Abstract
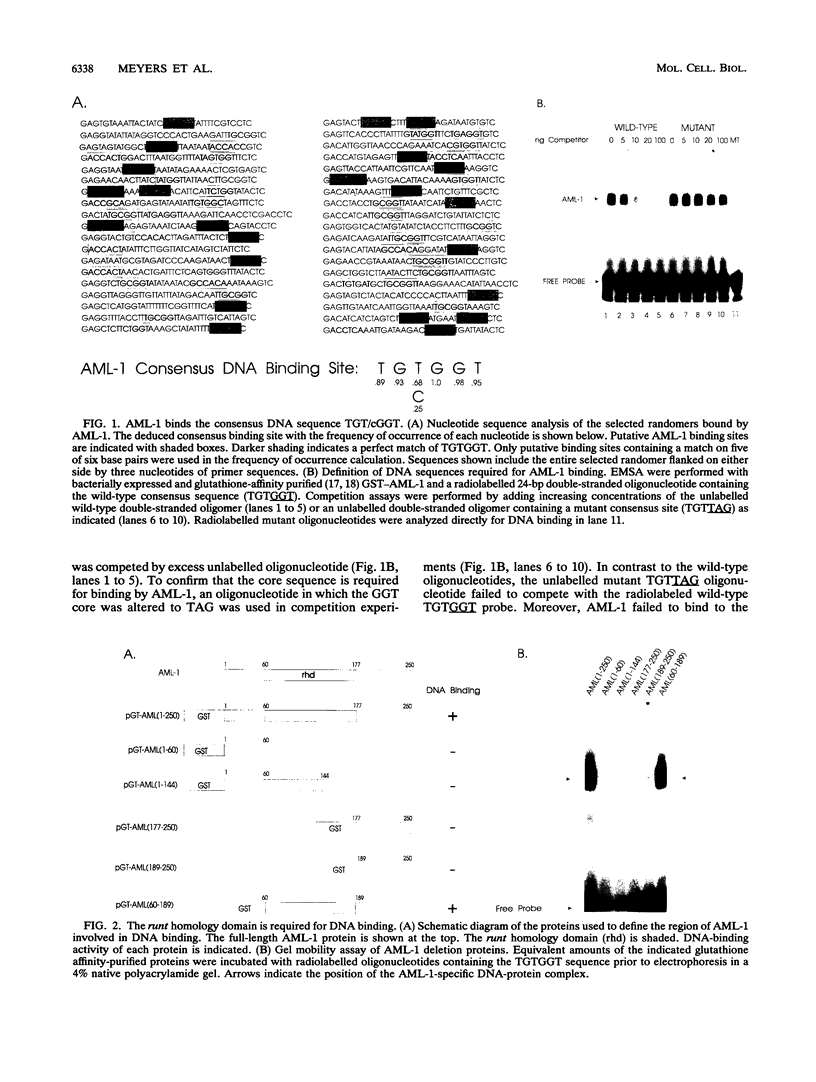
The AML1 gene on chromosome 21 is disrupted in the (8;21)(q22;q22) translocation associated with acute myelogenous leukemia and encodes a protein with a central 118-amino-acid domain with 69% homology to the Drosophila pair-rule gene, runt. We demonstrate that AML-1 is a DNA-binding protein which specifically interacts with a sequence belonging to the group of enhancer core motifs, TGT/cGGT. Electrophoretic mobility shift analysis of cell extracts identified two AML-1-containing protein-DNA complexes whose electrophoretic mobilities were slower than those of complexes formed with AML-1 produced in vitro. Mixing of in vitro-produced AML-1 with cell extracts prior to gel mobility shift analysis resulted in the formation of higher-order complexes. Deletion mutagenesis of AML-1 revealed that the runt homology domain mediates both sequence-specific DNA binding and protein-protein interactions. The hybrid product, AML-1/ETO, which results from the (8;21) translocation and retains the runt homology domain, both recognizes the AML-1 consensus sequence and interacts with other cellular proteins.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. M., Forbush K. A., Perlmutter R. M. Functional dissection of the lck proximal promoter. Mol Cell Biol. 1992 Jun;12(6):2758–2768. doi: 10.1128/mcb.12.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. C., Yamaguchi-Iwai Y., Ogawa E., Maruyama M., Inuzuka M., Kagoshima H., Shigesada K., Satake M., Ito Y. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993 Mar;8(3):809–814. [PubMed] [Google Scholar]
- Boral A. L., Okenquist S. A., Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989 Jan;63(1):76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A., Tighe J. E., Calabi F. Leukaemia/Drosophila homology. Nature. 1992 Apr 9;356(6369):484–484. doi: 10.1038/356484b0. [DOI] [PubMed] [Google Scholar]
- Downing J. R., Head D. R., Curcio-Brint A. M., Hulshof M. G., Motroni T. A., Raimondi S. C., Carroll A. J., Drabkin H. A., Willman C., Theil K. S. An AML1/ETO fusion transcript is consistently detected by RNA-based polymerase chain reaction in acute myelogenous leukemia containing the (8;21)(q22;q22) translocation. Blood. 1993 Jun 1;81(11):2860–2865. [PubMed] [Google Scholar]
- Duffy J. B., Gergen J. P. The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes Dev. 1991 Dec;5(12A):2176–2187. doi: 10.1101/gad.5.12a.2176. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Odell G. M., Schubiger G. A genetic switch, based on negative regulation, sharpens stripes in Drosophila embryos. Dev Genet. 1989;10(3):124–142. doi: 10.1002/dvg.1020100303. [DOI] [PubMed] [Google Scholar]
- Erickson P., Gao J., Chang K. S., Look T., Whisenant E., Raimondi S., Lasher R., Trujillo J., Rowley J., Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992 Oct 1;80(7):1825–1831. [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987 Nov;1(9):981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Gao J., Erickson P., Gardiner K., Le Beau M. M., Diaz M. O., Patterson D., Rowley J. D., Drabkin H. A. Isolation of a yeast artificial chromosome spanning the 8;21 translocation breakpoint t(8;21)(q22;q22.3) in acute myelogenous leukemia. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4882–4886. doi: 10.1073/pnas.88.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J. P., Butler B. A. Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes Dev. 1988 Sep;2(9):1179–1193. doi: 10.1101/gad.2.9.1179. [DOI] [PubMed] [Google Scholar]
- Gill L. L., Zaninetta D., Karjalainen K. A transcriptional enhancer of the mouse T cell receptor delta gene locus. Eur J Immunol. 1991 Mar;21(3):807–810. doi: 10.1002/eji.1830210339. [DOI] [PubMed] [Google Scholar]
- Gottschalk L. R., Leiden J. M. Identification and functional characterization of the human T-cell receptor beta gene transcriptional enhancer: common nuclear proteins interact with the transcriptional regulatory elements of the T-cell receptor alpha and beta genes. Mol Cell Biol. 1990 Oct;10(10):5486–5495. doi: 10.1128/mcb.10.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer A., Garson O. M., Kondo K. Fourth International Workshop on Chromosomes in Leukemia 1982: Translocation (8;21)(q22;q22) in acute nonlymphocytic leukemia. Cancer Genet Cytogenet. 1984 Mar;11(3):284–287. doi: 10.1016/s0165-4608(84)80007-2. [DOI] [PubMed] [Google Scholar]
- Hiebert S. W., Blake M., Azizkhan J., Nevins J. R. Role of E2F transcription factor in E1A-mediated trans activation of cellular genes. J Virol. 1991 Jul;65(7):3547–3552. doi: 10.1128/jvi.65.7.3547-3552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert S. W., Chellappan S. P., Horowitz J. M., Nevins J. R. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992 Feb;6(2):177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- Hiebert S. W., Lamb R. A. Cell surface expression of glycosylated, nonglycosylated, and truncated forms of a cytoplasmic protein pyruvate kinase. J Cell Biol. 1988 Sep;107(3):865–876. doi: 10.1083/jcb.107.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert S. W., Lipp M., Nevins J. R. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci U S A. 1989 May;86(10):3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y., Ogawa E., Asano M., Ishida S., Murakami Y., Satake M., Ito Y., Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990 Oct;64(10):4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania M. A., Bonner A. S., Duffy J. B., Gergen J. P. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990 Oct;4(10):1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- Kappes D. J., Browne C. P., Tonegawa S. Identification of a T-cell-specific enhancer at the locus encoding T-cell antigen receptor gamma chain. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2204–2208. doi: 10.1073/pnas.88.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Lacalli T. C. Modeling the Drosophila pair-rule pattern by reaction-diffusion: gap input and pattern control in a 4-morphogen system. J Theor Biol. 1990 May 22;144(2):171–194. doi: 10.1016/s0022-5193(05)80317-0. [DOI] [PubMed] [Google Scholar]
- LoSardo J. E., Boral A. L., Lenz J. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J Virol. 1990 Apr;64(4):1756–1763. doi: 10.1128/jvi.64.4.1756-1763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y., Nogawa H., Hashimoto Y., Kishi J., Hayakawa T. Accumulation of collagen III at the cleft points of developing mouse submandibular epithelium. Development. 1988 Sep;104(1):51–59. doi: 10.1242/dev.104.1.51. [DOI] [PubMed] [Google Scholar]
- Nisson P. E., Watkins P. C., Sacchi N. Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells. Cancer Genet Cytogenet. 1992 Oct 15;63(2):81–88. doi: 10.1016/0165-4608(92)90384-k. [DOI] [PubMed] [Google Scholar]
- Nucifora G., Birn D. J., Erickson P., Gao J., LeBeau M. M., Drabkin H. A., Rowley J. D. Detection of DNA rearrangements in the AML1 and ETO loci and of an AML1/ETO fusion mRNA in patients with t(8;21) acute myeloid leukemia. Blood. 1993 Feb 15;81(4):883–888. [PubMed] [Google Scholar]
- Nucifora G., Birn D. J., Espinosa R., 3rd, Erickson P., LeBeau M. M., Roulston D., McKeithan T. W., Drabkin H., Rowley J. D. Involvement of the AML1 gene in the t(3;21) in therapy-related leukemia and in chronic myeloid leukemia in blast crisis. Blood. 1993 May 15;81(10):2728–2734. [PubMed] [Google Scholar]
- Rabbitts T. H. Translocations, master genes, and differences between the origins of acute and chronic leukemias. Cell. 1991 Nov 15;67(4):641–644. doi: 10.1016/0092-8674(91)90057-6. [DOI] [PubMed] [Google Scholar]
- Redondo J. M., Hata S., Brocklehurst C., Krangel M. S. A T cell-specific transcriptional enhancer within the human T cell receptor delta locus. Science. 1990 Mar 9;247(4947):1225–1229. doi: 10.1126/science.2156339. [DOI] [PubMed] [Google Scholar]
- Redondo J. M., Pfohl J. L., Hernandez-Munain C., Wang S., Speck N. A., Krangel M. S. Indistinguishable nuclear factor binding to functional core sites of the T-cell receptor delta and murine leukemia virus enhancers. Mol Cell Biol. 1992 Nov;12(11):4817–4823. doi: 10.1128/mcb.12.11.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo J. M., Pfohl J. L., Krangel M. S. Identification of an essential site for transcriptional activation within the human T-cell receptor delta enhancer. Mol Cell Biol. 1991 Nov;11(11):5671–5680. doi: 10.1128/mcb.11.11.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J. D. Identification of the constant chromosome regions involved in human hematologic malignant disease. Science. 1982 May 14;216(4547):749–751. doi: 10.1126/science.7079737. [DOI] [PubMed] [Google Scholar]
- Rowley J. D. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973 Jun;16(2):109–112. [PubMed] [Google Scholar]
- Rubin C. M., Larson R. A., Anastasi J., Winter J. N., Thangavelu M., Vardiman J. W., Rowley J. D., Le Beau M. M. t(3;21)(q26;q22): a recurring chromosomal abnormality in therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 1990 Dec 15;76(12):2594–2598. [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Miyoshi H., Kozu T., Nagata J., Enomoto K., Maseki N., Kaneko Y., Ohki M. Consistent disruption of the AML1 gene occurs within a single intron in the t(8;21) chromosomal translocation. Cancer Res. 1992 Dec 15;52(24):6945–6948. [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991 Jan;65(1):42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M., Sánchez L. The segmentation gene runt is needed to activate Sex-lethal, a gene that controls sex determination and dosage compensation in Drosophila. Genet Res. 1992 Jun;59(3):189–198. doi: 10.1017/s0016672300030470. [DOI] [PubMed] [Google Scholar]
- Vardiman J., Catovsky D., Flandrin G. R., Pierre R. Fourth International Workshop on Chromosomes in Leukemia 1982: Correlation between morphology and karyotype. Cancer Genet Cytogenet. 1984 Mar;11(3):275–281. [PubMed] [Google Scholar]
- Wang S. W., Speck N. A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992 Jan;12(1):89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Yaspo M. L., Theophile D., Aurias A., Créte N., Créau-Goldberg N., Bastard C., Suberville A. M., Valensi F., Viguier F., Berger R. Molecular analysis of 12 patients with the t(8;21) translocation and M2 acute myelogenous leukemia. Genes Chromosomes Cancer. 1992 Sep;5(2):166–177. doi: 10.1002/gcc.2870050211. [DOI] [PubMed] [Google Scholar]