Abstract
Aminoglycosides (AGs) are broad-spectrum antibiotics whose constant use and presence in growth environment has led bacteria to develop resistance mechanisms to aid in their survival. A common mechanism of resistance to AGs is their chemical modification (nucleotidylation, phosphorylation, or acetylation) by AG-modifying enzymes (AMEs). Through evolution, fusion of two AME-encoding genes has resulted in bifunctional enzymes with broader spectrum of activity. Serratia marcescens, a human enteropathogen, contains such a bifunctional enzyme, ANT(3″)-Ii/AAC(6′)-IId. To gain insight into the role, effect, and importance of the union of ANT(3″)-Ii and AAC(6′)-IId in this bifunctional enzyme, we separated the two domains and compared their activity to that of the full-length enzyme. We performed a thorough comparison of the substrate and cosubstrate profiles as well as kinetic characterization of the bifunctional ANT(3″)-Ii/AAC(6′)-IId and its individually expressed components.
Keywords: Acetyltransferase, Aminoglycoside antibiotics, Bacterial resistance, Bifunctional enzyme, Nucleotidyltransferase
1. Introduction
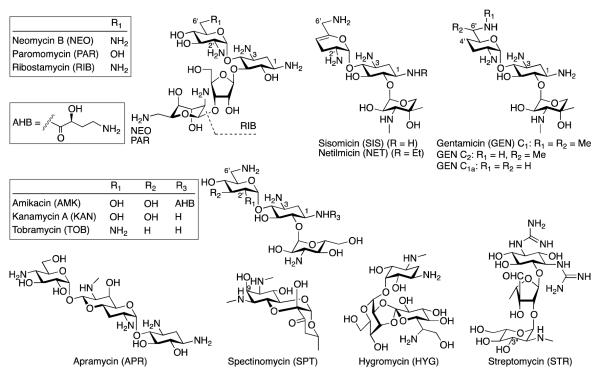
Aminoglycosides (AGs) (Fig. 1) are broad-spectrum antibiotics to which bacteria have developed resistance [1] mainly through mutations of their target, the ribosome, and through the evolution of AG-modifying enzymes (AMEs) that covalently alter the AG scaffolds by nucleotidylation (ANTs), acetylation (AACs), or phosphorylation (APHs) [2, 3]. AGs kill bacteria by disrupting protein synthesis [4]; the covalent chemical modifications of AGs render them inert with respect to these functions. In addition to the evolution of AMEs capable of performing a single chemical modification to inactivate the drug, bacteria have evolved bifunctional enzymes with the ability to perform two chemical modifications on either the same or different AG substrates. These fused enzymes include ANT(3″)-Ii/AAC(6′)-IId from Serratia marcescens [5, 6], AAC(6′)-Ie/APH(2″)-Ia responsible for high-level resistance in Staphylococci and Enterococci [7-9], and AAC(3)-Ib/AAC(6′)-Ib’ [10, 11] and AAC(6′)-30/AAC(6′)-Ib [12, 13] from Pseudomonas aeruginosa. Current theories on the existence of these dual-function enzymes propose that (i) a single modification on an AG scaffold only partially inactivates the AG and, therefore, a second modification is needed to achieve complete inactivation of the drug, and (ii) bifunctional enzymes broaden the spectrum of their AG substrates thereby expanding the resistance profile for the bacteria containing these dual-purpose enzymes. Multiple studies that demonstrate the dual modification of AGs and the retention of activity for some mono-acetylated AG antibiotics support the first theory [14-16]. The second theory is reinforced by the existence of ANT(3″)-Ii/AAC(6′)-IId, since the ANT domain of this fused enzyme modifies only streptomycin (STR) and spectinomycin (SPT), while its AAC(6′) domain targets a majority of the AGs having a 2-deoxystreptamine (DOS) core [5].
Fig. 1.
Structures of the aminoglycosides (AGs) discussed in this study. Note: ANT(3″)-Ii adds the (d)NMPs to position 9 of SPT.
With the increasing number of bacterial species gaining antibiotic resistance, comprehension of the evolution and mechanism of action of AMEs is paramount. In an effort to understand both the evolution of bifunctional enzymes and the purpose for their formation, we examined the difference in activity between the dual-domain ANT(3″)-Ii/AAC(6′)-IId and its individually expressed components, ANT(3″)-Ii and AAC(6′)-IId. In addition, we explore the substrate and cosubstrate profiles of the bifunctional full-length enzyme and of its individual domains and kinetically characterized the enzymes to clarify the evolutionary rationale for the dual-domain AMEs.
2. Materials and methods
2.1. Materials and instrumentation
Chemically competent E. coli TOP10 and BL21(DE3) were from Invitrogen. DNA primers (Table S1) were from Integrated DNA Technologies. The pET22b and pET28a vectors were from Novagen and the Int-pET19b-pps vector [17] was obtained from Dr. Tapan Biswas (University of Michigan). DNA sequencing was done at the University of Michigan DNA sequencing core. Restriction endonucleases, T4 DNA ligase, and Phusion DNA polymerase were from New England Biolabs. 4,4′-dithiodipyridine (DTDP), malachite green, ammonium molybdate, acyl-CoAs (acetyl-, n-propionyl-, acetoacetyl-, benzoyl-, n-butyryl-, β-hydroxybutyryl-, iso-butyryl-, crotonyl-, decanoyl-, glutaryl-, hexanoyl, lauroyl-, malonyl-, octanoyl-, palmitoyl-, and succinyl-), (deoxy)nucleotide triphosphates (ATP, dATP, CTP, dCTP, GTP, dGTP, ITP, dITP, TTP, sodium triphosphate (TP), UTP, and dUTP), and AGs (Fig. 1) (amikacin (AMK), apramycin (APR), gentamicin (GEN), hygromycin (HYG), kanamycin A (KAN), neomycin B (NEO), ribostamycin (RIB), sisomicin (SIS), STR, SPT, and tobramycin (TOB)) were from Sigma-Aldrich. Netilmicin (NET) and paromomycin (PAR) were from AK Scientific. Spectrophotometric assays were monitored on a SpectraMax M5 plate reader using flat-bottomed 96-well plates. Mass spectra were recorded on a Shimadzu LCMS-2019EV.
2.2. Cloning, overexpression, and purification of ANT(3″)-Ii, AAC(6′)-IId, and ANT(3″)-Ii/AAC(6′)-IId in pET22b, pET28a, and Int-pET19b-pps
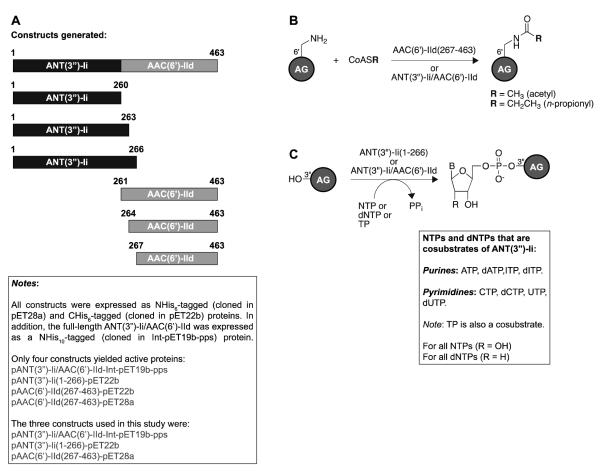
The primers used for PCR amplification are listed in Table S1. All primers introduced NdeI and XhoI restriction sites. The primers labeled as (NHis) were used for insertion of the genes into pET28a or Int-pET19b-pps NdeI/XhoI linearized vectors to generate NHis6-tagged and NHis10-tagged proteins, respectively. The primers labeled as (CHis) were used for insertion of the genes into the pET22b NdeI/XhoI linearized vector to generate CHis6-tagged proteins. Phusion DNA polymerase was used for the PCR reactions as described by NEB. Serratia marcesens genomic DNA (provided by Dr. Paul H. Roy, Université Laval, Canada) was used as a template in the PCR amplification of the gene encoding the full-length ANT(3″)-Ii/AAC(6′)-IId. The amplified ant(3″)-Ii/aac(6′)-IId was inserted into the linearized pET28a, Int-pET19b-pps, and pET22b vectors via the corresponding NdeI/XhoI restriction sites to afford plasmids pANT(3″)-Ii/AAC(6′)-IId-pET28a, pANT(3″)-Ii/AAC(6′)-IId-Int-pET19b-pps, and pANT(3″)-Ii/AAC(6′)-IId-pET22b. The pANT(3″)-Ii/AAC(6′)-IId-pET28a plasmid was used as a template for the subsequent PCR amplification of gene fragments ant(3″)-Ii(1-260), ant(3″)-Ii(1-263), ant(3″)-Ii(1-266), aac(6′)-IId(261-463), aac(6′)-IId(264-463), and aac(6′)-IId(267-463), which were inserted into pET28a and pET22b NdeI/XhoI linearized vectors to afford plasmids pANT(3″)-Ii(1-260)-pET28a, pANT(3″)-Ii(1-260)-pET22b, pANT(3″)-Ii(1-263)-pET28a, pANT(3″)-Ii(1-263)-pET22b, pANT(3″)-Ii(1-266)-pET28a, pANT(3″)-Ii(1-266)-pET22b, pAAC(6′)-IId(261-463)-pET28a, pAAC(6′)-IId(261-463)-pET22b, pAAC(6′)-IId(264-463)-pET28a, pAAC(6′)-IId(264-463)-pET22b, pAAC(6′)-IId(267-463)-pET28a, and pAAC(6′)-IId(267-463)-pET22b. The constructs generated in this study are shown in Fig. 2. All constructs were sequenced and verified.
Fig. 2.
A. Depiction of the various ANT(3″)-Ii/AAC(6′)-IId, ANT(3″)-Ii, and AAC(6′)-IId constructs generated in this study. B. Overview of acetylation activities observed with the constructs presented in panel A. C. Overview of nucleotidylation activities observed with the constructs presented in panel A.
For overexpression and purification, 2×500 mL of Terrific Broth supplemented with 100 μg/mL ampicillin (pET22b and Int-pET19b-pps constructs) or 50 μg/mL KAN (pET28a constructs) were each inoculated with 5-mL overnight cultures of E. coli BL21(DE3) transformed with the plasmid harboring the gene to be overexpressed. Cells were grown (200 rpm, 37 °C). Protein expression was induced at OD600 = 0.8 with 0.5 mM IPTG (final concentration) and the cells were grown (15 °C) for an additional 18-20 h. Cells harvested via centrifugation (6,000 rpm, 10 min) were resuspended in HEPES (50 mM, pH 7.5 adjusted at rt) and 200 mM NaCl (Buffer A) and lysed (10,000-15,000 psi) using an Avestin Emulsiflex-C3 homogenizer. The enzymes were purified by NiII-NTA affinity chromatography, eluting with Buffer A in a stepwise imidazole gradient (10 mL of 2 (1×) and 5 (1×) mM imidazole followed by 5 mL of 20 (1×), 40 (1×), 60 (1×), 200 (2×), and 500 (2×) mM imidazole). Fractions containing pure protein, as analyzed by SDS-PAGE, were combined and dialyzed in 50 mM HEPES (pH 7.5) and 10% (v/v) glycerol at 4 °C. Protein concentrations were determined using a NanoDrop® ND-1000 spectrophotometer. Proteins were flash frozen in liquid N2 and stored at −80 °C (Fig. S1). The 55.0-kDa NHis10-tagged ANT(3″)-Ii/AAC(6′)-IId, the 30.3-kDa CHis6-tagged ANT(3″)-Ii(1-266), and the 24.0-kDa NHis6-tagged AAC(6′)-IId(267-463) were obtained in yields of 5.8, 7.5, and 1.4 mg of enzyme per liter of culture, respectively.
2.3. Measurement of adenylation and acetylation activities for ANT(3″)-Ii/AAC(6′) and its individual domains
Previously reported spectrophotometric assays [18, 19] were used to monitor activities of ANT(3″)-Ii/AAC(6′)-IId, AAC(6′)-IId, and ANT(3″)-Ii.
2.3.1. Cosubstrate and substrate specificity profiles for ANT(3″)-Ii and AAC(6′) for the full-length bifunctional enzyme and for the domains purified separately
The acetyltransferase activity of the AAC(6′)-IId domain of ANT(3″)-Ii/AAC(6′)-IId was monitored at 324 nm through the production of 4-thiopyridone from the reaction of DTDP [18] with the CoA released upon acylation of AGs. To establish the substrate and cosubstrate profiles of the AAC(6′)-IId domain, reactions (200 μL) containing acyl-CoA (200 μM), DTDP (2 mM), and AG (100 μM) were performed in HEPES buffer (50 mM, pH 7.5). Reactions were initiated with AAC(6′)-IId or ANT(3″)-Ii/AAC(6′)-IId (0.4 μM) and incubated at 25 °C for 30 min taking measurements every 30 sec.
The nucleotidyltransferase activity of the ANT(3″)-Ii domain was monitored through the formation of a complex between the molybdate/malachite green reagent and the Pi generated by inorganic pyrophosphatase cleavage of the PPi released during the ANT(3″)-Ii catalyzed reaction [19]. To establish the substrate and cosubstrate profiles of the ANT(3″)-Ii domain, reactions (160 μL) containing Buffer B [Tris-HCl (50 mM, pH 7.5) MgCl2 (10 mM), KCl (50 mM), inorganic pyrophosphatase (0.2 U/mL)], AG (0.2 mM), and (d)NTP (1 mM) were performed at 25 °C. The reactions were initiated by addition of ANT(3″)-Ii or ANT(3″)-Ii/AAC(6′)-IId (0.25 μM), incubated for 20, 40, 60, 80, and 100 sec, and quenched with the molybdate/malachite green reagent (40 μL) to terminate the reaction. After 15 min of color development, the liberated Pi concentration was measured by reading absorbance at 600 nm.
2.4. Determination of kinetic parameters
2.4.1. Determination of kinetic parameters for AAC(6′)-IId
The kinetic parameters for the AAC(6′)-IId(267-463) enzyme alone and the AAC(6′)-IId portion of the ANT(3″)-Ii/AAC(6′)-IId full-length bifunctional enzyme were determined in an identical manner. Steady-state kinetic measurements as a function of AG concentration were performed in reactions (200 μL) containing varying concentrations of AG (0-100 or 0-200 or 0-300 μM depending on the AG) and a constant concentration of acetyl-CoA (500 μM) in HEPES (50 mM, pH 7.5), DTDP (2 mM), and enzyme (0.25 μM). Acetyl-CoA kinetic parameters were determined in a similar manner by varying the concentration of acetyl-CoA (0-200 μM for AAC(6′)-IId(267-463) or 0-500 μM for ANT(3″)-Ii/AAC(6′)-IId) while keeping the concentration of AG constant (1 mM NEO for AAC(6′)-IId(267-463) and 1 mM KAN for ANT(3″)-Ii/AAC(6′)-IId). Reactions were initiated by the addition of the component for which the concentration was varied and monitored at 324 nm taking measurements every 20 sec for 20 min. The initial rates were calculated using the first 2-5 min of the reaction. The Michaelis-Menten parameters, Km and kcat, were determined by Kaleidagraph 4.1 curve fitting software or Lineweaver-Burk analysis for the combinations indicated by an asterisk in Table 1 (Figs. S2-3).
Table 1.
Steady-state kinetic characterization of full-length and separated ANT(3”)-Ii/AAC(6′)-IId
| Activity | Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1s−1) | kcatfull / kcatfrag | kcat/Kmfull / kcat/Kmfrag |
|---|---|---|---|---|---|---|
| NHis6-tagged AAC(6′)-IId | ||||||
| Acetyltransferase | AMK* | 127 ± 6 | 7.09 ± 0.53 | 6.24 ± 0.53 × 104 | 0.412 | 84.1 |
| GEN | 14.3 ± 2.3 | 3.33 ± 0.14 | 2.34 ± 0.39 × 105 | 0.333 | 0.940 | |
| KAN | 16.0 ± 1.7 | 2.13 ± 0.06 | 1.33 ± 0.15 × 105 | 1.79 | 14.2 | |
| NEO | 32.1 ± 11.1 | 11.1 ± 1.0 | 3.46 ± 1.24 × 105 | 0.816 | 1.14 | |
| NET | 3.87 ± 0.90 | 3.21 ± 0.20 | 8.29 ± 2.00 × 105 | 0.595 | 1.30 | |
| RIB | 69.3 ± 4.5 | 5.70 ± 0.12 | 8.22 ± 0.56 × 104 | 0.681 | 2.96 | |
| SIS | 4.92 ± 1.14 | 5.35 ± 0.27 | 1.09 ± 0.26 × 106 | 0.548 | 1.22 | |
| TOB | 14.8 ± 3.4 | 3.73 ± 0.25 | 2.52 ± 0.60 × 105 | 1.03 | 1.92 | |
| AcCoA | 147 ± 36 | 12.1 ± 1.6 | 8.20 ± 2.29 ×104 | 0.132 | 0.102 | |
|
| ||||||
| NHis10-tagged ANT(3”)-Ii/AAC(6′)-IId | ||||||
| Acetyltransferase | AMK | 0.556 ± 0.130 | 2.92 ± 0.10 | 5.25 ± 1.25 × 106 | ||
| GEN | 5.04 ± 0.65 | 1.11 ± 0.03 | 2.20 ± 0.29 × 105 | |||
| KAN | 2.02 ± 0.41 | 3.81 ± 0.15 | 1.89 ± 0.39 × 106 | |||
| NEO | 23.0 ± 4.8 | 9.06 ± 0.4 | 3.94 ± 0.84 × 105 | |||
| NET | 1.77 ± 0.41 | 1.91 ± 0.08 | 1.08 ± 0.25 × 106 | |||
| RIB | 16.0 ± 3.9 | 3.88 ± 0.29 | 2.43 ± 0.63 × 105 | |||
| SIS | 2.20 ± 0.60 | 2.93 ± 0.16 | 1.33 ± 0.37 × 106 | |||
| TOB | 7.92 ± 0.53 | 3.84 ± 0.07 | 4.85 ± 0.34 × 105 | |||
| AcCoA* | 215 ± 25 | 1.62 ± 0.02 | 8.38 ± 1.09 × 103 | |||
| Nucleotidyltransferase | SPT | 0.159 ± 0.007 | 0.748 ± 0.007 | 4.70 ± 0.21 × 106 | ||
| ATP | 27.9 ± 4.1 | 0.753 ± 0.040 | 2.70 ± 0.42 × 104 | |||
| dATP | 20.9 ± 1.6 | 0.405 ± 0.010 | 1.94 ± 0.16 × 104 | |||
| CTP* | 131 ± 28 | 0.531 ± 0.064 | 4.04 ± 0.99 × 103 | |||
| dCTP* | 166 ± 19 | 1.11 ± 0.01 | 6.70 ± 0.76 × 103 | |||
| ITP | 30.6 ± 2.6 | 0.148 ± 0.004 | 4.83 ± 0.43 × 103 | |||
| UTP* | 105 ± 6 | 0.526 ± 0.014 | 5.02 ± 0.32 × 103 | |||
| dUTP | 74.7 ± 6.5 | 0.535 ± 0.022 | 7.16 ± 0.69 × 103 | |||
|
| ||||||
| CHis6-tagged ANT(3”)-Ii | ||||||
| Nucleotidyltransferase | SPT | 0.123 ± 0.015 | 5.13 ± 0.11 | 4.15 ± 0.51 × 107 | 0.147 | 0.113 |
| ATP | 8.50 ± 1.46 | 1.15 ± 0.07 | 1.35 ± 0.25 × 105 | 0.655 | 0.200 | |
| dATP | 4.48 ± 1.15 | 0.508 ± 0.035 | 1.13 ± 0.30 × 105 | 0.797 | 0.172 | |
| CTP | 33.3 ± 5.6 | 0.709 ± 0.040 | 2.13 ± 0.38 × 104 | 0.749 | 0.190 | |
| dCTP* | 121 ± 6 | 0.667 ± 0.007 | 5.48 ± 0.27 × 103 | 1.66 | 1.22 | |
| ITP | 12.6 ± 1.5 | 0.481 ± 0.013 | 3.81 ± 0.45 × 104 | 0.308 | 0.127 | |
| UTP | 26.2 ± 5.7 | 0.603 ± 0.047 | 2.30 ± 0.53 × 104 | 0.872 | 0.218 | |
| dUTP | 46.1 ± 6.4 | 0.565 ± 0.031 | 1.23 ± 0.18 × 104 | 0.947 | 0.582 | |
Indicates that the kinetic parameters were in these rare instances determined by Lineweaver-Burk instead of Michaelis-Menten.
Full = ANT(3”)-Ii/AAC(6′)-IId
Frag = ANT(3”)-Ii or AAC(6′)-IId
2.4.2. Generation of a standard curve for Pi concentrations using the molybdate/malachite green reagent
To generate a standard curve of Pi concentrations to be used for determination of the kinetic parameters for ANT(3″)-Ii, the molybdate/malachite green reagent (40 μL) was added to NaH2PO4 (0.06, 0.12, 0.24, 0.48, 0.95, 1.91, and 3.82 μM) in Buffer B, SPT (100 μM), and ANT(3″)-Ii(1-266) (0.02 μM)] (160 μL). After 15 min of color development, the Pi concentration was measured as above. We observed a linear response up to 3.82 μM Pi, which was best fit to a line y = 54.75x, where x = [Pi] (in μM) and y = absorbance at 600 nm (Fig. S4).
2.4.3. Determination of kinetic parameters for ANT(3″)-Ii
The kinetic parameters for the ANT(3″)-Ii(1-266) enzyme alone and the ANT(3″) domain of the ANT(3″)-Ii/AAC(6′)-IId bifunctional enzyme were determined in an identical manner. The SPT kinetic parameters were established in reactions (160 μL) containing SPT (0-2.5 μM for ANT(3″)-Ii/AAC(6′)-IId or 0-10 μM for ANT(3″)-Ii(1-266)) and a constant concentration of ATP (200 μM in Buffer B. Reactions were initiated by addition of enzyme (0.02 μM) at 25 °C. Reactions were quenched by addition of a malachite green solution (40 μL) as previously reported [20]. Measurements were taken every 15 sec for 75 sec. The kinetic parameters for (d)NTPs were determined in an analogous manner using varying concentrations of (d)NTPs (0-50 or 0-100 or 0-150 or 0-1000 μM depending on the (d)NTP) and a constant concentration of SPT (750 μM). Reactions were initiated by addition of enzyme (0.25 μM) and quenched as described above. The kinetic parameters Km and kcat were determined by data fitting to a classical Michaelis-Menten equation with Kaleidagraph 4.1 software or by Lineweaver-Burk plots for the combinations indicated by an asterisk in Table 1 (Figs. S5-6).
2.5. Mass spectrometric analysis of acetylation and nucleotidylation reactions
To monitor acetylation by mass spectrometry, reactions (10 μL) containing AGs (2 mM), acetyl-CoA (3 mM) and ANT(3″)-Ii/AAC(6′)-IId (6.5 μM or 0.36 mg/mL) in HEPES (50 mM, pH 7.5) were incubated overnight at rt. To monitor nucleotidylation by mass spectrometry, reactions (30 μL) containing SPT (0.67 mM), (d)NTP (13 mM) and ANT(3″)-Ii/AAC(6′)-IId (5 μM or 0.28 mg/mL) in Tris (50 mM, pH 7.5), MgCl2 (10 mM), and KCl (40 mM) were incubated overnight at rt. The protein was precipitated by addition of an equal volume of ice-cold methanol (10 or 30 μL) and the reaction mixture was kept at −20 °C for at least 10 min before centrifugation (rt, 13,000 rpm, 10 min). The supernatant (20 μL) was added to H2O (25 μL) and at least 15 μL of the sample was analyzed by LCMS using 0.1% formic acid in H2O under the positive ionization mode (Table 2 and Fig. S7).
Table 2.
Mass analysis of AGs acetylated and SPT nucleotidylated using the bifunctional enzyme ANT(3”)-Ii/AAC(6′)-IId
| Acetylation | Nucleotidylation | ||||
|---|---|---|---|---|---|
| AG | Calc [M+H]+ | Obs [M+H]+ | Cosubstrate | Calc [M+H]+ | Obs [M+H]+ |
| AMK | 628.65 | 628.25 | ATP | 662.22 | 662.10 |
| GEN C1 | 520.64 | Not observed | dATP | 646.22 | 646.00 |
| GEN C2 | 506.61 | 506.25 | ITP | 663.20 | 663.20 |
| GEN C1a | 492.59 | 492.30 | dITP | 647.21 | 670.95 (+Na) |
| KAN | 527.54 | 527.15 | UTP | 639.19 | 639.05 |
| NEO | 657.69 | 657.35 | dUTP | 623.20 | 623.00 |
| NET | 518.62 | 518.25 | TP | 413.31 | 435.95 (+Na) |
| RIB | 497.52 | 497.20 | |||
| SIS | 490.57 | 490.10 | |||
| TOB | 510.56 | 510.30 | |||
3. Results and discussion
3.1. Cloning, expression, and purification of various constructs of full-length ANT(3″)-Ii/AAC(6′)-IId and its individual domains
Despite decades of research, our understanding of the role and structure of bifunctional AMEs remains limited. In early attempts to understand the role and impact of domain interactions of bifunctional AMEs on antibiotic resistance, Wright and co-workers demonstrated that a connecting α-helix between the two domains of the fused AAC(6′)-Ie/APH(2″)-Ia was critical to maintain both correct structure and function of both domains [21]. More recently, by using small-angle X-ray scattering to model the structure of this bifunctional enzyme, Berghuis and co-workers concluded that AAC(6′)-Ie/APH(2″)-Ia adopts a rigid conformation in solution, which likely contributes to improvement of the enzymatic activities of the individual enzymes [22]. By revisiting the nucleotide and AG substrate specificity of this bifunctional enzyme, Vakulenko and co-workers recently reported that, contrary to previous beliefs, GTP is the exclusive phosphate donor in cells for this enzyme [9].
Inspired by these studies, we aimed to determine if the findings for the AAC(6′)-Ie/APH(2″)-Ia would hold true for ANT(3″)-Ii/AAC(6′)-IId, originally studied exclusively in its full-length form [5]. To first investigate if the ANT(3″)-Ii and AAC(6′)-IId domains were structurally integrated, in addition to cloning the full-length enzyme we split the ant(3″)-Ii/aac(6′)-IId gene at three positions to generate the ANT(3″)-Ii(1-260), ANT(3″)-Ii(1-263), ANT(3″)-Ii(1-266), AAC(6′)-IId(261-463), AAC(6′)-IId(264-463), and AAC(6′)-IId(267-463) enzymes (Fig. 2). All enzymes were cloned into both pET28a and pET22b vectors to afford NHis6- and CHis6-tagged enzymes, respectively. The full-length ANT(3″)-Ii/AAC(6′)-IId was additionally cloned into the Int-pET19b-pps vector to generate the NHis10-tagged version of the enzyme. Interestingly, even though all proteins were expressed in a soluble form, only four constructs yielded active enzymes. These four constructs were pANT(3″)-Ii/AAC(6′)-IId-Int-pET19b-pps, pANT(3″)-Ii(1-266)-pET22b, pAAC(6′)-IId(267-463)-pET22b, and pAAC(6′)-IId(267-463)-pET28a. It is to note that out of the two AAC(6′)-IId(267-463) enzymes, only the AAC(6′)-IId(267-463)(NHis) was used for the remainder of this study. In our case, this extremely precise cut between the two active domains is distinct from what was previously observed for the two domains of AAC(6′)-Ie/APH(2″)-Ia overlapping by 20 amino acid residues. This phenomenon may suggest a different mode of evolution for the ANT(3″)-Ii/AAC(6′)-IId than that involved with the formation of AAC(6′)-Ie/APH(2″)-Ia. We previously showed that the acetyltransferase activities of both domains of the bifunctional AAC(3)-Ib/AAC(6′)-Ib’ were necessary for total inactivation of GEN [14]. The complete lack of overlap between ANT(3″)-Ii and AAC(6′)-IId may also be indicative of limited interactions between the two domains and could potentially explain the fact that they have different AG substrates.
3.2. Substrate and cosubstrate specificity profiles of ANT(3″)-Ii and AAC(6′)-IId
To further explore the differences between the full-length ANT(3″)-Ii/AAC(6′)-IId and its individually expressed components, we determined the substrate and cosubstrate specificity profiles for all enzymes. We tested the acyltransferase activity of AAC(6′)-IId(267-463) and ANT(3″)-Ii/AAC(6′)-IId against a panel of 13 AGs. We found AMK, GEN, KAN, NEO, NET, RIB, SIS, and TOB to be good substrates of the acetyltransferase in both enzymes, while we did not observe any acetylation of APR, HYG, PAR, SPT, and STR. We confirmed all AG acetylations by mass spectrometry (Table 2). We previously demonstrated that some AACs display broad cosubstrate promiscuity [18] while others, like Eis [23], are characterized by a narrow cosubstrate profile. To investigate the cosubstrate profile of AAC(6′)-IId alone or in the presence of ANT(3″), we tested 16 acyl-CoA derivatives. Only acetyl-CoA and n-propionyl-CoA were found to display reasonable acylation activity. We next tested the nucleotidyltransferase activity of ANT(3″)-Ii(1-266) and ANT(3″)-Ii/AAC(6′)-IId against SPT and STR in combination with all biologically relevant (d)NTPs. By using a UV-Vis colorimetric assay, we observed that out of the eleven (d)NTPs tested, only TTP, GTP, and dGTP were not accepted as cosubstrates of the bifunctional enzyme and ANT(3″)-Ii(1-266) alone. These results were confirmed by mass spectrometry (Table 2 and Fig. S7). As with ANT(4′) [20], TP also resulted in a productive reaction yielding phosphorylated AGs as confirmed by MS analysis (Table 2 and Fig. S7).
3.3. Kinetic characterization of ANT(3″)-Ii, ANT(3″)-Ii/AAC(6′)-IId, and AAC(6′)-IId
With the activity of the bifunctional ANT(3″)-Ii/AAC(6′)-IId and of its monofunctional counterparts confirmed with a majority of the AGs and (d)NTPs tested as well as with two acyl-CoAs, we determined the kinetic parameters for the three active enzymes (Table 1). We first studied the acetyltransferase activities of AAC(6′)-IId(267-463) and ANT(3″)-Ii/AAC(6′)-IId. Acetyl-CoA was found to bind to both enzymes with similar affinity. Overall, the AGs studied were found to bind with lower affinities to the AAC(6′)-IId(267-463) domain than to the bifunctional enzyme. These data suggest that the ANT(3″)-Ii domain might play a role in altering the overall structure of the bifunctional enzyme to allow tighter binding of the AGs in the active site of the AAC(6′)-IId domain. With a Km value of 4.92 ± 1.14 μM and a kcat value of 5.35 ± 0.27 s−1, SIS was established as the best AG substrate for AAC(6′)-IId(267-463), while AMK, with a Km value of 0.556 ± 0.130 μM and a kcat value of 2.92 ± 0.10 s−1, was found to be the best AG substrate for the bifunctional enzyme. By examining the ratios of the kcat values for the full-length enzyme over those for the individually expressed AAC(6′)-IId(267-463), we established that, with the exception of KAN and TOB, all AGs have lower turnover rate constants with the bifunctional enzyme. However, the catalytic efficiencies for acetylation by AAC(6′)-IId(267-463) and ANT(3″)-Ii/AAC(6′)-IId are in general very similar, with the exception of KAN and AMK that display much higher kcat/Km values with the bifunctional enzyme. When compared to some previously published data for AMK, GEN, KAN, and NET with the full-length ANT(3″)-Ii/AAC(6′)-IId, we found that in our hands, lower Km and higher kcat values were generally obtained for these AGs.
We next studied the nucleotidyltransferase activity of ANT(3″)-Ii(1-266) and ANT(3″)-Ii/AAC(6′)-IId. All (d)NTPs were found to bind 1.4- to 4.7-fold better to the ANT(3″)-Ii(1-266) domain alone than to the same domain in the bifunctional enzyme, suggesting that the AAC(6′)-IId domain somewhat interferes with binding of (d)NTPs. Despite the difference in binding constants, the catalytic turnover constants (kcat) for each (d)NTP were similar. SPT bound nearly identically to both enzyme constructs. However, the kcat value for SPT was 7-fold higher with ANT(3″)-Ii(1-266) (5.13 ± 0.11 s−1) than with the bifunctional enzyme (0.748 ± 0.007 s−1). With a Km values of 8.50 ± 1.46 μM with ANT(3″)-Ii(1-266) and 27.9 ± 4.1 μM with ANT(3″)-Ii/AAC(6′)-IId as well as kcat values of 1.15 ± 0.07 s−1 with ANT(3″)-Ii(1-266) and 0.753 ± 0.040 s−1 with ANT(3″)-Ii/AAC(6′)-IId, ATP was found to be the best cosubstrate, followed closely by dATP (Table 1). For any given nucleotide, the NTPs and their dNTP counterparts have similar catalytic efficiencies with both enzymes. By examining the ratios of the kcat and kcat/Km values for the full-length enzyme relative to those for the individually expressed ANT(3″)-Ii(1-266), we established that for all triphosphates, with the exception of dCTP, the catalytic turnovers and catalytic efficiencies are higher for the ANT(3″)-Ii(1-266) domain alone. These data suggest that removing the AAC(6′)-IId domain allows the triphosphates to bind more thightly to the nucleotidyltransferase, speeds up product formation, and improves the enzyme efficiency.
To better understand the domain-domain interaction of ANT(3″)-Ii/AAC(6′)-IId, crystal structures of the bifunctional enzyme and of its individual components should be determined and compared for structural differences. Unfortunately, the only ANT structures available to date are that of ANT(4′) from Staphylococcus aureus [24] (PDB: 1KNY) and ANT(4′)-IIb from Pseudomonas aeruginosa (PDB: 4EBJ) and no conclusions can be drawn from these structures since ANT(3″)-Ii has a completely different substrate profile than those of ANT(4′)s [25, 26] and display only 13-16% sequence identity with these ANT(4′)s. There are, however, several crystal structures of AAC(6′)s. Based on the crystal structure of AAC(6′)-Ib [27] (PDB: 2QIR), for which the amino acid sequence is nearly identical (only 2 amino acid residues differ in the core sequence) to that of AAC(6′)-IId(267-463), the binding site of KAN is not particularly close to the N-terminus of the enzyme. This suggests that the ANT(3″)-Ii domain located at the N-terminus of the bifunctional enzyme most likely does not interact directly with the AG binding site. Rather, its presence at the N-terminus probably slightly alters the protein structure, enabling the AGs to bind more tightly. This theory is further corroborated by the crystal structure of AAC(6′)-Iy [28] (PDB: 1S32), which has poor sequence homology with AAC(6′)-IId, but for which the structure also demonstrates that the AG binding site is located far from the N-terminus of the enzyme.
In summary, we demonstrated that in contrary to the structural integration of both portions of AAC(6′)-Ie/APH(2″)-Ia essential for proper acetyltransferase and phosphotransferase activities, no structural overlap of the two domains of ANT(3″)-Ii/AAC(6′)-IId is required for proper nucleotidyltransferase and acetyltransferase activities. We showed that while AAC(6′)-IId appears to somewhat hinder (d)NTP binding to ANT(3″)-Ii, the latter domain appears to play a critical role in dictating the overall structure of the bifunctional enzyme for tighter binding of AGs to the acetyltransferase domain. We also demonstrated that while ANT(3″)-Ii substrate specificity is limited to STR and SPT, this enzyme displays broad cosubstrate promiscuity, accepting equally well eight of the eleven (d)NTPs tested. Conversely, we observed AAC(6′)-IId to display broad substrate promiscuity, accepting eight of the thirteen AGs tested, and high cosubstrate selectivity with only acetyl- and n-propionyl-CoA being preferentially transferred to 4,6-disubstituted-DOS AG scaffolds over 4,5-disubstituted-DOS AG cores. Unlike AAC(6′)-Ie/APH(2″)-Ia, ANT(3″)-Ii/AAC(6′)-IId can be spilt into two independent domains indicating the lack of structural overlap between the two domains, suggesting an alternative mode of evolution of the bifuntional ANT(3″)-Ii/AAC(6′)-IId. Crystal structures of ANT(3″)-Ii/AAC(6′)-IId and its individual domains are needed to shed more light onto this bifunctional enzyme. Efforts aimed at determining these structures are currently underway in our group. Methods to sidestep or overcome antibiotic resistance resulting from the acquisition of AMEs are also currently under investigation in our group.
Supplementary Material
Research highlights.
The activity of each domain of ANT(3″)/AAC(6′) is independent of the other domain.
ANT(3″) plays a critical role in binding of aminoglycosides to AAC(6′).
Many (d)NTPs are accepted equally well as cosubstrates of ANT(3″)/AAC(6′).
CoA analogs are preferentially transferred to 4,6-disubstituted aminoglycosides.
Acknowledgements
This work was supported by a grant from the Firland Foundation (S.G.-T.), a BSF grant 2008017 (S.G.-T.), and a National Institute of Health (NIH) grant AI090048 (S.G.-T.) We thank Vanessa Porter for cloning and preliminary experimental work. We thank Dr. Paul H. Roy for the Serratia marcescens genomic DNA and Dr. Tapan Biswas for the Int-pET19b-pps vector. We thank Oleg V. Tsodikov for critical reading and insightful comments on the manuscript.
Footnotes
Supplementary data. A table of primers used in this study (Table S1) and figures showing the purified ANT(3″)-Ii/AAC(6′)-IId, ANT(3″)-Ii(1-266), and AAC(6′)-IId(267-463) proteins (Fig. S1), kinetic curves (Figs. S2-3/S5-6), standard curve for Pi concentrations (Fig. S4), and representative mass spectra for acetylation of various AGs and nucleotidylation of SPT with various NTPs, dNTPs, and TP (Fig. S7) are provided.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jana S, Deb JK. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol. 2006;70:140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- [2].Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Houghton JL, Green KD, Chen WJ, Garneau-Tsodikova S. The Future of Aminoglycosides: The End or Renaissance? Chembiochem. 2010;11:880–902. doi: 10.1002/cbic.200900779. [DOI] [PubMed] [Google Scholar]
- [4].Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- [5].Kim C, Hesek D, Zajicek J, Vakulenko SB, Mobashery S. Characterization of the bifunctional aminoglycoside-modifying enzyme ANT(3″)-Ii/AAC(6′)-IId from Serratia marcescens. Biochemistry. 2006;45:8368–8377. doi: 10.1021/bi060723g. [DOI] [PubMed] [Google Scholar]
- [6].Centron D, Roy PH. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Daigle DM, Hughes DW, Wright GD. Prodigious substrate specificity of AAC(6′)-APH(2″), an aminoglycoside antibiotic resistance determinant in enterococci and staphylococci. Chem Biol. 1999;6:99–110. doi: 10.1016/S1074-5521(99)80006-4. [DOI] [PubMed] [Google Scholar]
- [8].Culebras E, Martinez JL. Aminoglycoside resistance mediated by the bifunctional enzyme 6′-N-aminoglycoside acetyltransferase-2″-O-aminoglycoside phosphotransferase. Front Biosci. 1999;4:D1–8. doi: 10.2741/culebras. [DOI] [PubMed] [Google Scholar]
- [9].Frase H, Toth M, Vakulenko SB. Revisiting the Nucleotide and Aminoglycoside Substrate Specificity of the Bifunctional Aminoglycoside Acetyltransferase(6′)-Ie/Aminoglycoside Phosphotransferase(2″)-Ia Enzyme. J Biol Chem. 2012 doi: 10.1074/jbc.M112.416453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim C, Villegas-Estrada A, Hesek D, Mobashery S. Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6′)-Ib’ from Pseudomonas aeruginosa. Biochemistry. 2007;46:5270–5282. doi: 10.1021/bi700111z. [DOI] [PubMed] [Google Scholar]
- [11].Dubois V, Poirel L, Marie C, Arpin C, Nordmann P, Quentin C. Molecular characterization of a novel class 1 integron containing bla(GES-1) and a fused product of aac3-Ib/aac6′-Ib’ gene cassettes in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2002;46:638–645. doi: 10.1128/AAC.46.3.638-645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang W, Fisher JF, Mobashery S. The bifunctional enzymes of antibiotic resistance. Curr Opin Microbiol. 2009;12:505–511. doi: 10.1016/j.mib.2009.06.013. [DOI] [PubMed] [Google Scholar]
- [13].Mendes RE, Toleman MA, Ribeiro J, Sader HS, Jones RN, Walsh TR. Integron carrying a novel metallo-beta-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistant gene aac(6′)-30/aac(6′)-Ib’: report from the SENTRY Antimicrobial Surveillance Program. Antimicrobial agents and chemotherapy. 2004;48:4693–4702. doi: 10.1128/AAC.48.12.4693-4702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Green KD, Chen W, Garneau-Tsodikova S. Effects of altering aminoglycoside structures on bacterial resistance enzyme activities. Antimicrob Agents Chemother. 2011;55:3207–3213. doi: 10.1128/AAC.00312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Azucena E, Grapsas I, Mobashery S. Properties of a bifunctional bacterial antibiotic resistance enzyme that catalyzes ATP-dependent 2″-phosphorylation and acetyl-CoA-dependent 6′-acetylation of aminoglycosides. J. Am. Chem. Soc. 1997;119:2317–2318. [Google Scholar]
- [16].Shaul P, Green KD, Rutenberg R, Kramer M, Berkov-Zrihen Y, Breiner-Goldstein E, Garneau-Tsodikova S, Fridman M. Assessment of 6′- and 6′″-N-acylation of aminoglycosides as a strategy to overcome bacterial resistance. Org Biomol Chem. 2011;9:4057–4063. doi: 10.1039/c0ob01133a. [DOI] [PubMed] [Google Scholar]
- [17].Biswas T, Tsodikov OV. Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. Febs J. 2008;275:3064–3071. doi: 10.1111/j.1742-4658.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- [18].Green KD, Chen WJ, Houghton JL, Fridman M, Garneau-Tsodikova S. Exploring the Substrate Promiscuity of Drug-Modifying Enzymes for the Chemoenzymatic Generation of N-Acylated Aminoglycosides. Chembiochem. 2010;11:119–126. doi: 10.1002/cbic.200900584. [DOI] [PubMed] [Google Scholar]
- [19].McQuade TJ, Shallop AD, Sheoran A, Delproposto JE, Tsodikov OV, Garneau-Tsodikova S. A nonradioactive high-throughput assay for screening and characterization of adenylation domains for nonribosomal peptide combinatorial biosynthesis. Anal Biochem. 2009;386:244–250. doi: 10.1016/j.ab.2008.12.014. [DOI] [PubMed] [Google Scholar]
- [20].Porter VR, Green KD, Zolova OE, Houghton JL, Garneau-Tsodikova S. Dissecting the cosubstrate structure requirements of the Staphylococcus aureus aminoglycoside resistance enzyme ANT(4 ′) Biochem Bioph Res Co. 2010;403:85–90. doi: 10.1016/j.bbrc.2010.10.119. [DOI] [PubMed] [Google Scholar]
- [21].Boehr DD, Daigle DM, Wright GD. Domain-domain interactions in the aminoglycoside antibiotic resistance enzyme AAC(6′)-APH(2″) Biochemistry. 2004;43:9846–9855. doi: 10.1021/bi049135y. [DOI] [PubMed] [Google Scholar]
- [22].Caldwell SJ, Berghuis AM. Small-angle X-ray scattering analysis of the bifunctional antibiotic resistance enzyme aminoglycoside (6′) acetyltransferase-ie/aminoglycoside (2″) phosphotransferase-ia reveals a rigid solution structure. Antimicrobial agents and chemotherapy. 2012;56:1899–1906. doi: 10.1128/AAC.06378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen W, Green KD, Garneau-Tsodikova S. Cosubstrate Tolerance of the Aminoglycoside Resistance Enzyme Eis from Mycobacterium tuberculosis. Antimicrobial agents and chemotherapy. 2012;56:5831–5838. doi: 10.1128/AAC.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pedersen LC, Benning MM, Holden HM. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry. 1995;34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- [25].Revuelta J, Vacas T, Torrado M, Corzana F, Gonzalez C, Jimenez-Barbero J, Menendez M, Bastida A, Asensio JL. NMR-based analysis of aminoglycoside recognition by the resistance enzyme ANT(4′): the pattern of OH/NH3(+) substitution determines the preferred antibiotic binding mode and is critical for drug inactivation. J Am Chem Soc. 2008;130:5086–5103. doi: 10.1021/ja076835s. [DOI] [PubMed] [Google Scholar]
- [26].Jacoby GA, Blaser MJ, Santanam P, Hachler H, Kayser FH, Hare RS, Miller GH. Appearance of amikacin and tobramycin resistance due to 4′-aminoglycoside nucleotidyltransferase [ANT(4′)-II] in gram-negative pathogens. Antimicrob Agents Chemother. 1990;34:2381–2386. doi: 10.1128/aac.34.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maurice F, Broutin I, Podglajen I, Benas P, Collatz E, Dardel F. Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep. 2008;9:344–349. doi: 10.1038/embor.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem Biol. 2004;11:565–573. doi: 10.1016/j.chembiol.2004.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.