Abstract
FAT10 is a new member of the ubiquitin-like protein family with yet-to-be defined biological functions in the heart. Our objective was to determine the role of FAT10 in the heart. FAT10 is expressed in the normal human and murine hearts, as detected by qPCR and Western blotting. Expression of FAT10 is increased in the heart at the border zone of myocardial infarction and in cultured neonatal rat cardiac myocytes (NRCM) subjected to hypoxia/reoxygenation (H/R) stress. Lentiviral-mediated overexpression of FAT10 in NRCM reduced p53 (TP53) and its target miR-34a levels, while BCL2 level, a target of miR-34a, was increased and BAX level, a pro-apoptotic protein, was reduced. These changes were associated with reduced apoptosis, detected by FACS analysis of annexin-V expression and TUNEL assay, in response to H/R injury. Knock down of FAT10 by shRNA targeting had the opposite effects. Likewise, lentiviral mediated expression of miR-34a was associated with reduced BCL2 and increased BAX levels in NRCM and also reversed changes in BCL-2 and BAX levels observed upon over-expression of FAT10. Treatment of NRCM with proteasome inhibitor MG132 increased p53 and miR-34a levels and reduced BLC2/BAX ratio. These changes were not reversed upon over-expression of FAT10.
Thus, FAT10 is upregulated in the heart and NRCM in response to H/R stress, which protects cardiac myocytes against apoptosis. The anti-apoptotic effects of FAT10 are associated with suppression of p53, probably through fatylation and proteasomal degradation, reduced miR-34a expression, and a shift in the BCL2/BAX proteins against apoptosis. Thus, FAT10 is a cardioprotective protein.
Keywords: FAT10, cardiac myocyte, apoptosis, p53, microRNA, Fatylation
1.0. INTRODUCTION
The ubiquitin-proteasome system (UPS) regulates degradation of most cellular proteins. [1] UPS is a key regulator of cardiac function under both physiological and pathological conditions. [2–4] Impaired cardiac UPS functions have been demonstrated in a variety of cardiac pathological states including myocardial ischemia, cardiomyopathies and heart failure. [4–6] Recently several ubiquitin-like proteins with structural and functional similarities to ubiquitin, such as SUMO and NEDD8 have been identified and implicated in targeting protein substrates for possible degradation. [7–9]
FAT10 encodes FAT10 (human leukocyte antigen F-associated transcript 10); also known as ubiquitin D (UbD) is an enigmatic ubiquitin-like protein modifier with poorly defined biological functions. [10, 11] It contains two tandem ubiquitin-like (UBL) domains with 30% sequence identity to ubiquitin that might form covalent conjugates with its substrates and target them for 26S proteasomal degradation. [12, 13] Expression of FAT10 is induced in response to inflammatory cytokines, such as TNF-α and suppressed by tumor suppressor protein p53 (TP53) [14, 15]. The role of FAT10 in apoptosis is unsettled. FAT10-deficient mice are prone to spontaneous apoptosis of lymphocytes. [16] Over-expression of FAT10 in cultured cells, however, is associated with apoptosis. [12]
Expression and potential biological functions of FAT10 in the heart are unknown. We show that FAT10 is expressed in cardiac myocytes and its expression protects the myocytes against apoptosis through suppression of p53 and miR-34a and increased expression of BCL2. These findings render FAT10 as a novel cardioprotective gene that is upregulated in response to hypoxic/ischemic injury.
2.0. MATERIALS AND METHODS
2.1. Animals and Surgical Procedures
All animal studies were approved by the Animal Ethics and Experimentation Committee of Nanchang University, and performed in accordance with the “Guide for the Care and Use of Laboratory animals” (revised 1996). The research protocol for human is reviewed and approved by the Ethical Committee at the Hospital of Nanchang University, and confirmed to the guidelines of the World Medical Assembly.
Myocardial infarction (MI) was produced in 20 male 10-week old Sprague Dawley anesthetized open-chest adult rats by ligating the left anterior descending artery (LAD). Regional ischemia was confirmed by the detection of ST-segment elevation on the electrocardiogram and the presence of a pale myocardium (Online Figure 1). Sham-operated (control) rats underwent an identical operation without ligation of the LAD.
2.2. Isolation of neonatal rat cardiac myocytes (NRCM) and Hypoxia/Reoxygenation (H/R) injury
Cultured cardiac myocytes were isolated from the ventricular myocardium of 2-day-old Sprague Dawley rats by enzymatic digestion according to a published protocol. 17,18 NRCM were sequentially exposed to hypoxia for 3 h (CO2/N2 at a 95:5 ratio) and reoxygenation (O2/CO2, a 95:5 ratio).
2.3. Immunoblotting
Myocardial tissue samples from hearts of adult males with no apparent cardiovascular diseases were used in immunoblotting. In brief, aliquots of 100 ug protein were subjected to polyacrylamide gel electrophoresis, transferred to a membrane and probed with antibodies against FAT10, p53, BCL2, and BAX.
2.4. RT-PCR, and qPCR
RT-PCR and qPCR were performed per conventional methods 19.
2.5. H&E staining and Immunohistochemical Study
One portion of the cardiac tissue was fixed in 10% buffered formalin, embedded in paraffin, sectioned. Deparaffinized sections were used to H&E staining, which applied to determine morphological changes. Another portion of fresh cardiac tissue, embedded in O.C.T. compound and snap-frozen in n-hexane cooled with a mixture of dry ice, was cut at 7µm on a cryostat. Frozen sections were used to detect Fat10. For staining of Fat10, sections were incubated with goat anti-Fat10 polyclonal antibody. Thereafter, the sections were incubated with biotinylated secondary antibody. After washing with phosphate-buffered saline containing 0.01% Tween, the sections were stained with 3,3’-diaminobenzidine solution and then counters stained with hematoxylin.
2.6. Construction of Fat10, si-Fat10 and miR-34a lentiviral constructs
Briefly, full-length Fat10 cDNA was synthetized and were subcloned into a lentivirus shuttle vector (pLenti6.3_MCS_IRES2-GFP). We designed oligonucleotides targeting rat Fat10 mRNA and the sequence complementary to miR-34a, cloned the designed oligonucleotides into lentivirus vector pcDNA™6.2-GW/EmGFP-miR. Then, lentivirus vectors transfected the packing cells (Invitrogen) for the generation of recombinant lentivirus.
2.7. Cell culture, lentiviral Fat10, si-Fat10 and miR-34a transfection
One day before transfection, NRCM were seeded in 6-well plate at density 1×105 cells/well. Confluence will reach approximately 70% at the time of transfection. NRCM were infected with lentiviral particles carrying Fat10, si-Fat10 and miR-34a at MOI of 50 for 48 hours. Medium were refreshed and cells were allow to grow for another 24hours, then cells were either lysed for protein/ RNA extraction for further analysis at the experimental groups indicated.
2.8. Cardiac myocyte apoptosis
Apoptosis was detected by flow cytometry after mixing the cell with fluorescein isothicyanate (FITC)-labeled annexin-V and propidium iodide.
In situ TUNEL assay was performed in accordance with the manufacturer’s protocol (In Situ Cell Death Detection Kit, POD; Roche diagnostics).
2.9. Treatment with proteasome inhibitor MG132
NRCM was transduced with recombinant lentiviruses for 72 h and then were treated with MG132 (15 µmol/L) for 4 h. Immunoblotting and qPCR were performed as described above.
2.10. Statistical analysis
Each experiment was performed for a minimum of three different myocyte isolations or three different animals and was repeated at least 3 times. Data were presented as Mean ± SEM and compared by one-way ANOVA and Student–Newman Keuls test, the latter for multiple group comparisons (Sigma-Stat software, San Francisco, CA).
3.0. RESULTS
3.1. FAT10 is expressed in normal myocardial tissue
We determined expression of FAT10 in the human, mouse and rat hearts by RT-PCR, immunoblotting and immunocytochemistry. As shown in Figure 1 (A and B), FAT10 is expressed in the human myocardial tissue as well as in mouse and rat hearts.
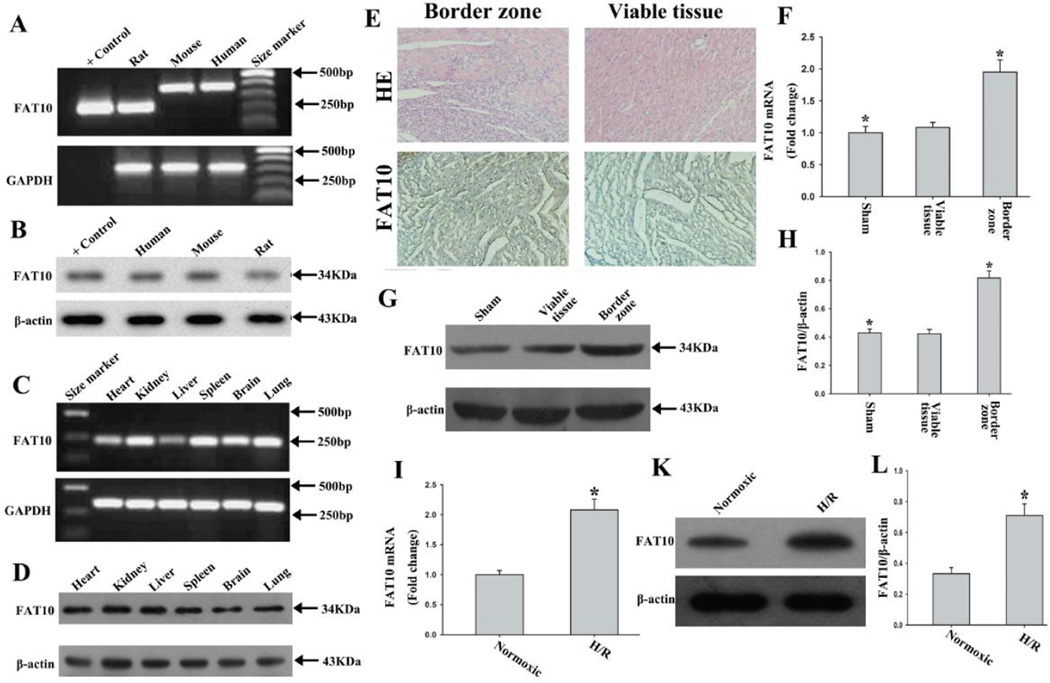
Figure 1. Expression of FAT10 in the heart and its up-regulation in response to ischemia.
A. Detection of expression of Fat10 mRNA by RT-PCR and agarose gel electrophoresis in human, mouse and rat hearts, as identified by the label in each lane. The expected products, based on the design of PCR primers, were 180bp for rat Fat10 cDNA, 360bp for mouse Fat10 cDNA and 366bp for human FAT10 cDNA. Positive control group used the construction of vector expressing rat Fat10 cDNA as template. GADPH served as an internal control.
B. FAT10 protein, detected in myocardial tissues from rat, mouse and human, by Western blotting. Positive control group used proteins from 293T cells after transfection the vector expressing rat Fat10 cDNA. β-actin is used as a control for loading conditions.
C-D. Expression of FAT10 mRNA in different rat tissues as detected by RT-PCR, and western blot showing expression of FAT10 protein. Tissue source of RNA and protein are indicated above the panel. GADPH is used as an internal control for mRNA expression and β-actin is used as a control for loading conditions for protein expression.
E. Representative images of rat heart sections stained with H&E and immunocytochemistry from the sham-operated group and the MI group respectively. FAT10 is distributed abundantly in the border zone of myocardial infraction. Bar: 10 µm
F. Bar graph showing Fat10 mRNA level was significantly increased in the border zone of myocardial infarction as compared with sham-operated rats. The data were compared to the Gapdh level and normalized to the mean value of controls, n=6, * P<0.05 compared to sham-operated group.
G. Expression of FAT10 protein in heart tissues of sham and MI rats, as detected by Western blotting. β-actin is used as a loading control.
H. Quantitative analysis of FAT10 protein level in control and ischemic border zone, (n=3 experiments for each groups). *P<0.05 vs. sham-group.
I -K. Illustrate changes in Fat10 mRNA, as detected by qPCR (I), and FAT10 protein level as detected by Western blotting (K) and quantitative data (H) in cultured cardiac myocytes exposed to hypoxia. β-actin is used as a control for loading conditions. Quantitative analysis is in right panels (n=3 experiments for each groups). *P < 0.05 compared to normoxic condition.
3.2. Fat10 is a ubiquitously expressed gene
We detected expression of the Fat10 gene in multiple rat tissues by RT-PCR and Western blotting analyses of total RNA and proteins, respectively, isolated from 6- to 8- week-old rats. The results (Figure 1 C and D) indicate that Fat10 mRNA and protein are expressed in the rat heart, kidney, liver, spleen, brain and lung.
3.3. Expression of FAT10 is increased in myocardial ischemia and in cardiac myocytes subjected to H/R
To begin investigating the functional role of FAT10 in the heart, we determined mRNA and protein levels of FAT10 in control and ischemic myocardium as well as in cardiac myocytes subjected to H/R. Total RNA and proteins were extracted from the border zones of MI and the viable myocardium remote from the site of MI. The results of qPCR and Western blotting showed about a 2-fold increase in the mRNA and protein levels of FAT10, respectively, in the infarct border zone as compared with the viable tissue or the myocardium from the sham-operated rat (Figure 1, E-H).
To determine the effects of H/R stress on FAT10 expression in cardiac myocytes, isolated NRCM were subjected to H/R and Fat10 mRNA and protein levels were quantified by qPCR and Immunoblotting, respectively. Fat10 mRNA and protein were increased by about 2.0 fold in NRCM subjected to H/R as compared to NRCM cultured under a normoxic condition (Figure 1, I-L).
3.4. Over-expression of FAT10 prevents hypoxia-induced myocyte apoptosis
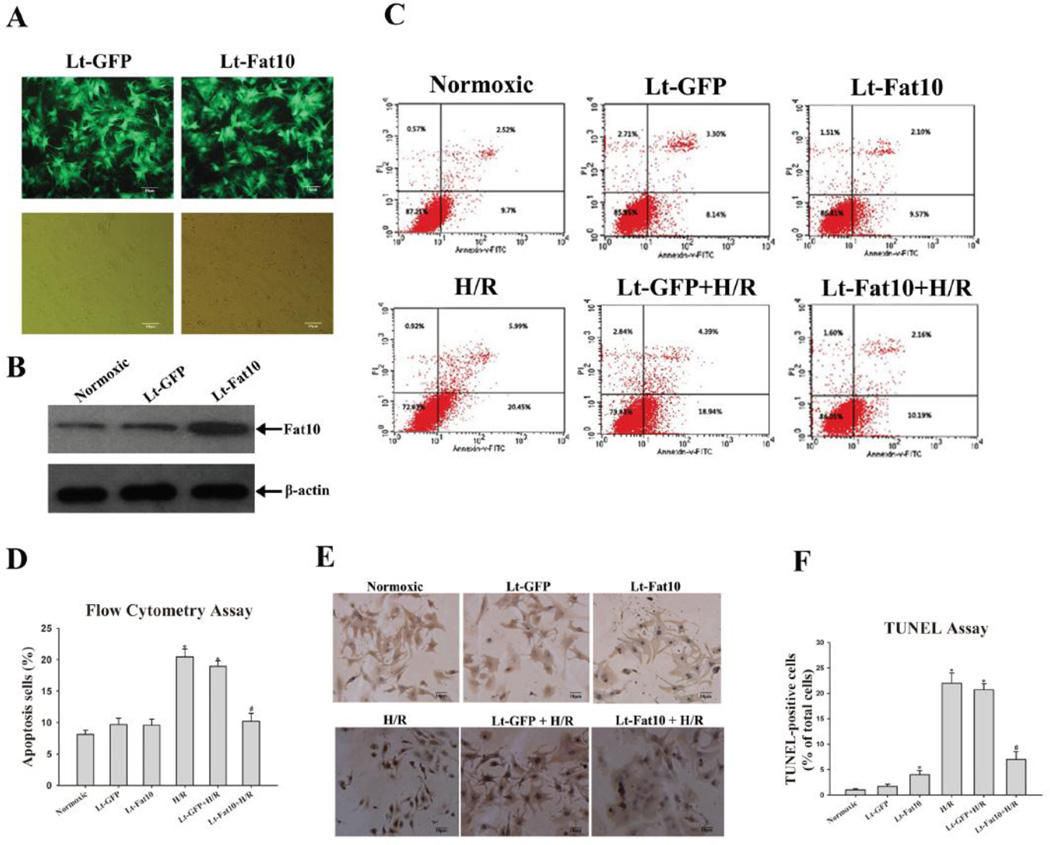
To explore the effects of FAT10 on cardiac myocytes apoptosis, NRCM were transduced with recombinant lentiviruses expressing FAT10 for 72 h and then exposed to H/R. As expected, H/R stress markedly increased the number of apoptotic NRCM. Over-expression of FAT10 attenuated myocyte apoptosis in response to H/R by ~ 65 %, as compared to control non-transduced myocytes, detected by FCM and by ~ 75 %, as detected by the TUNEL assay (Figure 2, A-F).
Figure 2. Effect of over-expression of FAT10 on cardiac myocyte apoptosis following H/R.
A Cultured cardiac myocytes were transfected with Lt-FAT10 (transfection of lentivirus containing Fat10 gene) as well as Lt-GFP (transfection of negative lentivirus). 72 h after transfection, expression of FAT10 was analyzed by Western blotting. Transfection efficiency of lentivirus was estimated to be about 80% as determined by the percent cells expressing GFP protein by fluorescent microscopy (magnification, x400).
B. Shows increased expression of FAT10 in cardiac myocytes transduced with Lt-FAT10 viruses.
C. Cardiac myocytes apoptosis was analyzed with flow cytometry and the percentage of apoptotic cells was determined according to cells expressing Annexin.
D. The bar graphs represented the Mean ± SEM of data from six replicate experiments.
E. Apoptosis was detected by TUNEL assay in the experimental groups. Cells were counter-stained with hematoxylin. A representative field is shown for each group.
F. Quantitative analysis of TUNEL-positive cells from 3 independent experiments (magnification, x200). Apoptosis in cardiac myocytes was quantified by the number of apoptotic nuclei in the total nuclei in 10 continuous microscopic fields.
*P<0.05 vs. normoxic; **P<0.05 vs. H/R group
3.5. FAT10 downregulates p53 and suppresses expression of miR-34a in cardiac myocytes
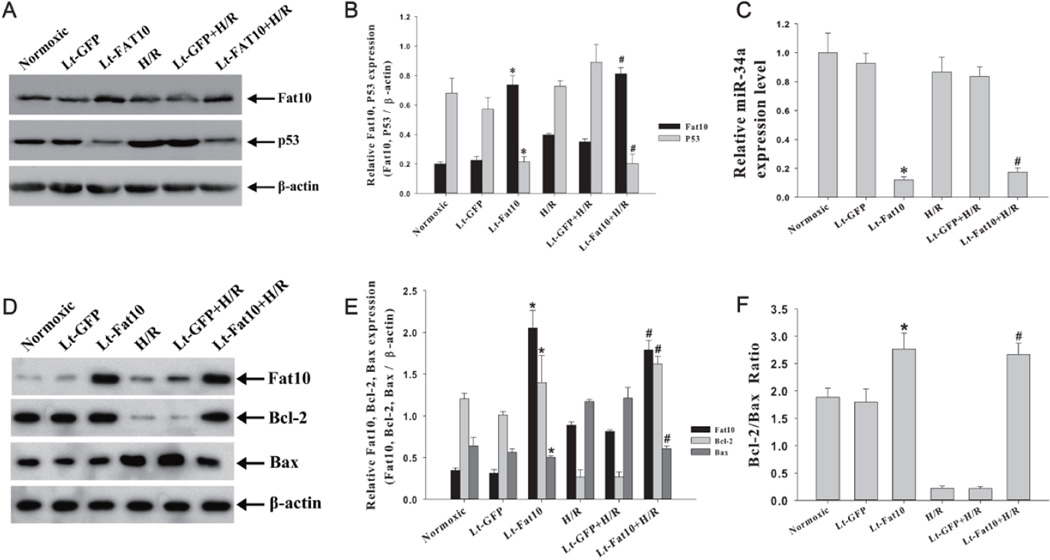
The p53 protein, which is known to be upregulated in cardiac myocytes in response to hypoxia [20, 21] has been implicated in regulation of FAT10 [15, 22]. To determine whether over-expression of FAT10 affected p53 level, we quantified p53 level in NRCM transduced with lentiviruses expressing FAT10. Expression of FAT10 was associated with a significant reduction in the p53 protein level under normoxic conditions and attenuated the increase in p53 protein level in response to H/R stress (Figure 3, A-B). H/R per se had no significant effect on miR-34a level (Figure 3C).
Figure 3. Molecular anti-apoptotic effects of over-expression of FAT10.
A. Western blot showing FAT10 and p53 levels in cardiac myocytes transduced with lentiviruses under normoxic condition and in response to H/R. β-actin is used as a control for loading conditions.
B. The bar graphs represented the Mean ± SEM of results from three replicate experiments. *p<0.05 vs. normoxic; # p<0.05 vs. H/R group.
C. Decreased miR-34a level in cardiac myocytes transduced with Lt-FAT under nomoxic and H/R conditions. Data were shown as Mean ± SEM in 3 tests from different experiments for each group. *p<0.05 vs. normoxic; # p<0.05 vs. H/R group.
D. Western blot showing over-expression of FAT10 attenuates H/R-induced suppression of BCL2 and up-regulation of BAX. β-actin is used as a control for loading conditions.
E. Quantitative data of three independent experiments per each group is shown. &P<0.05, *p<0.05 vs. control in normoxic groups and #p<0.05 vs. control in the H/R groups;.
F. BCL2/BAX ratio in cardiac myocytes exposed to H/R. Each bar presents the Mean ± SEM of 3 tests from different experiments. *p<0.05 vs. normoxic; # p<0.05 vs. H/R group.
Because p53 is a known transactivator of miR-34a [23, 24], which is known to target anti-apoptotic protein BCL2 [23, 25], we quantified miR-34a level by qPCR. As shown in Figure 3C), over-expression of FAT10 in NRCM was associated with a significant down-regulation of miR-34a level under normoxic estate as well as in NRCM exposed to H/R stress.
In conjunction with reduced expression level of p53 and miR-34a, BCL2 level was increased. Over-expression of FAT10 rescued the H/R-induced suppression of BCL2 level (Figure 3 D,E, F).
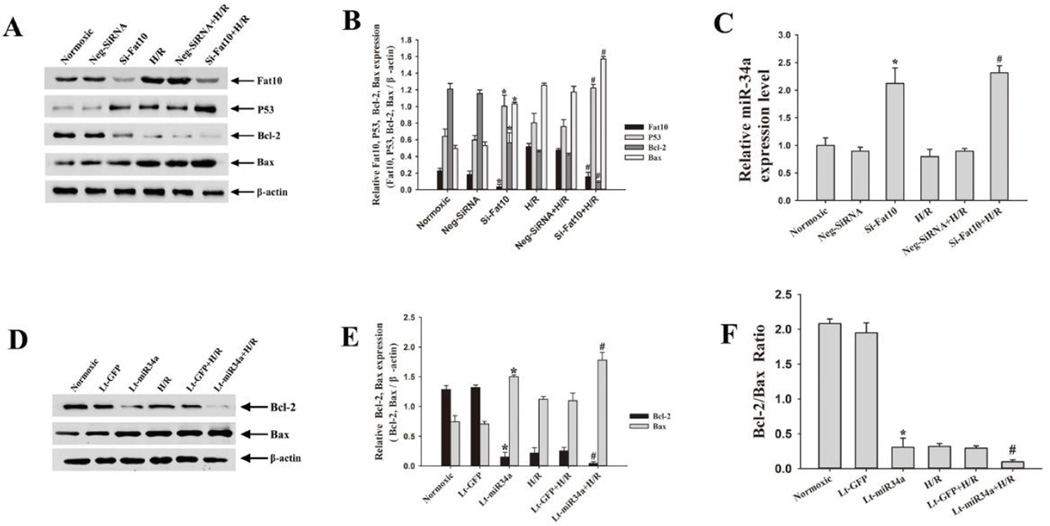
3.6. FAT10 knock down is associated with up-regulation of p53 and miR-34a and down-regulation of BCL2
To further substantiate the effects of FAT10 on p53 protein, we targeted Fat10 by RNAi (Figure 4). Suppression of FAT10 level was associated with increased p53 and miR-34a levels and reduced BCL2 level (Figure 4A-C). Likewise, suppression of FAT10 level accentuated hypoxia-mediated up-regulation of p53 and down-regulation of BCL2 (Figure 4A-B).
Figure 4. Lentiviral shRNA-mediated knock-down of FAT10 and over-expression of miR-34a induce a pro-apoptotic profile.
A. Western blot and quantitative data showing knock down of FAT10 is associated with increased p53 and down-regulation of BCL2 levels in NRCM under normoxic condition and in response to H/R. β-actin is used as a control for loading conditions.
B. The bar graphs represent the Mean ± SEM of results from 3 replicate experiments. *P<0.05 vs. normoxic group. #P<0.05 vs. H/R group.
C. BCL2/BAX ratio in shRNA knockdown of FAT10 in cardiac myocytes exposed to H/R. Each bar presents the Mean ± SEM of 3 tests from different experiments. *P<0.05 vs. normoxic group; #P<0.05 vs. H/R group.
D-F. Lentiviral mediated over-expression of miR-34a induces a pro-apoptotic profile, as shown by reduced expression of BCL2 and increased level of BAX (Panels D, E), and reduced ratio of BCL2/BAX under normoxic conditions. β-actin is used as a control for loading conditions. The pro-apoptotic molecular profile is accentuated under H/R condition. Data were shown as Mean ± SEM in 3 tests from different experiments for each group. *p<0.05 vs. normoxic group; #p<0.05 vs. H/R group.
To determine whether BCL2 and BAX were direct targets of FAT10, we detected co-immunoprecipitation (Co-IP) of BCL2 and BAX with FAT10 and repeated the experiments in the reverse order. Under the experimental conditions used in these experiments, we could not detect fatylated BCL2 or BAX, suggesting that FAT10 did not directly bind to BCL2 or BAX (data not shown).
3.7. Over-expression of miR-34a is associated with suppression of BCL2 and up-regulation of BAX
To determine whether miR-34a target BCL2, we transduced NRCM with lentiviruses expressing miR-34a. Expression of miR34-a was associated with reduced BCL2 and increased BAX levels under normoxic as well as hypoxic conditions (Figure 4D-F).
3.8. Expression of miR-34a revokes the anti-apoptotic molecular profile of FAT10
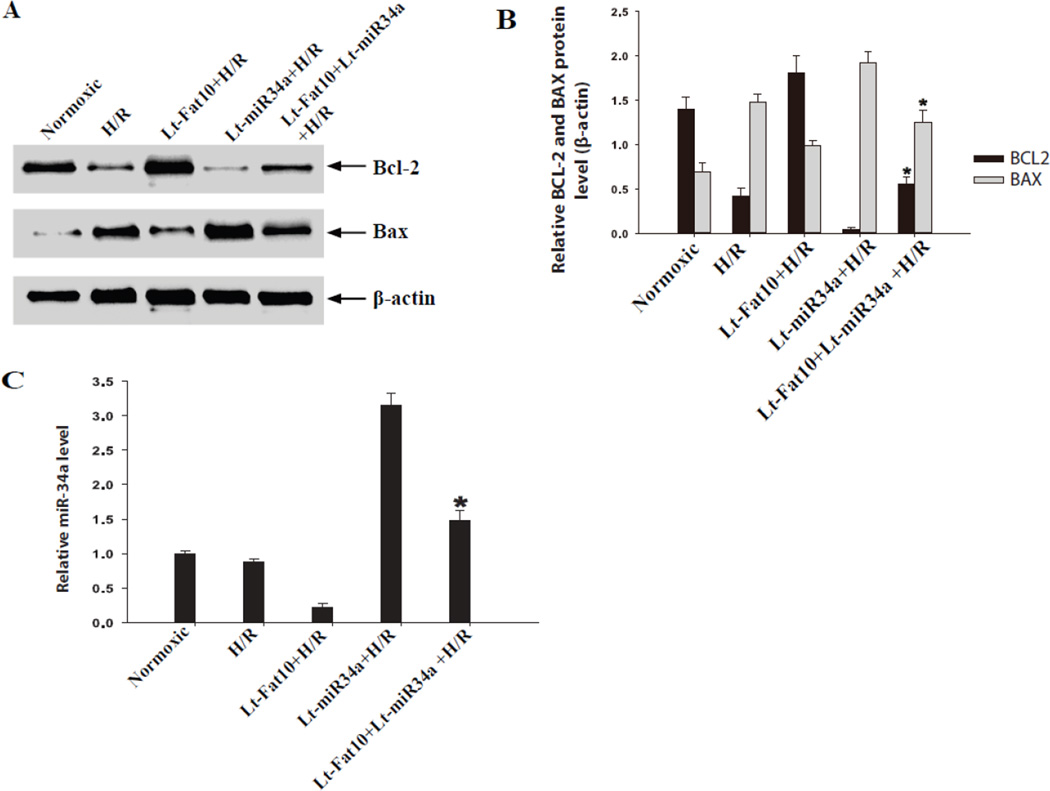
To determine whether miR-34a was a major effector of FAT10 in exerting an anti-apoptotic profile, NRCM were co-transduced with recombinant lenti-viruses (MOI of 100) expressing FAT10 and miR-34a. Over-expression of FAT10 exerted an anti-apoptotic and that of miR-34a pro-apoptotic effects on BCL-2 and BAX levels. However, over-expression of miR-34a in the background of over-expression of FAT10 largely nullifies the anti-apoptotic effects of FAT10 on the BCL2/BAX ratio (Figure 5, panels A-C). These findings identify miR-34a a major effector of FAT10 in NRCM.
Figure 5. Over-expression of miR-34a abrogates the anti-apoptotic effects of over-expression of FAT10.
A. Western blots showing BCL-2 and BAX proteins in cardiac myocytes in the experimental groups. H/R reduced BCL-2 and increased BAX levels. Transduction of the NRCM, subjected to H/R, with the recombinant viruses (at a MOI of 100) expressing FAT10 had the opposite effects. In contrast, over-expression of miR-34a produced a pro-apoptotic molecular profile (reduced BCL-2 and increased BAX levels). Likewise, over-expression of miR-34a in the background of over-expression of FAT10 also was associated with reduced BCL2 and increased BAX levels. Quantitative data of the protein levels are shown in Panel B and relative miR-34a levels in Panel C. * p<0.05
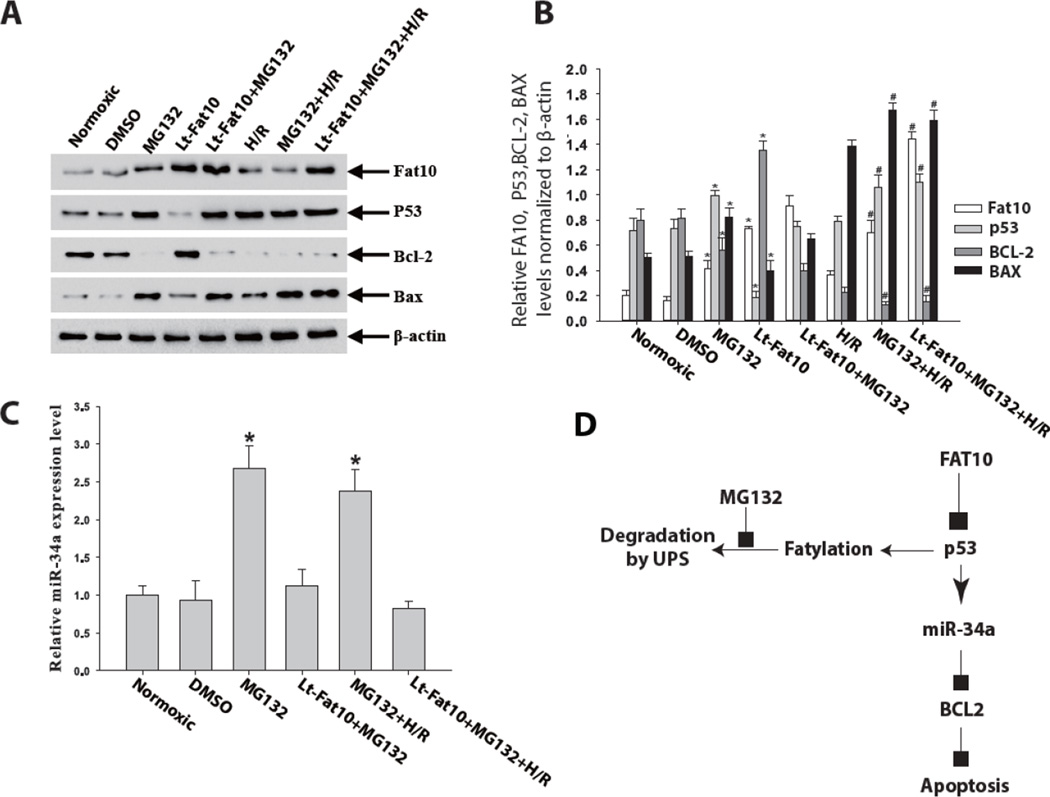
3.9. Inhibition of the Ubiquitin-Proteasome System (UPS) abrogates the anti-apoptotic effects of FAT10
Treatment of NRCM with MG132, an inhibitor of proteasome, was associated with increased p53 and miR-34a level and a decreased BCL2/BAX ratio, indicative of a pro-apoptotic profile. Over-expression of FAT10 in NRCM suppressed p53 and increased BCL2 levels in NRCM under normoxia and in the absence of MG132, as expected. However, in the presence of MG132, over-expression of FAT10 did not significantly attenuate p53 level or restore BCL2 level under the normoxic condition. In response to H/R stress, FAT10 and BCL2 levels were decreased while p53 and BAX levels were increased in NRCM not treated with MG132. However, over-expression of FAT10 in NRCM treated with MG132 did not significantly change p53, BCL2, BAX or miR-34a levels (Figure 5A-C). These findings suggest that FAT10 exerts its anti-apoptotic effects through UPS-mediated degradation of p53.
4.1. DISCUSSION
FAT10 is a relatively new member of the UBL family, with poorly understood biological functions, particularly in the heart. We provide the first set of data on expression and potential biological role of FAT10 in the heart. We show that FAT10 is expressed in cardiac myocytes and its expression is up-regulated in response to H/R stress and myocardial ischemia. Functionally, over-expression of FAT10 reduced H/R-induced apoptosis in cultured cardiac myocytes, as determined by two complementary methods of Flow Cytometry and TUNEL assay. At the mechanistic level, over-expression of FAT10 was associated with suppressed expression of p53, which is a known transactivator of miR-34a. Consequently, overexpression of FAT10 was associated with decreased miR-34a level and increased level of BCL2, which is a known miR-34a target. In contrast, level of pro-apoptotic protein BAX, which is upregulated by p53 was reduced [28]. These changes shifted the BCL2/BAX ratio toward protection from apoptosis. Thus, our data implicate the FAT10/p53/miR-34a/BCL2 pathway as a mechanism responsible for the anti-apoptotic effects of FAT10 on cardiac myocytes (Figure 5D). The anti-apoptotic molecular profile of FAT10 was effectively blocked through inhibition of the UPS with MG132, which implicated FAT10 exerts its anti-apoptotic effects through fatylation of p53 and its degradation by the UPS. The findings offer the first line of evidence for the potential biological role of FAT10 in the heart and set the stage for additional studies to delineate other biological functions of FAT10 in the heart and elucidate additional responsible mechanisms.
Over-expression of FAT10 was associated with reduced expression of p53. This finding is in accord with data showing that FAT10 Fatylates p53 and co-localizes with p53 in the nucleus.[22] Fatylation of p53 is expected to target the protein to the proteasome for degradation. We could not detect direct interactions between FAT10 and p53 or FAT10 and BCL2 by co-immunoprecipitation studies, which might reflect the experimental conditions. Nevertheless, treatment with proteasomal inhibitor MG132 mitigated the effects of FAT10 on p53 levels, which suggest fatylation and UPS-mediated degradation of p53 as a potential mechanism for the anti-apoptotic effects of FAT10. In spite of these data, interactions between FAT10 and p53 remain to be fully defined and might be complex, as p53 was also shown to suppress FAT10 expression [15]. Indirect evidence also supports interactions between FAT10 and p53, as the former is upregulated in various cancers, probably as a consequence of loss of p53. While the findings implicate FAT10/p53/miR-34a/BCL2 pathway as a responsible mechanism for the anti-apoptotic effect of FAT10, additional and alternative mechanisms remain to be explored.
H/R and ischemia can lead to myocyte death by apoptosis and necrosis.[29] Extrinsic death receptor and intrinsic mitochondrial pathway both contribute to ischemic-induced apoptosis.[29] In the present studies, over-expression of FAT10 significantly promoted expression of BCL2 and suppressed expression of BAX under H/R condition, leading to an increased BCL2/BAX ratio and reduced hypoxic-induced apoptosis. These data suggest that FAT10 modulates ischemic-induced apoptosis via the intrinsic apoptotic pathway. However, these changes were most notable in myocytes exposed to H/R stress and less so in cardiac myocytes under normoxic condition. Thus, the findings suggest that over-expression of FAT10 attenuated sensitivity of cardiac myocytes to apoptosis under hypoxic stress but might not constitutively activate the anti-apoptotic pathway in cultured cardiac myocytes.
In summary, data in the present study show that over-expression of FAT10 leads to reduced p53 level in cardiac myocytes exposed to H/R, which results in reduced miR-34a and increased BCL2 levels. The latter in conjunction with reduced BAX level shifts the cardiac myocyte survival homeostasis against H/R-induced apoptosis. These results underscore the potential significance of FAT10 in regulating myocardial survival during ischemia and imply that FAT10 up-regulation might serve as a therapeutic target in ischemic heart disease.
Supplementary Material
Figure 6. Inhibition of the Ubiquitin-Proteasome System (UPS) abrogates the anti-apoptotic effects of FAT10.
A. Western blots showing levels of p53, BCL2 and BAX proteins in cardiac myocytes transduced with recombinant lentiviruses expressing FAT10 before and after treatment with MG132 for 4 h under normoxic conditions as well as before and after exposure to H/R. β-actin was used as a control for loading conditions. Over-expression of FAT10 is associated with reduced p53 and BAX levels and increased BCL2 level. Treatment with MG132 attenuates the molecular anti-apoptotic effects observed upon over-expression of FAT10.
B. Quantitative analysis of p53, BCL2 and BAX proteins levels in the experimental groups. *p<0.05 under normoxic, and #p<0.05 vs. normoxic groups.
C. qPCR analysis of miR-34a level in the experimental group. Data were shown as Mean ± SEM in 3 tests from different experiments for each group. *p<0.05 vs normoxic group.
D. A proposed mechanism for the anti-apoptotic role of FAT10 in NRCM. FAT10 through fatylation and UPS-mediated degradation reduced level of p53 and its trans-regulated miR, miR34-a, which in turn results in increased expression of anti-apoptotic protein BCL2 and reduced apoptosis.
Highlights.
FAT10 is an ubiquitin-like protein family whose functions in the heart are unknown
FAT10 is expressed in the human and murine hearts
Ischemia and hypoxia increase expression of FAT10
FAT10 suppresses p53 and miR-34a levels but increases BCL2 level
FAT10 protects against cardiac myocyte apoptosis
ACKNOWLEDGEMENT
The authors wish to acknowledge supports from the Ministry of Chinese Education Innovation Team Development Plan (IRT1141,HK); National Basic Research Program of China [973 Program: 2007CB512002 and 2008CB517305], the National Natural Science Foundation of China [81070148]; NHLBI (R01-HL088498 and R34HL105563); NIA (R21 AG038597-01), Burroughs Wellcome Award in Translational Research (#1005907), TexGen Fund from Greater Houston Community Foundation and George and Mary Josephine Hamman Foundation.
Non-standard Abbreviations and Acronyms
- NRCM
neonatal rat cardiac myocytes
- H/R
hypoxia/reoxygenation
- UPS
ubiquitin-proteasome system
- MI
Myocardial infarction
- LAD
left anterior descending artery
- FITC
fluorescein isothicyanate
- Co-IP
co-immunoprecipitation
- Lt-FAT10
lentivirus containing Fat10 gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
None
REFERENCES
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nature reviews Molecular cell biology. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Powell SR. The ubiquitin-proteasome system in cardiac physiology and pathology. American journal of physiology Heart and circulatory physiology. 2006;291:H1–H19. doi: 10.1152/ajpheart.00062.2006. [DOI] [PubMed] [Google Scholar]
- 3.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta. 2008;1782:749–763. doi: 10.1016/j.bbadis.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Schlossarek S, Carrier L. The ubiquitin-proteasome system in cardiomyopathies. Current Opinion in Cardiology. 2011;26:190–195. doi: 10.1097/HCO.0b013e32834598fe. [DOI] [PubMed] [Google Scholar]
- 5.Carrier L, Schlossarek S, Willis MS, Eschenhagen T. The ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy. Cardiovascular Research. 2010;85:330–338. doi: 10.1093/cvr/cvp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Xu Z, He XR, Michael LH, Patterson C. CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. American journal of physiology Heart and circulatory physiology. 2005;288:H2836–H2842. doi: 10.1152/ajpheart.01122.2004. [DOI] [PubMed] [Google Scholar]
- 7.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature reviews Molecular cell biology. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 8.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. The Journal Of Biological Chemistry. 2004;279:49091–49098. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]
- 10.Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Molecular and Cellular Biology. 2005;25:3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchsbaum S, Bercovich B, Ciechanover A. FAT10 is a proteasomal degradation signal that is itself regulated by ubiquitination. Molecular biology of the cell. 2012;23:225–332. doi: 10.1091/mbc.E11-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raasi S, Schmidtke G, Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. The Journal Of Biological Chemistry. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- 13.Schmidtke G, Kalveram B, Weber E, Bochtler P, Lukasiak S, Hipp MS, et al. The UBA domains of NUB1L are required for binding but not for accelerated degradation of the ubiquitin-like modifier FAT10. The Journal Of Biological Chemistry. 2006;281:20045–20054. doi: 10.1074/jbc.M603063200. [DOI] [PubMed] [Google Scholar]
- 14.Lukasiak S, Schiller C, Oehlschlaeger P, Schmidtke G, Krause P, Legler DF, et al. Proinflammatory cytokines cause FAT10 up-regulation in cancers of liver and colon. Oncogene. 2008;27:6068–6074. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- 15.Zhang DW, Jeang KT, Lee CG. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene. 2006;25:2318–2327. doi: 10.1038/sj.onc.1209220. [DOI] [PubMed] [Google Scholar]
- 16.Canaan A, Yu X, Booth CJ, Lian J, Lazar I, Gamfi SL, et al. FAT10/diubiquitin-like protein-deficient mice exhibit minimal phenotypic differences. Molecular and Cellular Biology. 2006;26:5180–5189. doi: 10.1128/MCB.00966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinecke H, Zhang M, Bartosek T, Murry CE. Survival, integration, and differentiation of cardiomyocyte grafts: a study in normal and injured rat hearts. Circulation. 1999;100:193–202. doi: 10.1161/01.cir.100.2.193. [DOI] [PubMed] [Google Scholar]
- 18.Tsybouleva N, Zhang L, Chen S, Patel R, Lutucuta S, Nemoto S, et al. Aldosterone, through novel signaling proteins, is a fundamental molecular bridge between the genetic defect and the cardiac phenotype of hypertrophic cardiomyopathy. Circulation. 2004;109:1284–1291. doi: 10.1161/01.CIR.0000121426.43044.2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circulation Research. 2011;109:1342–1353. doi: 10.1161/CIRCRESAHA.111.255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long X, Boluyt MO, Hipolito ML, Lundberg MS, Zheng JS, O'Neill L, et al. p53 and the hypoxia-induced apoptosis of cultured neonatal rat cardiac myocytes. The Journal Of Clinical Investigation. 1997;99:2635–2643. doi: 10.1172/JCI119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Santockyte R, Yu S, Shen RF, Tekle E, Lee CG, et al. FAT10 modifies p53 and upregulates its transcriptional activity. Archives of biochemistry and biophysics. 2011;509:164–169. doi: 10.1016/j.abb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Current biology : CB. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 24.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Molecular Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewings KE, Wiggins CM, Cook SJ. Bim and the pro-survival BCL2 proteins: opposites attract, ERK repels. Cell Cycle. 2007;6:2236–2240. doi: 10.4161/cc.6.18.4728. [DOI] [PubMed] [Google Scholar]
- 27.Crow MT, Mani K, Nam YJ, Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circulation Research. 2004;95:957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- 28.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Current opinion in cell biology. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Kung G, Konstantinidis K, Kitsis RN. Programmed necrosis, not apoptosis, in the heart. Circulation Research. 2011;108:1017–1036. doi: 10.1161/CIRCRESAHA.110.225730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.