Abstract
Global dengue virus spread in tropical and sub-tropical regions has become a major international public health concern. It is evident that DENV genetic diversity plays a significant role in the immunopathology of the disease and that the identification of polymorphisms associated with adaptive responses is important for vaccine development. The investigation of naturally occurring genomic variants may play an important role in the comprehension of different adaptive strategies used by these mutants to evade the human immune system. In order to elucidate this role we sequenced the complete polyprotein-coding region of thirty-three DENV-3 isolates to characterize variants circulating under high endemicity in the city of São José de Rio Preto, Brazil, during the onset of the 2006-07 epidemic. By inferring the evolutionary history on a local-scale and estimating rates of synonymous (dS) and nonsynonimous (dN) substitutions, we have documented at least two different introductions of DENV-3 into the city and detected 10 polymorphic codon sites under significant positive selection (dN/dS > 1) and 8 under significant purifying selection (dN/dS < 1). We found several polymorphic amino acid coding sites in the envelope (15), NS1 (17), NS2A (11), and NS5 (24) genes, which suggests that these genes may be experiencing relatively recent adaptive changes. Furthermore, some polymorphisms correlated with changes in the immunogenicity of several epitopes. Our study highlights the existence of significant and informative DENV variability at the spatio-temporal scale of an urban outbreak.
Introduction
Dengue viruses (genus flavivirus, family Flaviviridae) are responsible for dengue fever and severe dengue (previously known as dengue hemorrhagic fever, DHF and dengue shock syndrome, DSS), they are some of the most important arthropod-borne viral diseases worldwide [1]–[4]. Increasing incidences of transmission in urban and semi-urban areas in the tropical and sub-tropical countries have promoted dengue to a major public health issue with a hefty economic burden [5]–[10].
Dengue epidemics are largely attributed to three factors: (i) Increased urbanization, (ii) global dissemination of the major mosquito vectors, Aedes aegypti and Aedes albopictus, and (iii) the interaction and evolution of the four DENV serotypes [11]. Inadequate sustained vector control may partially explain unsuccessful disease prevention in developing countries however variables related to the viruses, the human host and the environment should also be considered [12]. Vaccine development is currently underway as a strategy for controlling this worldwide health threat [13]–[15]. Due to the complexity of DENV dynamics, prevailing serotypes and differences in distribution of viruses within different regions of the globe, serotype and lineage replacement events may play an important role in the evolution of DENV viruses and, consequently must be addressed during vaccine development [16].
Gene genealogy-based DENV evolutionary studies helped characterize the genetic diversity and distribution of different serotypes/genotypes in space and time [17]–[22]. Also, they are important for exploring the role of selection on particular dengue proteins [23]–[29]. Given that serotype-specific neutralizing antibodies confer limited, if any, protection from subsequent infection by the other three serotypes, the appropriate choice of nucleotide residues in the variable genomic regions is critical for the design of effective tetravalent vaccines [30], [31].
Brazil is the South American country with the highest economic impact of dengue and also accounts for the majority of reported cases in the continent as evidenced by a 27 year-long study [8], [32]. The highest dengue incidence was among young adults from 2000 to 2007. However, in 2006 the highest incidence rate of severe dengue increased dramatically among children and remained high during severe epidemics in 2007 and 2008 in the state of Rio de Janeiro [32]–[34]. These circumstances highlight the urgent need for continued studies on dengue molecular epidemiology in order to help determine appropriate policies and effective public health campaigns.
The municipality of São José de Rio Preto (SJRP), one of the 645 municipalities within the State of São Paulo according to the Brazilian Institute of Geography and Statistics (Instituto Brasileiro de Geografia e Estatística, IBGE), has a tropical savanna climate (Köppen climate classification), with an annual rainfall of 1410 mm and mean annual temperature of 23.6°C, making it a favorable breeding ground for Aedes mosquitoe vectors. Our previous study on dengue in SJRP [35] using a 399 bp-long portion of the NS5 gene, suggested that DENV-3 was introduced into the city in 2005. Since SJRP is one of the municipalities in São Paulo with the highest number of dengue cases reported yearly according to the Epidemiological Surveillance Center of the state (Centro de Vigilância epidemiológica Alexandre Vranjac, CVE). We chose to further characterize the isolated DENV-3 strains from SJRP from 2006 to 2007 to gain insights into the viral epidemiology that may help control disease.
Materials and Methods
Area of study
The city of SJRP is located in the northwestern region of the state of São Paulo, Brazil (20°49′11″ S and 49°22′46″ W), with a total area of 431 km2 and an estimated population of 415,769 inhabitants in 2012 (Data obtained from the IBGE). The infestation by Aedes mosquitoes was reported for the first time in 1985 and the first autochthonous cases were reported in 1990, with the introduction of DENV-1 in 1996 (Chiaravalloti-Neto, Superintendence of Endemic Disease Control, SUCEN: personal communication). Unpublished reports by the Instituto Adolfo Lutz (Brazilian Public Health Laboratory) point to the introduction of DENV-2 in 1998. Dengue viruses have been endemic in the municipality for almost ten years [36].
Sequencing genomes
A total of 33 plasma samples were collected from dengue patients during the 2006 and 2007 outbreaks as described elsewhere [35]. Viral RNA was extracted directly from plasma using the QIAamp Viral RNA mini kit (Qiagen). cDNA was synthetized in a 20 µl reverse transcription reaction containing: 1 µl of Superscript III Reverse Transcriptase (Invitrogen), 1 µl of random hexamers (50 ng/µl stock), 1 µL of specific reverse primer 5′AGAACCTGTTGATTCAACAGCAC3′ (10 µM stock) and 5 µl of template RNA. Viral cDNA were diluted to 800 µl in DEPC-treated water and 20 µl were used as template for a 96 specific PCR reactions. Each 10 µl PCR reaction contained: 3 µl of template, 0.03 µl of pfuUltra II polymerase 1 (5 U/µl) (Stratagene), 100 mM dNTPs (Applied Biosystems) and 4 µl of a mixture of forward and reverse primers (0.5 µM stock) (Table S1). Primers were synthesized with M13 sequence tags (forward primers with 5′GTAAAACGACGGCCAGT3′ and reverse primers with 5′CAGGAAACAGCTATGACC3′) so that PCR amplicons could be sequenced using universal M13 forward and reverse primers. Each PCR reaction produced 96 overlapping amplicons, each 500–900 nucleotides in length, which were subsequently sequenced bidirectionally using the Big Dye chemistry on ABI3730xl DNA sequencers (Applied Biosystems). The 33 obtained genomic sequences were submitted to the Broad Institute genome project and deposited in GenBank (Table S2 shows all GenBank accession numbers and name of all samples). (http://www.broadinstitute.org/annotation/viral/Dengue/).
Phylogenetic Inference
The data set includes the obtained 33 genomic sequences from SJRP that were aligned together with 33 DENV-3 worldwide genomic sequences deposited in GenBank (Table 1) using the Muscle 3.8.31 program [37], [38]. Visual inspection of the alignment and manual editing were done with the Se-Al v2.0 program (http://tree.bio.ed.ac.uk/software/seal/). A Bayesian maximum clade credibility tree was inferred from a set of plausible trees sampled at the stationary phase of four independent Markov Chain Monte Carlo (MCMC) runs with 40 million generations each using a general time reversible (GTR) model of nucleotide substitution [39], a gamma distributed rate variation and a proportion of invariable sites (GTR + Γ + I). A relaxed (uncorrelated lognormal) molecular clock [40] and a Bayesian Skyline coalescent tree were used as priors [41]. Analyses were done with Beast v1.6.1 [42]. Convergence of parameters was assessed using Tracer v1.5 program (http://tree.bio.ed.ac.uk/software/tracer/) until all parameters estimates showed Effective Sample Size (ESS) values over 100.
Table 1. GenBank and SJRP sequences used for phylogenetic inference.
| Country | Year | GenBank Accesion | Genotype |
| French Polynesia (PF) | 1989 | AY744678 | I |
| French Polynesia (PF) | 1994 | AY744685 | I |
| Indonesia (ID) | 2004 | AY858048 | I |
| Indonesia (ID) | 1982 | DQ401690 | II |
| Thailand (TH) | 1973 | DQ863638 | II |
| Thailand (TH) | 2001 | FJ810414 | II |
| Taiwan (TW) | 1998 | DQ675531 | II |
| Vietnam (VN) | 2006 | EU660411 | II |
| Brazil/Acre | 2004 | EF629367 | III |
| Brazil/Acre | 2004 | EF629366 | III |
| Brazil/Central West | 2001 | FJ913015 | III |
| Brazil/Northern | 2003 | FJ850079 | III |
| Brazil/Northern | 2004 | FJ850083 | III |
| Brazil/Northern | 2005 | FJ850086 | III |
| Brazil/Northern | 2006 | FJ850089 | III |
| Brazil/Northern | 2007 | FJ850092 | III |
| Brazil/Northern | 2008 | FJ850094 | III |
| Brazil/Rio de Janeiro | 2002 | EF629369 | III |
| Brazil/RP | 2006 | GQ868548 | III |
| Brazil/RP | 2006 | GQ868547 | III |
| Brazil/RP | 2006 | GQ868546 | III |
| Brazil | 2001 | FJ898446 | III |
| Brazil | 2001 | FJ177308 | III |
| Brazil | 2003 | FJ898447 | III |
| Colombia (CO) | 2007 | GQ868578 | III |
| Mozambique (MZ) | 1985 | FJ882575 | III |
| Nicaragua (NI) | 1994 | FJ882576 | III |
| Puerto Rico (PR) | 2006 | EU529692 | III |
| Sri Lanka (LK) | 1983 | GQ199889 | III |
| Sri Lanka (LK) | 1997 | GQ252674 | III |
| Trinidad & Tobago (TT) | 2002 | GQ868617 | III |
| Venezuela (VE) | 2007 | GQ868587 | III |
| Brazil/Rondonia (BR) | 2002 | EF629370 | V |
| Brazil/SJRP | 2006 | RP06161 | III |
| Brazil/SJRP | 2006 | RP06391 | III |
| Brazil/SJRP | 2006 | RP06411 | III |
| Brazil/SJRP | 2006 | RP06611 | III |
| Brazil/SJRP | 2006 | RP06641 | III |
| Brazil/SJRP | 2006 | RP06781 | III |
| Brazil/SJRP | 2006 | RP06801 | III |
| Brazil/SJRP | 2006 | RP06921 | III |
| Brazil/SJRP | 2006 | RP06951 | III |
| Brazil/SJRP | 2006 | RP061901 | III |
| Brazil/SJRP | 2006 | V065491 | III |
| Brazil/SJRP | 2006 | RP06171 | III |
| Brazil/SJRP | 2006 | RP06231 | III |
| Brazil/SJRP | 2006 | RP06561 | III |
| Brazil/SJRP | 2006 | RP06651 | III |
| Brazil/SJRP | 2006 | RP06791 | III |
| Brazil/SJRP | 2006 | V061231 | III |
| Brazil/SJRP | 2006 | RP06541 | III |
| Brazil/SJRP | 2007 | RP07071 | III |
| Brazil/SJRP | 2007 | RP07111 | III |
| Brazil/SJRP | 2007 | RP07031 | III |
| Brazil/SJRP | 2007 | RP07141 | III |
| Brazil/SJRP | 2007 | RP07181 | III |
| Brazil/SJRP | 2007 | RP07191 | III |
| Brazil/SJRP | 2007 | RP07251 | III |
| Brazil/SJRP | 2007 | RP07371 | III |
| Brazil/SJRP | 2007 | RP07411 | III |
| Brazil/SJRP | 2007 | RP07531 | III |
| Brazil/SJRP | 2007 | RP07601 | III |
| Brazil/SJRP | 2007 | RP07611 | III |
| Brazil/SJRP | 2007 | RP07661 | III |
| Brazil/SJRP | 2007 | RP07751 | III |
| Brazil/SJRP | 2007 | RP07081 | III |
Selection detection analysis
The SJRP dataset was used in a selection analysis to determine whether amino acid positions were subject to negative or positive selection pressures. Comparisons were made with the dengue reference strain Philippines H87/57, following Barrero and Mistchenko [43] general approach. Both genealogy-based, codon-site models Single Likelihood Ancestor Counting (SLAC) and the Random-Effects (REL) available in the HyPhy package (www.datamonkey.org) were used to estimate the nonsynonymous (dN) and synonymous (dS) rates of substitution [44]–[46]. Polymorphisms were analyzed with the program DNASP v5 [47] and subjected to the Tajima D statistic test, as proposed by Tajima [48] to evaluate deviations from the neutral expectation of molecular evolution [49].
Epitope and immunogenicity prediction
The amino acid sequences coding for the capsid, envelope, NS1, NS2a and NS5 proteins were aligned (http://multalin.toulouse.inra.fr/multalin/multalin.html) and a consensus sequence of each protein was submitted to B (http://www.cbs.dtu.dk/services/BepiPred/) and T (http://www.cbs.dtu.dk/services/NetCTL/) epitope prediction algorithms. The Allele Frequency Net Database (http://www.allelefrequencies.net/) was used to select the HLA classes most predominant in the Southeast of Brazil and used to set up the NetCTL server. The mean value of the epitope propensity scores for each sequence was then classified and plotted according to its potential immunogenicity.
Results and Discussion
Complete genome analyzes define distinct DENV3 lineages circulating in SJRP
The availability of high throughput, parallel DNA sequencing allows for comprehensive information on population genetics at the complete viral genome level. In this study, we determined the sequence of 3390 codon-sites of the complete polyprotein from 33 DENV-3 strains isolated during the 2006 and 2007 outbreaks.
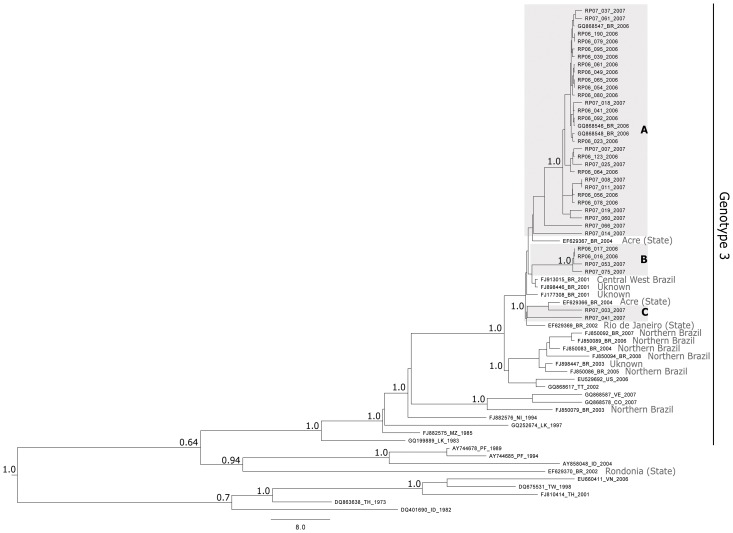
The inferred phylogenetic relationship among DENV-3 strains was summarized in a Bayesian maximum credibility tree (MCC) shown in Figure 1. The majority of Brazilian samples grouped with genotype III, as did most taxa from Latin America isolated since 1994 [20], [50]–[52]. Likewise, the Brazilian strain EF629370 grouped within genotype I, as previously described by Schreiber et al. [53]. DENV-3 isolates, other than genotype III, have been reported in Colombia [54] and in the Brazilian states of Minas Gerais and Rondonia, likely imported from Asia [55], [56]. The successive circulation of different genotypes is of public health concern given that distinct combinations could have different epidemic potentials and risk burdens [57].
Figure 1. Maximum clade credibility (MMC) tree showing the phylogenetic relationships among SRJP clades (light gray boxes) and other Brazilian isolates.
Posterior probability values are shown for main nodes only. Light grey text represents localities (sources) for other Brazilian isolates.
The samples from SJRP all fell within genotype III. The MCC tree indicates that SJRP sequences group with Brazilian isolates from different regions in highly supported clades (Figure 1). Clade A included most of SJRP isolates and is more closely related to an isolate from Acre in 2004 (EF629367_BR_2004). Clade B includes four SJRP isolates grouping with (i) a sample from Central West Brazil (FJ913015_BR_2001) and (ii) the Brazilian sample FJ898446_BR_2001 of unknown exactly provenance. Clade C includes two SJRP isolates from 2007 that cluster with a sequence from Rio de Janeiro (EF629369_BR_2002) sampled in 2002. Moreover, in clades A and B there was no temporal separation, suggesting that the 2007 isolates apparently originated from previously circulating 2006 lineages. These results suggest distinct virus introductions into SJRP with one lineage becoming more prevalent and experiencing in situ evolution (clade A). Co-circulation of multiple DENV-1 lineages has been documented in Asian and other Brazilian cities [58], [59].
Succession of DENV-3 genotypes in Brazil
Our data also provides information on the movement of DENV-3 (genotype III) in South America, because sample FJ850079_BR_2002 obtained in the northern part of Brazil grouped with Colombian and Venezuelan sequences suggesting a plausible transmission route.
DENV-3 (genotype III) was isolated for the first time in Brazil from an autochthonous case of DF in December 2000 from Nova Iguaçu, a city located in Rio de Janeiro (RJ) State, which is the second largest metropolitan area in the country, in 2001- 02, a large outbreak of DENV-3 occurred in the neighboring city of Rio de Janeiro [60], [61]. Following its introduction into RJ, the virus spread to the neighboring State of São Paulo (SP), and eventually to the municipality of SJRP. A previous introduction of DENV-3 took place in Brazil in Rondonia State (Amazon region), but sample EF629370_BR_2002 position in the MCC tree indicates it belongs to genotype V with its sister taxa coming from Asia. Nevertheless, most genotype III isolates from the north of Brazil grouped with the Caribbean strains (samples from Puerto Rico and Trinidad and Tobago), which suggests the existence of preferential routes of spread into Brazil, possibly facilitated by anthropogenic factors [51], [62].
The dynamics of viral change
Sequence comparisons using the reference strain Philippines H87/57 (accession no. M93130) as an outgroup, revealed a total of 893 nucleotide changes, of which 132 corresponded to non-synonymous substitutions. The nucleotide diversity π (that is the average number of nucleotide differences per site between two sequences) [63] within SJRP strains was 0.0052 ± 0.001. Nevertheless, this π value was one order of magnitude lower than that for the complete data set used for phylogenetic inference (n = 0.038 ± 2×10−5), which suggests a genetic bottleneck following the introduction into a new human population, with the virus experiencing rapid in situ evolution afterwards. This is a likely scenario given that we sampled from the first documented DENV-3 outbreak in a city where the local human population is expected to be largely susceptible to this serotype. Likewise, the average value of Tajimas' D for all genes was −1.823 (p<0.05) indicating an excess of low frequency polymorphisms, which would be the case in viral populations expanding following a selective sweep.
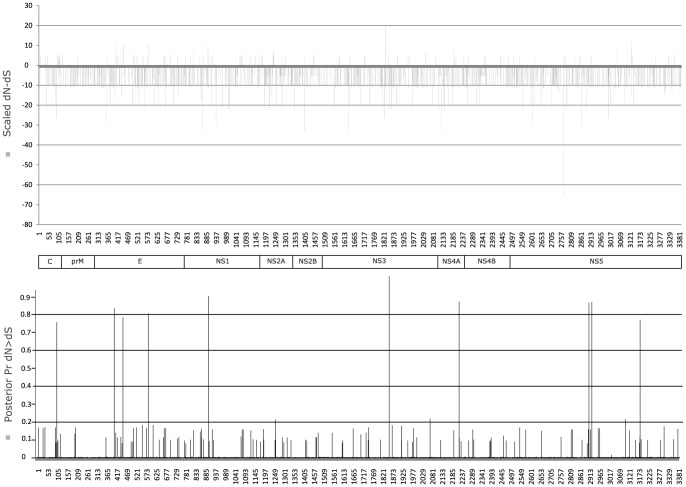
The Tajima's D value near −2 also implies the outcome of negative selection, most likely due to functional constraints imposed on the viral proteins. In agreement with this hypothesis, the overall dN/dS value for the entire polyprotein coding region was 0.068 (estimated 95% CI from 0.057 to 0.080) that implied purifying (stabilizing) selection. Zanotto et al. [64] have argued that at a coarse-grain, the most common pressure acting on DENV is purifying selection with little evidence of recent adaptive evolution when comparing distantly related sequences in space and time, which would be expected due to synonymous change saturation at the serotype level. Moreover, given that arboviruses have adapted to diverse cell types it is generally considered that most mutations produced are likely to be deleterious [24]. Accordingly, the per-site SLAC dN/dS analyses revealed that only eight codons were non-neutral and negatively-selected (p<0.05) (Table 2). Nevertheless, the presence of low frequency polymorphisms could account for adaptive changes, such as the 10 sites under significant positive selection (Bayes factor >50) that we found with the per-site REL dN/dS analyses.
Table 2. Non-neutral codons according to the assayed methods (SLAC and REL) at the specified significance levels (p = 0.05 and Bayes factor = 50, respectively).
| Region | Codon | Residue | Scaled dN-dS (SLAC) | Bayes Factor (REL) | Selection |
| Endoplasmic reticulum anchor for the protein C | 106 | V24M10 | 265.737 | Positive | |
| Glycoprotein E: central and dimerization domains | 355 | P34 | −26.589 | Negative | |
| Glycoprotein E: central and dimerization domains | 404 | P26L7S1 | 422.066 | Positive | |
| Glycoprotein E: central and dimerization domains | 449 | T32I1A1 | 310.144 | Positive | |
| Glycoprotein E: immunoglobulin-like domain | 581 | T33L1 | 349.007 | Positive | |
| Nonstructural NS1 | 865 | D34 | −35.355 | Negative | |
| Nonstructural NS1 | 893 | K33L1 | 738.801 | Positive | |
| Nonstructural NS1 | 935 | V34 | −30.491 | Negative | |
| Nonstructural NS2B | 1401 | D34 | −35.355 | Negative | |
| NS3 serine Protease: Domain S7 | 1634 | Y34 | −35.355 | Negative | |
| NS3 serine Protease: ATP binding domain | 1829 | A27V6S1 | 80226.4 | Positive | |
| Nonstructural NS4A: Cytoplasmic domain | 2124 | H34 | −35.355 | Negative | |
| Nonstructural NS4A: Lumenal domain | 2191 | E29D1A1 | 553.026 | Positive | |
| AdoMet-Mtase | 2605 | P34 | −30.491 | Negative | |
| NS5: Cytoplasmic domain | 2862 | V34 | −30.491 | Negative | |
| NS5: Cytoplasmic domain | 2864 | E20G14 | 527.608 | Positive | |
| NS5: Cytoplasmic domain | 2879 | K32R2 | 548.683 | Positive | |
| NS5: Catalytic domain | 3129 | P32L1S1 | 286.641 | Positive |
Selected sites were located in both structural (S) and nonstructural (NS) proteins (Figure 2). Among the four non-neutral sites within the envelope protein, codon 355 coding for A Proline located in the central and dimerization domains, was under negative selection suggesting its structural importance. Likewise, positively-selected codons 404 and 581 in the envelope are near sites associated with disulfide bonds, suggesting adaptive changes near structurally important residues. Interestingly, Barrero and Mistchenko [43] also identified codons 581 and 893 (within the nonstructural NS1 gene) as positively-selected. King et al. [65] detected a statistically significant positively-selected site in the NS1 gene of DENV-3 genotype II. They also detected selective pressure in the C and NS4B genes of genotype I and in the E and NS3 genes of genotype II. We also found additional positively-selected codon sites; 106 (Endoplasmatic reticulum anchor for the protein Capsid), 2191 (Lumenal domain of NS4A) and 3129 (catalytic domain of the RNA-directed RNA polymerase) that fell within domains that are important for membrane interactions/rearrangements and virus replication.
Figure 2. Per-site and per-gene dN/dS plot.
The calculated SLAC dN/dS and REC posterior probability for positive selection (dN>dS) are plotted for each site in the upper and lower part of the polyprotein respectively.
Polymorphism analysis of individual genomic regions indicated that the NS5 and the envelope proteins varied the most with 24 and 22 replacement changes respectively, followed by NS1 (17), NS2A (11), NS4B (8) and Capsid (8) (Table 3). Changes found in the glycoprotein M, the AdoMet-MTase region and the NS3 serine protease were mostly conservative, while replacements in the capsid were less conservative. Twiddy et al. [66] found codon sites 169 and 380 to be under positive selection in DENV-3, with the latter being located within the central and dimerization domains of the glycoprotein, which is involved in cell tropism. Likewise, Rodpothong and Auewarakul [25] reported one codon site in the prM gene and twelve residues in the E gene to be under positive selection. In summary, our findings along with others, suggest that genes other than the envelope may experience adaptive changes following introduction into a susceptible population, which implies a possible role for either host-immunity interaction or vector adaptations that require further study.
Table 3. Genetic analysis per genomic region.
| Region | Codon Position | Sites | Segregating sites | ETA* | Pi** | K*** | NS sites | S Sites | TajimaD |
| Capsid Protein C | 13–342 | 330 | 26 | 26 | 0.009 | 2.806 | 8 | 18 | −1.952 |
| Polyprotein Propeptide | 355–612 | 258 | 18 | 18 | 0.005 | 1.166 | 3 | 15 | −2.472 |
| Envelope Glycoprotein M | 616–840 | 225 | 19 | 19 | 0.008 | 1.829 | 0 | 19 | −2.052 |
| Glycoprotein E: central and dimerization domains | 841–1722 | 882 | 88 | 88 | 0.010 | 8.513 | 15 | 73 | −2.257 |
| Glycoprotein E: immunoglobulin-like domain | 1726–2013 | 288 | 31 | 31 | 0.011 | 3.234 | 7 | 24 | −2.033 |
| NS1 | 2323–3387 | 1065 | 87 | 88 | 0.008 | 8.011 | 17 | 71 | −2.344 |
| NS2A | 3406–4029 | 624 | 66 | 66 | 0.010 | 6.054 | 11 | 55 | −2.311 |
| NS2B | 4039–4419 | 381 | 33 | 34 | 0.007 | 2.765 | 5 | 29 | −2.382 |
| NS3 Serine Protease | 4459–4923 | 465 | 30 | 30 | 0.006 | 2.749 | 3 | 27 | −2.211 |
| DEXDc domain | 4987–5811 | 825 | 59 | 60 | 0.008 | 2.717 | 8 | 52 | −2.241 |
| NS4A | 6286–6720 | 435 | 40 | 40 | 0.010 | 4.146 | 6 | 34 | −2.078 |
| NS4B | 6727–7455 | 729 | 66 | 66 | 0.009 | 6.496 | 8 | 58 | −2.210 |
| AdoMet-MTase | 7633–8133 | 501 | 34 | 34 | 0.008 | 3.972 | 1 | 33 | −1.864 |
| RNA-directed RNA polymerase | 8218–10158 | 1941 | 161 | 162 | 0.008 | 15.130 | 24 | 138 | −2.340 |
| Full coding region | 1–10170 | 10170 | 888 | 893 | 0.008 | 83.398 | 132 | 761 | −2.373 |
Total number of mutations.
Nucleotide diversity.
Average number of nucleotide differences.
Short-term evolution of DENV-3 genotype III
Studies documenting selectively driven evolution in DENV are limited and mostly available for serotype 2 [25], [27]–[29], [43], [66], further studies to better characterize possible specific adaptive strategies of each serotype are needed. Therefore we looked at allele replacement in DENV 3 (genotype III).
The allele replacement process, from a population genetics standpoint, has an explicit genealogic representation, since frequency increase of an allele will be manifested by appearing deeper as a synapomorphy in a viral genealogy [67]. Therefore, a synapomorphy indicates a fixed allele in the clade defined by it. To clarify this, we coded each of the ten positively-selected residues as a set of terminal unordered character states, represented as a single capital letter. The most parsimonious reconstruction sets of changes at each internal state in the phylogeny were calculated with MacClade v4.07 [68] using the MCC tree rooted with the reference strain Philippines H87/57. The reconstructed changes are illustrated in Figure 3. They indicate that positively-selected codons (404, 449, 581, 2191, 2879 and 3129) mapped to internal nodes as synapomorphies defining the genotype III. For example, codon 106 Met to Val, (within Protein C, the most non-conservative region in our study) characterized the SJRP clade A and codon 404 Pro to Leu characterized the SJRP clade B. Codon 893 appeared as an autapomorphy for the reference strain H87/57 (M93130). The synapomorphic changes differentiating clades A and B that we observed could reflect some aspects of the intra-genotypic interactions exposed to the human population [22].
Figure 3. Positively-selected codon transformations tracked over a clade credibility (MMC) tree.
Light gray boxes represent SJRP clades. Numbers indicates codon position and one-letter symbols indicate amino-acids. Transformations for each character can be evaluated as follows: • Unique transformation all over the tree; ▪ character state fixed along that branch; ◂ same transformation occurs in another branch (homoplasy); ▸ transformation(s) in the same character occurs along that branch; ⧫ same transformation occurs in another branch (homoplasy) and transformation(s) in the same character occur along that branch.
Mondini et al. [35] described the pattern of spread of two DENV lineages circulating in SJRP, Brazil, during 2006. Based on 82 NS5 sequences, the phylogenetic analysis indicated that all samples were of DENV-3 and related to strains circulating in Martinique during 2000–2001. We have considerably extended that analysis by further showing that different lineages, closely relate to different Brazilian isolates, were co-circulating while experiencing distinct adaptive changes as indicated by their positively-selected synapomorphies. This finding is of relevance, since in several other viral systems, positively-selected changes are adaptive in nature and indicative of adaptive changes imposed by vectors, animal reservoirs or human hosts (i.e., immune escape and herd immunity) [69]–[73].
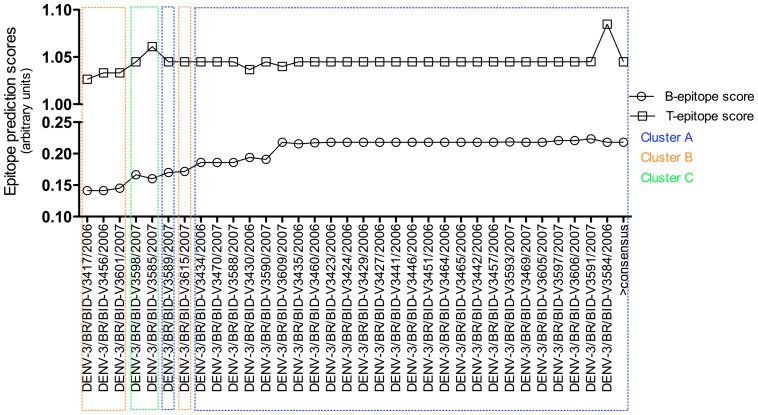
When we observed that DENV-3 sequences, previously clustered in clades A, B and C (Figure 3), we conducted in silico analysis to verify the influence of amino acids substitutions in the immunogenicity of capsid, envelope, NS1, NS2a and NS5 proteins. We found that amino acid substitutions diminished the B/T epitope propensity scores mainly in the DENV-3 sequences clustered in clades B and C (Figure 4). These analyses suggest that amino acid substitutions in sequences grouped in clade A increases its immunogenicity. As a consequence, DENV-3 strains grouped in clade A should expose more antigenic epitopes and more potentially recognized/accessed by the human immune system. We expected that the most immunogenic strains would be negatively selected in a given population. In this particular study we found exactly the opposite. Thus, one can hypothesize that the ADE phenomenon, which would be responsible for an increased pathogenicity and replication in host, can also affect the viral dynamics in a population level. We wonder whether this increased immunogenicity of clade A can induce an antibody dependent enhancement (ADE)-like response in the individual leading to high titers of the virus, which can facilitate the viral transmission to the mosquitoes and, consequently, increase this strain basic reproductive rate. Our findings are of relevance given that ADE may be manifested by challenges imposed by increasing viral immunogenic diversity.
Figure 4. Rank of immunogenicity of DENV-3 sequences.
Thirty-three DENV-3 sequences were submitted to B and T epitope prediction using bioinformatics servers and further classified according to their immunogenic potential. Sequences previously clustered in B and C clades have epitopes, in envelope, NS1, NS2a and NS5, putatively less immunogenic when compared to the same epitopes of sequences clustered in clade A, except for sequence ACY70777.1. Substitutions in the capsid were irrelevant.
While most studies document the broad geographic expansion of dengue, urban outbreaks dynamics deserves investigation because they contribute to a better understanding of the genetic diversity of strains during local transmission; knowledge that could be exploited for antiviral and vaccine development. In our analyses of 33 complete polyprotein-coding regions we revealed distinct lineages and detected polymorphisms sites that correlated with changes in the immunogenicity of several epitopes. The study provides fine-grain information about the molecular epidemiology of dengue infection.
Supporting Information
Primers used for amplification and sequencing.
(DOC)
GenBank accession numbers and name of all samples.
(XLSX)
Funding Statement
CJVA holds a FAPESP scholarship (2011/17071-2); PMAZ and MLN hold a CNPq-PQ scholarship, AM holds an INCT - Dengue scholarship. This work was funded by FAPESP process number 10/19059-7 (PMAZ), by INCT-Dengue and PRONEX-Dengue (CNPq to MLN) and FAPESP Grants # 2012/11733-6 and 2011/10458-9 to MLN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gubler DJ (1998) Dengue and dengue hemorrhagic fever. Clinical microbiology reviews 11: 480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanotto PMAGEA (2002) Molecular epidemiology, evolution and dispersal of the genus flavivirus; T L, editor. Norwell: Kluwer Academic Publishers. 167–195 p. [Google Scholar]
- 3.Guzman M, Halstead S, Artsob H (2010) Dengue: a continuing global threat. Nature Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons CP, Farrar JJ, Chau N, Van Vinh Chau, Wills B (2012) Dengue. The New England Journal of Medicine: 1423–1432. [Google Scholar]
- 5. Garg P, Nagpal J, Khairnar P, Seneviratne SL (2008) Economic burden of dengue infections in India. Transactions of the Royal Society of Tropical Medicine and Hygiene 102: 570–577. [DOI] [PubMed] [Google Scholar]
- 6. Halasa Ya, Shepard DS, Zeng W (2012) Economic Cost of Dengue in Puerto Rico. American Journal of Tropical Medicine and Hygiene 86: 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luz PM, Grinsztejn B, Galvani aP (2009) Disability adjusted life years lost to dengue in Brazil. Tropical medicine & international health: TM & IH 14: 237–246. [DOI] [PubMed] [Google Scholar]
- 8. Shepard DS, Coudeville L, Halasa Ya, Zambrano B, Dayan GH (2011) Economic impact of dengue illness in the Americas. The American journal of tropical medicine and hygiene 84: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beauté J, Vong S (2010) Cost and disease burden of dengue in Cambodia. BMC public health 10: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castañeda-Orjuela C, Díaz H, Alvis-Guzman N, Olarte A, Rodriguez H, et al. (2012) Burden of Disease and Economic Impact of Dengue and Severe Dengue in Colombia, 2011. Value in Health Regional Issues 1: 123–128. [DOI] [PubMed] [Google Scholar]
- 11. Gubler DJ (2011) Dengue, Urbanization and Globalization: The Unholy Trinity of the 21(st) Century. Tropical medicine and health 39: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kyle JL, Harris E (2008) Global spread and persistence of dengue. Annual review of microbiology 62: 71–92. [DOI] [PubMed] [Google Scholar]
- 13. Guy B, Almond JW (2008) Towards a dengue vaccine: progress to date and remaining challenges. Comparative immunology, microbiology and infectious diseases 31: 239–252. [DOI] [PubMed] [Google Scholar]
- 14. Harris E, Ennis FA (2012) Defeating dengue: a challenge for a vaccine. Nature medicine 18: 1622–1623. [DOI] [PubMed] [Google Scholar]
- 15. Halstead SB (2012) Dengue vaccine development: a 75% solution? Lancet 380: 1535–1536. [DOI] [PubMed] [Google Scholar]
- 16. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, et al. (2012) Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380: 1559–1567. [DOI] [PubMed] [Google Scholar]
- 17. Costa RL, Voloch CM, Schrago CG (2012) Comparative evolutionary epidemiology of dengue virus serotypes. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 12: 309–314. [DOI] [PubMed] [Google Scholar]
- 18. Rico-Hesse R (1990) Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174: 479–493. [DOI] [PubMed] [Google Scholar]
- 19. Zanotto P, Gould EA, Gao GF, Harvey PH, Holmes EC (1996) Population dynamics of flaviviruses revealed by molecular phylogenies. Proceedings of the National Academy of Sciences of the United States of America 93: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kochel T, Aguilar P, Felices V, Comach G (2008) Molecular epidemiology of dengue virus type 3 in Northern South America: 2000–2005. Infection, Genetics and: 2000–2005. [DOI] [PubMed] [Google Scholar]
- 21. Araújo JMG, Nogueira RMR, Schatzmayr HGH, Zanotto PMDa, Bello G (2009) Phylogeography and evolutionary history of dengue virus type 3, Genetics and Evolution. 9: 716–725. [DOI] [PubMed] [Google Scholar]
- 22.Villabona-Arenas CJ, Zanotto PMDA (2011) Evolutionary history of Dengue virus type 4: Insights into genotype phylodynamics. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases: 1–8. [DOI] [PubMed] [Google Scholar]
- 23. Zanotto PM, Gibbs MJ, Gould Ea, Holmes EC (1996) A reevaluation of the higher taxonomy of viruses based on RNA polymerases. Journal of virology 70: 6083–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holmes EC (2003) Patterns of intra-and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. Journal of virology 77: 11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodpothong P, Auewarakul P (2012) Positive selection sites in the surface genes of dengue virus: phylogenetic analysis of the interserotypic branches of the four serotypes. Virus genes 44: 408–414. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt DJ, Pickett BE, Camacho D, Comach G, Xhaja K, et al.. (2011) A phylogenetic analysis using full-length viral genomes of South American dengue serotype 3 in consecutive Venezuelan outbreaks reveals novel NS5 mutation. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennett SNS, Holmes EEC, Chirivella M, Rodriguez DM, Beltran M, et al. (2003) Selection-driven evolution of emergent dengue virus. biology and evolution 20: 1650–1658. [DOI] [PubMed] [Google Scholar]
- 28. Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, et al. (2006) Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. The Journal of general virology 87: 885–893. [DOI] [PubMed] [Google Scholar]
- 29. Twiddy S (2002) Phylogenetic Relationships and Differential Selection Pressures among Genotypes of Dengue-2 Virus. Virology 298: 63–72. [DOI] [PubMed] [Google Scholar]
- 30. Khan AM, Miotto O, Nascimento EJM, Srinivasan KN, Heiny aT, et al. (2008) Conservation and variability of dengue virus proteins: implications for vaccine design. PLoS neglected tropical diseases 2: e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Danecek P, Lu W, Schein CH (2010) PCP consensus sequences of flaviviruses: correlating variance with vector competence and disease phenotype. Journal of molecular biology 396: 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. San Martín JL, Brathwaite O, Zambrano B, Solórzano JOJO, Bouckenooghe A, et al. (2010) The epidemiology of dengue in the americas over the last three decades: a worrisome reality. The American journal of tropical medicine and hygiene 82: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saúde SdVe (2007) Informe Epidemiológico da Dengue. Janeiro a Dezembro 2007. Available at: http://portal.saude.gov.br/portal/arquivos/pdf/boletim_dengue_010208.pdf. Accessed 24 Janeiro 2013.
- 34.Saúde SdVe (2008) Informe Epidemiológico da Dengue. Janeiro a Novembro 2008. Available at: http://portal.saude.gov.br/portal/arquivos/pdf/boletim_dengue_janeiro_novembro.pdf. Accessed 24 Janeiro 2013.
- 35. Mondini A, de Moraes Bronzoni RV, Nunes SHP, Chiaravalloti Neto F, Massad E, et al. (2009) Spatio-temporal tracking and phylodynamics of an urban dengue 3 outbreak in Sao Paulo, Brazil. PLoS neglected tropical 3: e448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mondini A, Neto FC, Sanches MGY, Lopes JCC (2005) Análise espacial da transmissão de dengue em cidade de porte médio do interior paulista. Rev Saúde Pública 39: 444–451. [DOI] [PubMed] [Google Scholar]
- 37. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez F, Oliver J, Marin A, Medina J (1990) The general stochastic model of nucleotide substitution. Journal of theoretical biology. [DOI] [PubMed] [Google Scholar]
- 40. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS biology 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond A, Rambaut A, Shapiro B (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and. [DOI] [PubMed] [Google Scholar]
- 42. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barrero PR, Mistchenko AS (2008) Genetic analysis of dengue virus type 3 isolated in Buenos Aires, Argentina. Virus research 135: 83–88. [DOI] [PubMed] [Google Scholar]
- 44.Pond SLK, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics (Oxford, England) 21: : 676–679. [DOI] [PubMed] [Google Scholar]
- 45. Pond S, Kosakovsky Pond SL, Frost SDW (2005) Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular biology and evolution 22: 1208–1222. [DOI] [PubMed] [Google Scholar]
- 46.Pond SLK, Frost SDW (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics (Oxford, England) 21: : 2531–2533. [DOI] [PubMed] [Google Scholar]
- 47.Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 25: : 1451–1452. [DOI] [PubMed] [Google Scholar]
- 48. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura M (1989) The neutral theory of molecular evolution: Cambridge University Press. 384 p. [Google Scholar]
- 50. Uzcategui NY (2003) Molecular epidemiology of dengue virus type 3 in Venezuela. Journal of General Virology 84: 1569–1575. [DOI] [PubMed] [Google Scholar]
- 51. Aquino VH, Anatriello E, Gonçalves PF, DA Silva EV, Vasconcelos PFC, et al. (2006) Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002–2004. The American journal of tropical medicine and hygiene 75: 710–715. [PubMed] [Google Scholar]
- 52. Villabona-Arenas CJ, Miranda-Esquivel DR, Jimenez REO (2009) Phylogeny of dengue virus type 3 circulating in Colombia between 2001 and 2007. Tropical medicine & international health: TM & IH 14: 1241–1250. [DOI] [PubMed] [Google Scholar]
- 53.Schreiber M, Holmes E, Ong S (2009) Genomic epidemiology of a dengue virus epidemic in urban Singapore. Journal of. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Usme-Ciro J, Mendez J, Tenorio A, Rey G (2008) Simultaneous circulation of genotypes I and III of dengue virus 3 in Colombia. Virol J 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Figueiredo L, Ferreira G (2008) Dengue virus 3 genotype 1 associated with dengue fever and dengue hemorrhagic fever, Brazil. Emerging Infectious 14: 314–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aquino VH, Amarilla AA, Alfonso HL, Batista WC, Figueiredo LTM (2009) New genotype of dengue type 3 virus circulating in Brazil and Colombia showed a close relationship to old Asian viruses. PloS one 4: e7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guzman MG, Alvarez A, Vazquez S, Alvarez M, Rosario D, et al. (2012) Epidemiological studies on dengue virus type 3 in Playa municipality, Havana, Cuba, 2001–2002. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases 16: e198–203. [DOI] [PubMed] [Google Scholar]
- 58. Raghwani J, Rambaut A, Holmes EC, Hang VT, Hien TT, et al. (2011) Endemic dengue associated with the co-circulation of multiple viral lineages and localized density-dependent transmission. PLoS pathogens 7: e1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. dos Santos FB, Nogueira FB, Castro MG, Nunes PC, de Filippis AMB, et al. (2011) First report of multiple lineages of dengue viruses type 1 in Rio de Janeiro, Brazil. Virology journal 8: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nogueira RM, Miagostovich MP, de Filippis aM, Pereira Ma, Schatzmayr HG (2001) Dengue virus type 3 in Rio de Janeiro, Brazil. Memórias do Instituto Oswaldo Cruz 96: 925–926. [DOI] [PubMed] [Google Scholar]
- 61. Casali CGC, Pereira MMRR, Santos L, Passos MNP, Fortes B, et al. (2004) A epidemia de dengue/dengue hemorrágico no município do Rio de Janeiro, 2001/2002. Rev Soc Bras Med 37: 296–299. [DOI] [PubMed] [Google Scholar]
- 62. Araújo JMG, Nogueira RMR, Schatzmayr HG, Zanotto PMDa, Bello G (2009) Phylogeography and evolutionary history of dengue virus type 3. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 9: 716–725. [DOI] [PubMed] [Google Scholar]
- 63.Nei M (1987) Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- 64. Zanotto P, Gould EAE, Gao GFG, Harvey PH, Holmes EC (1996) Population dynamics of flaviviruses revealed by molecular phylogenies. Proceedings of the 93: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. King C-C, Chao D-Y, Chien L-J, Chang G-JJ, Lin T-H, et al. (2008) Comparative analysis of full genomic sequences among different genotypes of dengue virus type 3. Virology journal 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Twiddy SS, Woelk CH, Holmes EC (2002) Phylogenetic evidence for adaptive evolution of dengue viruses in nature. The Journal of general virology 83: 1679–1689. [DOI] [PubMed] [Google Scholar]
- 67. Zanotto PM, Kallas EG, de Souza RF, Holmes EC (1999) Genealogical evidence for positive selection in the nef gene of HIV-1. Genetics 153: 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maddison DM, WP (2005) MacClade 4: Analysis of Phylogeny and Character Evolution. Version 4.08a. Available at: http://macclade.org. Accessed 24 Janeiro 2013.
- 69.Ross H, Rodrigo A (2002) Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. Journal of virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferguson NM, Donnelly Ca, Anderson RM (1999) Transmission dynamics and epidemiology of dengue: insights from age-stratified sero-prevalence surveys. Philosophical transactions of the Royal Society of London Series B, Biological sciences 354: 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sheridan I, Pybus OG, Holmes EC, Klenerman P (2004) High-Resolution Phylogenetic Analysis of Hepatitis C Virus Adaptation and Its Relationship to Disease Progression. Journal of Virology 78: 3447–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen R, Holmes EC (2008) The evolutionary dynamics of human influenza B virus. Journal of molecular evolution 66: 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Suzuki Y (2008) Positive selection operates continuously on hemagglutinin during evolution of H3N2 human influenza A virus. Gene 427: 111–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for amplification and sequencing.
(DOC)
GenBank accession numbers and name of all samples.
(XLSX)