. Using a recessive statistical model, an intronic variant in ESR1 (rs9322336) was associated with increased risk of musculoskeletal toxicity-related exemestane discontinuation (HR 5.0 (95% CI 2.1–11.8), p<0.0002).
Conclusion
An inherited variant potentially affecting estrogen signaling may be associated with exemestane-associated toxicity, which could partially account for intra-patient differences in AI tolerability. Validation of this finding is required.
Keywords: breast cancer, aromatase inhibitor, single nucleotide polymorphism, treatment discontinuation, toxicity
Introduction
The third-generation aromatase inhibitors (AI), anastrozole, exemestane, and letrozole, decrease risk of breast cancer recurrence compared to tamoxifen in postmenopausal women with hormone receptor positive breast cancer.[7] Extended adjuvant AI therapy after 5 years of tamoxifen further decreases recurrence rates compared to placebo.[9] In addition to the selective estrogen receptor modulators (SERM), exemestane has been found to be superior to placebo for prevention of breast cancer in postmenopausal women at increased risk of disease.[8] Since there are now multiple endocrine therapy options for treatment and prevention of breast cancer in postmenopausal women, physicians and patients must weigh the risks and benefits of each therapeutic option when making decisions about choice of therapy.
AIs have a different risk profile than SERMs. In addition to the increased risk of bone fractures and cardiovascular disease, AIs are also associated with bothersome side effects that can lead to intolerance and subsequent discontinuation of treatment.[1, 13] Cross-trial and direct comparisons have demonstrated that all AIs have similar toxicities, especially musculoskeletal and menopausal side effects.[3, 10] These observations suggest the side effects are likely due to a class effect from aromatase inhibition. However, since several reports have suggested that patients who are intolerant to one AI can tolerate a different one, host factors may make a substantial contribution to drug tolerance.[2, 11] The most common toxicity leading to premature discontinuation of AI therapy is the AI-associated musculoskeletal syndrome (AIMSS), which has been reported in up to 25% of patients.[11] Prior studies have implicated multiple clinical factors in development of AIMSS, including age, body mass index, prior taxane chemotherapy, and prior tamoxifen.[5, 11, 18, 25]
In addition to clinical factors, inherited or somatic genetic variants may impact benefit or toxicity from a medication.[28] For example, a possible association between a single nucleotide polymorphism (SNP) in the aromatase gene (CYP19A1) and response to treatment with letrozole in metastatic breast cancer has been identified.[4] Likewise, investigators have reported potential polymorphisms associated with presence of AIMSS, including a SNP in the gene TCL1A identified in a genome-wide association study (GWAS) as well as a variant in CYP19A1.[16, 19, 21] None of these associations has been validated in an independent cohort.
The Consortium on Breast Cancer Pharmacogenomics conducted a prospective randomized clinical trial of exemestane versus letrozole in postmenopausal women with HR positive breast cancer who were initiating adjuvant AI therapy. We prospectively collected whole blood for isolation of germ line DNA, as well as non-cancer clinical endpoints, including patient-reported reasons for treatment discontinuation.[13] For this exploratory endpoint, we hypothesized that we could identify or further assess associations between AI treatment discontinuation due to intolerable symptoms and inherited genetic variants in candidate genes identified because of their potential for involvement in biologically-relevant pathways or through review of the literature.
Materials and Methods
Patients
Postmenopausal women who had hormone receptor (HR)-positive stage 0–III breast cancer and were planning to initiate adjuvant AI therapy were enrolled in the Exemestane and Letrozole Pharmacogenetics (ELPh) clinical trial (clinicaltrials.gov NCT00228956) between August 2005 and July 2009. Detailed eligibility criteria have previously been published.[13] In brief, all recommended surgery, neoadjuvant or adjuvant chemotherapy, and adjuvant radiation therapy were completed prior to enrollment. Prior tamoxifen was permitted, but prior AI therapy was not allowed. The clinical trial was approved by the Institutional Review Boards at all three participating institutions (Indiana University, Johns Hopkins University, University of Michigan), and all enrolled subjects provided written informed consent.
Following enrollment, subjects were randomly assigned to exemestane 25 mg orally daily or letrozole 2.5 mg orally daily. Three subjects withdrew and were not randomized (Figure 1). Randomization was stratified based on prior tamoxifen, chemotherapy, and bisphosphonate therapy. At baseline and after 1, 3, 6, 12, and 24 months of therapy, subjects underwent serial clinical assessments. If subjects discontinued initial AI therapy prior to the 24 month study visit for any reason, reasons for study discontinuation were prospectively recorded on a case report form by the study coordinators.[11]
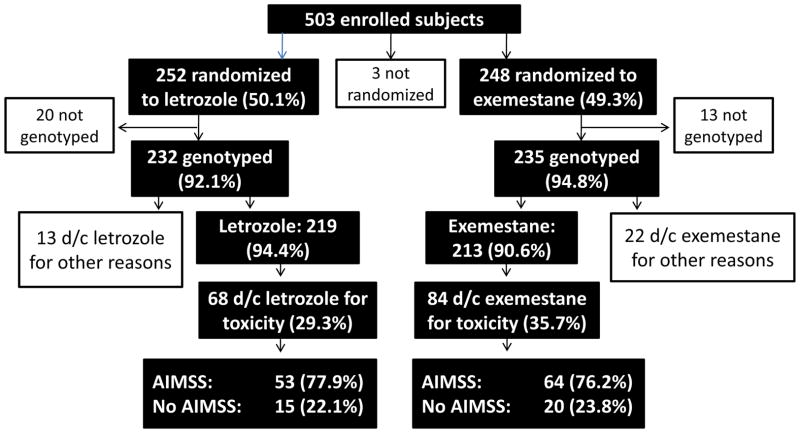
Figure 1.
Consort diagram of patient flow in the study.
Sample processing and genotyping
Whole blood was collected at the baseline study visit from each study participant. DNA was extracted from whole blood using Qiafilter Blood DNA Maxi kit (Qiagen, Inc., Valencia, CA). Germ line DNA was available from 467 (93.4%) eligible subjects (Figure 1). DNA was not available for analysis from the remaining 33 subjects because of technical errors (n=2), inability to obtain blood at baseline (n=8), or insufficient quantity of DNA (n=23).
At the time of protocol development, candidate gene variants were identified that are involved in AI drug metabolism (CYP2A6, CYP3A5), estrogen metabolism (ARVCF, COMT, CYP19A1), estrogen receptor (ER) signaling (ESR1, ESR2, PGR), co-regulation of ER (EP300, EZH2, NCOA1-3, NCOR1-2, NRIP, PELP1), and neurotransmitter and neuropeptide signaling (HTR1A, HTR2A, SCL6A4, HCRT, HCRTR1, HCRTR2). SNPs were included based on their potential functional significance and on mapping of tag SNPs. Prior to analyzing DNA, additional candidate SNPs near the TCL1A gene and in CYP19A1 were identified through ongoing review of the literature.[16, 19] In total, 178 candidate variants in 24 individual genes were identified. Genotype quality control was performed before analysis of the genetic associations. The 37 variants with minor allele frequencies less than 5%, two that did not meet Hardy-Weinberg equilibrium, and one for which genotype could be determined in fewer than 80% of subjects were excluded from analysis. A total of 138 variants in 24 genes were included in the final analysis. The reference SNP numbers, minor allele frequencies, and genotype frequencies for each analyzed SNP are listed in Supplemental Table 1.
Genotyping for all SNPs, except for the TCL1A, CYP3A5, CYP2A6, and two of the CYP19A1 SNPs, was performed using the BioTrove OpenArray™ platform (Applied Biosystems, Inc, Foster City, CA). The TCL1A SNPs were genotyped using individual Taqman® genotyping assays (Applied Biosystems, Inc, Foster City, CA). The assay numbers were C___1927667_10 (rs11849538), C___1927662_10 (rs7159713), C___1927663_20 (rs2369049), and C__29078024_10 (rs7158782).[16] CYP3A5 *3 was genotyped using the Taqman assay (C__26201809_30). CYP2A6 genotyping was performed as previously described.[6] The CYP19A1 TTTA repeat (rs60271534) and TCT deletion (rs11575899) were determined using PCR and direct sequencing, as previously described.[29] For quality control purposes, approximately 10% of the samples were randomly selected and genotyped in duplicate using the same assay, and the overall concordance rate was 97%.
Statistical analysis
The primary endpoint of the ELPh trial was the correlation between change in breast density with 2 years of AI therapy and inherited genetic variants in CYP19, the gene that encodes aromatase. These data will be reported separately. In the current study, the main exploratory objective was the correlation between early treatment discontinuation of AI therapy due to any patient-reported side effect and SNPs in candidate genes. Treatment discontinuation because of toxicity could have resulted from either request by the patient or recommendation of the treating physician. Secondary objectives of this study included (a) correlation between early treatment discontinuation of AI therapy due to AIMSS and SNPs in candidate genes, (b) correlation between early treatment discontinuation of specific AI medications due to any side effect and SNPs in candidate genes, and (c) correlation between early treatment discontinuation of specific AI medications due to AIMSS and SNPs in candidate genes. Discontinuation due to AIMSS was defined as premature discontinuation of AI therapy because of patient-reported intolerable arthralgias, myalgias, joint pain or stiffness, tendinitis, numbness or tingling, and/or carpal tunnel syndrome.
Discontinuation because of either any toxicity or AIMSS was analyzed as a time to event outcome. Step-wise regression was used to estimate the associations between the discontinuation because of either any toxicity or AIMSS and SNPs and other clinical variables. The associations between the SNPs and discontinuation because of either any toxicity or AIMSS were analyzed either without or with justifying for the other variables. Three genetic models, specifically dominant, recessive, and additive, were used to test for the associations between SNPs and treatment discontinuation. Statistical significance was defined as a p value <0.00036 based on Bonferroni correction. Manhattan plots were presented to illustrate the statistical significance and the effect size of the SNPs. All the data analyses were performed in R. The data from this prospective trial are reported according to the REMARK guidelines.[20]
Results
Patient and sample flow
Five hundred and three subjects were enrolled on the ELPh trial (Figure 1). Five hundred were randomized to exemestane (n=248) or letrozole (n=252), and the remaining three subjects withdrew prior to treatment initiation. Germ line DNA was available from 467 (93.4%) eligible subjects. Thirty-five eligible subjects with available DNA discontinued AI therapy for reasons unrelated to toxicity (letrozole, 13 [5.6%] of 252 patients; exemestane, 22 [9.4%] of 248 patients) and were excluded from these analyses.[11] Baseline characteristics for the 432 subjects included in these analyses are listed in Table 1. The reference SNP numbers, minor allele frequencies, and genotype frequencies for each analyzed SNP are listed in Supplemental Table 1.
Table 1.
Baseline patient characteristics for all randomized, genotyped patients, by treatment allocation and by treatment discontinuation.
| All enrolled, genotyped patients (n=467) | Randomized to letrozole (n=232) | Randomized to exemestane (n=235) | Discontinued AI because of symptoms (n=152) | Continued AI (n=280) | |
|---|---|---|---|---|---|
| Median age, yrs (range) | 59 (35–89) | 59 (38–89) | 59 (35–83) | 57.5 (37–83) | 60 (35–84) |
| Race | |||||
| - White | 413 (88.4%) | 203 (87.5%) | 210 (89.4%) | 138 (91.5%) | 247 (88.2%) |
| - Black | 42 (9.0%) | 24 (10.3%) | 18 (7.7%) | 12 (7.4%) | 25 (8.9%) |
| - Other | 12 (2.6%) | 5 (2.2%) | 7 (3.0%) | 2 (1.2%) | 8 (2.9%) |
| Weight, kg (SD) | 79.5 (17.6) | 79.4 (17.9) | 79.7 (17.2) | 78.5 (16.3) | 80.7 (18.0) |
| BMI (SD) | 30.0 (6.4) | 30.1 (6.7) | 29.9 (6.2) | 29.6 (6.1) | 30.4 (6.5) |
| Prior tamoxifen | 169 (36.2%) | 85 (36.6%) | 84 (35.7%) | 59 (38.9%) | 96 (34.3%) |
| Prior HRT | 227 (48.6%) | 105 (45.3%) | 122 (51.9%) | 75 (48.8%) | 145 (51.8%) |
| Prior chemotherapy | 212 (45.4%) | 104 (44.8%) | 108 (46.0%) | 68 (44.4%) | 124 (44.3%) |
| Prior taxane | 153 (32.8%) | 77 (33.2%) | 76 (32.3%) | 55 (35.8%) | 85 (30.4%) |
| Mean time since chemotherapy, yrs (SD) | 1.7 (1.9) | 1.6 (1.8) | 1.8 (1.9) | 1.6 (1.7) | 1.7 (2.0) |
| Assigned AI | |||||
| - Letrozole | 232 (49.7%) | 232 | 0 | 68 (44.7%) | 151 (53.9%) |
| - Exemestane | 235 (50.3%) | 0 | 235 | 84 (55.3%) | 129 (46.1%) |
AI=aromatase inhibitor; HRT=hormone replacement therapy; Kg=kilograms; N=number; SD =Standard deviation; yrs=years
We previously reported treatment discontinuation rates for this entire patient cohort.[11] Of those with available germ line DNA, 152 (32.5%) subjects discontinued the AI because of toxicity. Of the 235 patients randomized to exemestane who had DNA available for genotyping, 84 (35.7%) discontinued therapy because of toxicity. Of the 232 patients randomized to letrozole who had DNA available, 68 (29.3%) discontinued therapy because of toxicity. There was a trend towards increased treatment discontinuation in exemestane-treated patients (p=0.08). Of the patients who discontinued therapy because of toxicity, 64 (76.2%) of exemestane-treated patients and 53 (77.9%) letrozole-treated patients discontinued therapy specifically because of AIMSS, a difference that was not statistically significant (p=0.21).
Associations between variants in candidate genes and discontinuation of AI therapy
Univariate analyses
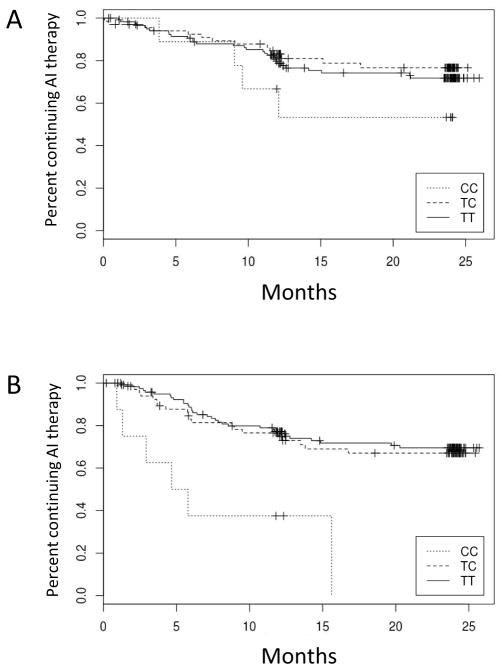
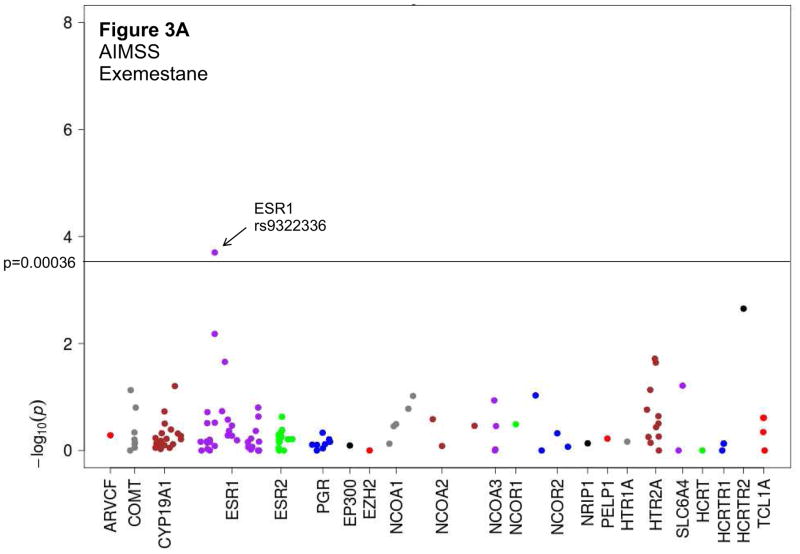
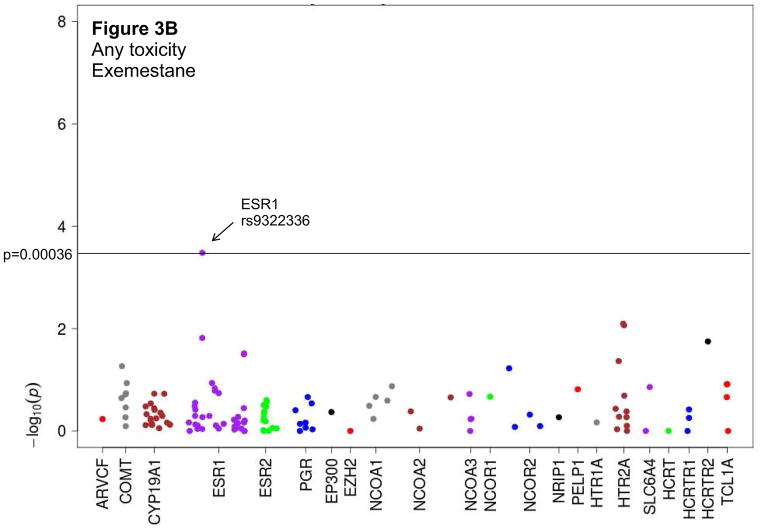
We evaluated associations between candidate SNPs and discontinuation of AI therapy because of either any toxicity or AIMSS (Supplemental Figure 1, Supplemental Table 2). When assessed using a recessive model, an intronic variant in the estrogen receptor ESR1 (rs9322336) was associated with increased risk of discontinuation of exemestane therapy because of AIMSS ((HR 5.0 (95% CI 2.2–11.8), p=0.0002); Table 2, Figures 2A and 3A). Analyses also demonstrated a trend towards an association between the same SNP in ESR1 and increased risk of discontinuation of exemestane therapy because of any toxicity ((HR 4.2 (95% CI 1.9–9.2), p=0.0003); Table 2, Figure 3B). A similar trend was not detected for letrozole, suggesting a drug-specific association (Table 2, Figure 2B). However, also after Bonferroni correction and using any of the genetic models, we failed to observe any statistically significant potential associations between the other candidate genetic variants and discontinuation of AI therapy because of either any toxicity or AIMSS (Supplemental Table 3).
Table 2.
Associations between ESR1 single nucleotide polymorphism rs9322336 and toxicity-related treatment discontinuation using a recessive statistical model.
| Drug | Reason for discontinuation | Genotype | Number patients discontinued AI/total with genotype | HR (95% CI) | P value |
|---|---|---|---|---|---|
| Both AIs | Any toxicity | All combined | 141/396 (35.6%) | 2.4 (1.3–4.5) | 0.005 |
| TT | 82/241 (34.0%) | ||||
| TC | 48/137 (35.0%) | ||||
| CC | 11/18 (61.1%) | ||||
| AIMSS | All combined | 107/396 (27.0%) | 3.0 (1.6–5.7) | 0.001 | |
| TT | 63/241 (26.1%) | ||||
| TC | 34/137 (24.8%) | ||||
| CC | 10/18 (55.6%) | ||||
| Letrozole | Any toxicity | All combined | 63/198 (31.8%) | 1.5 (0.5–4.1) | 0.44 |
| TT | 39/121 (32.2%) | ||||
| TC | 20/68 (29.4%) | ||||
| CC | 4/9 (44.4%) | ||||
| AIMSS | All combined | 48/198 (24.2%) | 2.0 (0.7–5.6) | 0.18 | |
| TT | 30/121 (24.8%) | ||||
| TC | 14/68 (20.6%) | ||||
| CC | 4/9 (44.4%) | ||||
| Exemestane | Any toxicity | All combined | 78/198 (39.4%) | 4.2 (1.9–9.2) | 0.0003 |
| TT | 43/120 (35.8%) | ||||
| TC | 28/69 (40.6%) | ||||
| CC | 7/9 (77.8%) | ||||
| AIMSS | All combined | 59/198 (29.8%) | 5.0 (2.1–11.8) | 0.0002 | |
| TT | 33/120 (27.5%) | ||||
| TC | 20/69 (29.0%) | ||||
| CC | 6/9 (66.7%) |
Number of patients in total cohort or divided by aromatase inhibitor (AI) who discontinued therapy because of toxicity is given by genotype. Hazard ratios (HR) and corresponding p values are given for both aromatase inhibitors (AI) combined and for each AI individually. AIMSS: AI-associated musculoskeletal syndrome; CI: confidence interval.
Figure 2. Kaplan Meier plot of associations between ESR1 rs9322336 SNP and aromatase inhibitor (AI) discontinuation because of AI-associated musculoskeletal syndrome.
Percentage of subjects continuing AI therapy are given over time for subjects with CC, TC, or TT genotypes. (A) Letrozole-treated subjects. (B) Exemestane-treated subjects. Analysis was performed using a recessive statistical model, and P values were adjusted for age > 55, AI medication, and prior taxane therapy.
Figure 3. Manhattan plots depicting associations between candidate SNPs and treatment discontinuation using a recessive statistical model.
Dashed line signifies level of statistical significance after Bonferroni correction (p=0.00036). AIMSS: Aromatase inhibitor-associated musculoskeletal syndrome; HR: hazard ratio. A. Treatment discontinuation because of AIMSS for patients treated with exemestane. B. Treatment discontinuation because of any toxicity for patients treated with exemestane.
Multivariate analyses
Step-wise regression was performed to identify clinical covariates that were associated with treatment discontinuation. As previously reported, age < 55 and AI medication were found to be statistically significantly associated with discontinuation due to any toxicity.[11] Similarly, age < 55, prior taxane chemotherapy, and AI medication were statistically significantly associated with discontinuation due to AIMSS in our study. After adjustment for the covariates, there remained a trend towards an association between the rs9322336 ESR1 SNP and treatment discontinuation because of AIMSS, with a hazard ratio of 3.2 (95% CI 1.6–6.2), p=0.0006. As shown in Supplemental Tables 4 and 5, no other genetic variants were statistically significantly associated with treatment discontinuation in the multivariate analysis.
Other candidate gene variants
Other authors have reported a non-significant putative association between 4 SNPs in linkage disequilibrium near the TCL1A gene, identified by GWAS, and increased musculoskeletal toxicity (odds ratios 2.09–2.21) in patients who were treated with anastrozole or exemestane on a prospective randomized controlled trial (MA27), using a gene-dose model.[16] We therefore analyzed the association between the SNPs near TCL1A and discontinuation of AI therapy in the ELPh trial (Supplemental Tables 3–5). Using a gene-dose model, in contrast to the findings in MA27 we observed just the opposite, a non-statistically significant trend towards an association between decreased likelihood of discontinuation of either AI because of toxicity and two of the SNPs, the imputed SNP (rs11849538) and a second SNP (rs2369049), with hazard ratios of 0.6 (95% CI 0.4–0.9, p=0.007) and 0.6 (95% CI 0.4–0.8, p=0.002), respectively. Using the same statistical model, there was a trend towards an association between decreased likelihood of discontinuation of exemestane because of toxicity and the same two SNPs (rs11849538 and rs2369049) with hazard ratios of 0.5 (95% CI 0.3–0.8, p=0.009) and 0.5 (95% CI 0.3–0.8, p=0.006), respectively. A similar trend was not seen for letrozole-treated patients.
Using data from a cross-sectional trial of women with AI-associated arthralgias, others reported an association between presence of at least one 8-repeat TTTAn allele in intron 4 of the aromatase gene (rs60271534) and decreased likelihood of reporting AI-associated arthralgias (adjusted odds ratio 0.41 (95% CI 0.21–0.79); p=0.008).[19] In contrast, we observed a non-statistically significant increase in AIMSS when at least one 8-repeat allele was present (HR 1.8 (95% CI 0.8–1.8); p=0.49). In the letrozole-treated subjects the hazard ratio was 1.8 (95% CI 0.96–3.3, p=0.065), whereas in the exemestane-treated subjects the hazard ratio was 0.8 (95% CI 0.4–1.5, p=0.44). Similarly, no associations were noted for other TTTA repeat lengths.
Discussion
In this study we identified a potentially clinically important association between an inherited germ line genetic variation in the gene encoding ER alpha and discontinuation of AI therapy primarily because of musculoskeletal toxicity in postmenopausal women with hormone receptor positive, early stage breast cancer. Musculoskeletal side effects have previously been found to contribute to non-persistence with AI therapy, which can negatively impact breast-cancer outcomes.[11] In addition, this association appears to be drug-specific, which could at least partially account for differences in inter-patient and intra-patient tolerability of the AIs.[2, 11]
Identification of patients who are more likely to experience intolerable AI-associated toxicity because of inherited genetic variants could permit better treatment-decision making for women with breast cancer who are starting adjuvant endocrine therapy. For example, if presence of a SNP predicts development of toxicity from exemestane, as may be the case for the SNP in ESR1, it could potentially be used to select treatment with a specific AI. In addition, if a patient population could be identified that is at high risk for development of a specific toxicity, such as AIMSS, this cohort could be targeted for interventional trials to prevent or reduce the burden of such toxicities. In this regard, we have reported a pilot phase II trial suggesting duloxetine reduces pain related to AIMSS by approximately 60%.[12] If validated, the association between AIMSS and the SNP in ESR1 could be used to personalize this strategy.
The mechanism by which the SNP identified in our study contributes to the development of AI-associated toxicity is unclear. We are only aware of a single report regarding this intronic ESR1 SNP (rs9322336): a possible association with increased risk of ovarian cancer.[23] However, estrogen receptors have been shown to be present in multiple cell types in the central nervous system, and estrogen has been shown to have antinociceptive effects.[17, 27] Estrogen also has anti-inflammatory effects, and pro-inflammatory cytokines can increase during menopause,[22] although studies of AIMSS to date have not demonstrated a direct effect of AI therapy on systemic inflammatory cytokine concentrations.[14] However, it is possible that estrogen is exerting an effect locally, such as within bone, joints, or the central nervous system. Since the effect that we identified appears to be primarily in exemestane-treated patients, it is also possible that the effect of the SNP could relate to an off-target effect of exemestane, rather than a direct effect on ER function.
Other associations between genetic variants in candidate genes and AI-associated musculoskeletal symptoms have been reported. In a cross-sectional study of patients receiving AI therapy, Mao et al explored potential associations between 5 polymorphisms in the aromatase gene (CYP19) and development of AI-associated arthralgias.[19] They found that carriers of at least one 8 (TTTA)n-repeat in intron 4 had decreased risk of musculoskeletal toxicity (adjusted odds ratio 0.41). We were unable to confirm this finding in our cohort. One key reason why we may have been unable to validate the previous finding is the difference in phenotypic endpoint, since our study prospectively evaluated treatment discontinuation due to AIMSS, whereas the endpoint in the study by Mao et al was a cross-sectional evaluation of patient-reported arthralgias. In addition, the possible effect observed in the ELPh trial was more prominent in letrozole-treated subjects. In contrast, the majority of subjects in the cross-sectional study were treated with anastrozole, and only 20% received letrozole. This possible drug-specific genetic association could also result in discordant findings between the two studies.
We also failed to validate the previously reported finding of a potential association between SNPs in TCL1A and lower toxicity.[16] Since the findings in neither study met the stringent criteria for statistical significance, both observations may represent the play of chance. In addition, the phenotypic endpoints were not identical. In the ELPh study we evaluated discontinuation of AI therapy because of intolerable toxicity, whereas in MA.27 the researchers evaluated a composite endpoint of CTCAE grade 3 or 4 musculoskeletal toxicity or treatment discontinuation because of musculoskeletal toxicity.[16] Futhermore, the genome-wide association study (GWAS) approach taken in the MA.27 study is more prone to false discovery than the more limited candidate gene strategy used in our study. Regardless, a recently reported analysis from the randomized Arimidex, Tamoxifen, Alone, or in Combination (ATAC) Trial, revealed no association between the rs11849538 TCL1A variant and CTCAE-graded musculoskeletal toxicity.[24] Taken together, these data do not strongly suggest that this SNP is involved in the development of AI-associated musculoskeletal toxicity.
The strength of our observations is supported by the derivation of the data from a prospective, randomized clinical trial of patients treated with AIs from two different classes (steroidal and non-steroidal). Furthermore, reasons for treatment discontinuation were prospectively recorded. We were therefore able to evaluate associations between inherited genetic variants involved in estrogen metabolism and activity, and reasonably accurate measures of discontinuation of AI therapy because of either any drug-associated toxicity or, specifically, AIMSS.
Nonetheless, given the large number of gene variants that we analyzed, we applied a stringent statistical threshold in order to account for multiple comparisons. These corrections in this relatively small data set (n=467) limit our ability to detect statistically significant associations.
An additional limitation is the lack of a standardized definition of AIMSS, and therefore absence of objective criteria on which to base decisions to continue or discontinue therapy. Pain is an inherently subjective measure and can have a myriad of etiologies. Therefore, defining the primary endpoint as change in pain score between baseline and on-treatment study visit can be confounded by multiple factors including recent breast cancer surgery or chemotherapy (at baseline), lymphedema exacerbations, and pre-existing arthritis. To minimize these coufounding factors, we chose to use persistence of patient-reported symptoms despite conservative symptom management as the criterion on which to base treatment discontinuation decisions.
Non-adherence to therapy could confound our findings. Patients were queried about adherence to therapy at each study visit, but no formal adherence data were obtained. The majority of patients reported high levels of adherence to therapy, which was provided free of charge by the study. Because of the predefined cross-over in the event of treatment-emergent side effects from one treatment to the other with continued clinical trial participation, as well as the patient-reported high adherence to therapy, we do not believe that poor adherence likely substantially impacts our results.
In summary, discontinuation of AI therapy is a clinically important problem for women with ER-positive breast cancer, since decreased persistence with or adherence to AI therapy leads to worse breast cancer outcomes.[15] The findings presented here suggest that variants in the biologically-relevant ESR1 gene may be associated with premature discontinuation of AI therapy because of musculoskeletal toxicity. Ability to identify patients using genetic testing who are at increased risk of developing toxicity may enable improved patient management. However, confirmation of this result in an independent cohort is important before it can be incorporated into clinical care.[26]
Supplementary Material
Acknowledgments
We thank the patients who participated in the study, and the treating physicians, research nurses and data managers at the three sites.
Supported in part by Pharmacogenetics Research Network Grant # U-01 GM61373 (DAF) and Clinical Pharmacology training grant: 5T32-GM08425 (DAF) from the National Institute of General Medical Sciences, National Institutes of Health (NIH), Bethesda, MD and by grant numbers M01-RR000042 (UM), M01-RR00750 (IU), and M01-RR00052 (JHU) from the National Center for Research Resources (NCRR), a component of the NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. NLH is a Damon Runyon-Lilly Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI#53-10). In addition, these studies were supported by grants from Pfizer, Inc. (DFH), Novartis Pharma AG (DFH), and the Fashion Footwear Association of New York/QVC Presents Shoes on Sale™ (DFH). Study medication was provided by Pfizer, Inc. and Novartis Pharma AG.
Footnotes
The results of this study were presented in part in a poster discussion session at the 2012 American Society of Clinical Oncology Annual Meeting, Chicago, IL, June 1–5, 2012.
Conflicts of Interest
NLH receives research funding from AstraZeneca, Eli Lilly, and Sanofi Aventis. DFH receives research funding from AstraZeneca, Novartis, Pfizer, Veridex, and Janssen. DAF receives research funding from Novartis and Pfizer. VS receives research funding from Abbott, Abraxis, Merck, Novartis and Pfizer. JMR received a research grant from Pfizer. TCS, JD, LL, KK, CG, ATN, ZD, SO, SP, JSC, and AMS reported no conflicts of interest.
References
- 1.Amir E, Seruga B, Niraula S, Carlsson L, Ocana A. Toxicity of adjuvant endocrine therapy in postmenopausal breast cancer patients: a systematic review and meta-analysis. J Natl Cancer Inst. 2011;103(17):1299–1309. doi: 10.1093/jnci/djr242. [DOI] [PubMed] [Google Scholar]
- 2.Briot K, Tubiana-Hulin M, Bastit L, Kloos I, Roux C. Effect of a switch of aromatase inhibitors on musculoskeletal symptoms in postmenopausal women with hormone-receptor-positive breast cancer: the ATOLL (articular tolerance of letrozole) study. Breast Cancer Res Treat. 2010;120(1):127–134. doi: 10.1007/s10549-009-0692-7. [DOI] [PubMed] [Google Scholar]
- 3.Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, Giordano SH, Hudis CA, Malin J, Mamounas EP, Rowden D, Solky AJ, Sowers MR, Stearns V, Winer EP, Somerfield MR, Griggs JJ. American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J Clin Oncol. 2010;28(23):3784–3796. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colomer R, Monzo M, Tusquets I, Rifa J, Baena JM, Barnadas A, Calvo L, Carabantes F, Crespo C, Munoz M, Llombart A, Plazaola A, Artells R, Gilabert M, Lloveras B, Alba E. A single-nucleotide polymorphism in the aromatase gene is associated with the efficacy of the aromatase inhibitor letrozole in advanced breast carcinoma. Clin Cancer Res. 2008;14(3):811–816. doi: 10.1158/1078-0432.CCR-07-1923. [DOI] [PubMed] [Google Scholar]
- 5.Crew KD, Greenlee H, Capodice J, Raptis G, Brafman L, Fuentes D, Sierra A, Hershman DL. Prevalence of joint symptoms in postmenopausal women taking aromatase inhibitors for early-stage breast cancer. J Clin Oncol. 2007;25(25):3877–3883. doi: 10.1200/JCO.2007.10.7573. [DOI] [PubMed] [Google Scholar]
- 6.Desta Z, Kreutz Y, Nguyen AT, Li L, Skaar T, Kamdem LK, Henry NL, Hayes DF, Storniolo AM, Stearns V, Hoffmann E, Tyndale RF, Flockhart DA. Plasma letrozole concentrations in postmenopausal women with breast cancer are associated with CYP2A6 genetic variants, body mass index, and age. Clin Pharmacol Ther. 2011;90(5):693–700. doi: 10.1038/clpt.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowsett M, Cuzick J, Ingle J, Coates A, Forbes J, Bliss J, Buyse M, Baum M, Buzdar A, Colleoni M, Coombes C, Snowdon C, Gnant M, Jakesz R, Kaufmann M, Boccardo F, Godwin J, Davies C, Peto R. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J Clin Oncol. 2010;28(3):509–518. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 8.Goss PE, Ingle JN, Ales-Martinez JE, Cheung AM, Chlebowski RT, Wactawski-Wende J, McTiernan A, Robbins J, Johnson KC, Martin LW, Winquist E, Sarto GE, Garber JE, Fabian CJ, Pujol P, Maunsell E, Farmer P, Gelmon KA, Tu D, Richardson H. Exemestane for breast-cancer prevention in postmenopausal women. N Engl J Med. 2011;364(25):2381–2391. doi: 10.1056/NEJMoa1103507. [DOI] [PubMed] [Google Scholar]
- 9.Goss PE, Ingle JN, Martino S, Robert NJ, Muss HB, Piccart MJ, Castiglione M, Tu D, Shepherd LE, Pritchard KI, Livingston RB, Davidson NE, Norton L, Perez EA, Abrams JS, Therasse P, Palmer MJ, Pater JL. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349(19):1793–1802. doi: 10.1056/NEJMoa032312. [DOI] [PubMed] [Google Scholar]
- 10.Goss PE, Ingle JN, Pritchard KI, Ellis MJ, Sledge GW, Budd GT, Rabaglio M, Ansari RH, Johnson DB, Tozer R, D’Souza DP, Chalchal H, Spadafora S, Stearns V, Perez EA, Liedke PE, Lang I, Elliott C, Gelmon KA, Chapman JA, Shepherd LE. Exemestane versus anastrozole in postmenopausal women with early breast cancer: NCIC CTG MA.27--a randomized controlled phase III trial. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.44.7805. epub Feb 4 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry NL, Azzouz F, Desta Z, Li L, Nguyen AT, Lemler S, Hayden J, Tarpinian K, Yakim E, Flockhart DA, Stearns V, Hayes DF, Storniolo AM. Predictors of aromatase inhibitor discontinuation due to treatment-emergent symptoms in early-stage breast cancer. J Clin Oncol. 2012;30(9):936–942. doi: 10.1200/JCO.2011.38.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry NL, Banerjee M, Wicha M, Van Poznak C, Smerage JB, Schott AF, Griggs JJ, Hayes DF. Pilot study of duloxetine for treatment of aromatase inhibitor-associated musculoskeletal symptoms. Cancer. 2011;117(24):5469–5475. doi: 10.1002/cncr.26230. [DOI] [PubMed] [Google Scholar]
- 13.Henry NL, Giles JT, Ang D, Mohan M, Dadabhoy D, Robarge J, Hayden J, Lemler S, Shahverdi K, Powers P, Li L, Flockhart D, Stearns V, Hayes DF, Storniolo AM, Clauw DJ. Prospective characterization of musculoskeletal symptoms in early stage breast cancer patients treated with aromatase inhibitors. Breast Cancer Res Treat. 2008;111(2):365–372. doi: 10.1007/s10549-007-9774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry NL, Pchejetski D, A’Hern R, Nguyen AT, Charles P, Waxman J, Li L, Storniolo AM, Hayes DF, Flockhart DA, Stearns V, Stebbing J. Inflammatory cytokines and aromatase inhibitor-associated musculoskeletal syndrome: a case-control study. Br J Cancer. 2010;103(3):291–296. doi: 10.1038/sj.bjc.6605768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershman DL, Kushi LH, Shao T, Buono D, Kershenbaum A, Tsai WY, Fehrenbacher L, Gomez SL, Miles S, Neugut AI. Early discontinuation and nonadherence to adjuvant hormonal therapy in a cohort of 8,769 early-stage breast cancer patients. J Clin Oncol. 2010;28(27):4120–4128. doi: 10.1200/JCO.2009.25.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, Chapman JA, Kubo M, Jenkins GD, Batzler A, Shepherd L, Pater J, Wang L, Ellis MJ, Stearns V, Rohrer DC, Goetz MP, Pritchard KI, Flockhart DA, Nakamura Y, Weinshilboum RM. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol. 2010;28(31):4674–4682. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggi A, Ciana P, Belcredito S, Vegeto E. Estrogens in the nervous system: mechanisms and nonreproductive functions. Annu Rev Physiol. 2004;66:291–313. doi: 10.1146/annurev.physiol.66.032802.154945. [DOI] [PubMed] [Google Scholar]
- 18.Mao JJ, Stricker C, Bruner D, Xie S, Bowman MA, Farrar JT, Greene BT, Demichele A. Patterns and risk factors associated with aromatase inhibitor-related arthralgia among breast cancer survivors. Cancer. 2009;115(16):3631–3639. doi: 10.1002/cncr.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao JJ, Su HI, Feng R, Donelson ML, Aplenc R, Rebbeck TR, Stanczyk F, Demichele A. Association of functional polymorphisms in CYP19A1 with aromatase inhibitor associated arthralgia in breast cancer survivors. Breast Cancer Res. 2011;13(1):R8. doi: 10.1186/bcr2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93(4):387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park IH, Lee YS, Lee KS, Kim SY, Hong SH, Jeong J, Lee H, Ro J, Nam BH. Single nucleotide polymorphisms of CYP19A1 predict clinical outcomes and adverse events associated with letrozole in patients with metastatic breast cancer. Cancer Chemother Pharmacol. 2011;68(5):1263–1271. doi: 10.1007/s00280-011-1615-y. [DOI] [PubMed] [Google Scholar]
- 22.Pfeilschifter J, Koditz R, Pfohl M, Schatz H. Changes in proinflammatory cytokine activity after menopause. Endocr Rev. 2002;23(1):90–119. doi: 10.1210/edrv.23.1.0456. [DOI] [PubMed] [Google Scholar]
- 23.Quaye L, Tyrer J, Ramus SJ, Song H, Wozniak E, DiCioccio RA, McGuire V, Hogdall E, Hogdall C, Blaakaer J, Goode EL, Schildkraut JM, Easton DF, Kruger-Kjaer S, Whittemore AS, Gayther SA, Pharoah PD. Association between common germline genetic variation in 94 candidate genes or regions and risks of invasive epithelial ovarian cancer. PLoS One. 2009;4(6):e5983. doi: 10.1371/journal.pone.0005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rae JM, Sestak I, Henry NL, Drury S, Hayes DF, Thibert JN, Lopez-Knowles E, Salter J, Pineda S, Cuzick J, Dowsett M. Correlation between gene variants in CYP19 (Aromatase) and TCL1A with disease and tolerability endpoints in the ATAC trial. Cancer Res. 2011;71(24 Suppl):P1-06-02. [Google Scholar]
- 25.Sestak I, Cuzick J, Sapunar F, Eastell R, Forbes JF, Bianco AR, Buzdar AU. Risk factors for joint symptoms in patients enrolled in the ATAC trial: a retrospective, exploratory analysis. Lancet Oncol. 2008;9(9):866–872. doi: 10.1016/S1470-2045(08)70182-7. [DOI] [PubMed] [Google Scholar]
- 26.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith YR, Stohler CS, Nichols TE, Bueller JA, Koeppe RA, Zubieta JK. Pronociceptive and antinociceptive effects of estradiol through endogenous opioid neurotransmission in women. J Neurosci. 2006;26(21):5777–5785. doi: 10.1523/JNEUROSCI.5223-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woods NF, Mitchell ES, Tao Y, Viernes HM, Stapleton PL, Farin FM. Polymorphisms in the estrogen synthesis and metabolism pathways and symptoms during the menopausal transition: observations from the Seattle Midlife Women’s Health Study. Menopause. 2006;13(6):902–910. doi: 10.1097/01.gme.0000227058.70903.9f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.