Abstract
The Morita-Baylis-Hillman reaction forms a carbon-carbon bond between the alpha carbon of a conjugated carbonyl compound and a carbon electrophile. The reaction mechanism involves Michael addition of a nucleophile catalyst at the carbonyl beta carbon, followed by bond formation with the electrophile and catalyst disassociation to release the product. We used Rosetta to design 48 proteins containing active sites predicted to carry out this mechanism, of which two show catalytic activity by mass spectrometry (MS). Substrate labeling measured by MS and site-directed mutagenesis experiments show that the designed active-site residues are responsible for activity, although rate acceleration over background is modest. To characterize the designed proteins, we developed a fluorescence-based screen for intermediate formation in cell lysates, carried out microsecond molecular dynamics simulations, and solved X-ray crystal structures. These data indicate a partially formed active site, and suggest several clear avenues for designing more active catalysts.
Introduction
Enzymes with the ability to catalyze commonly used synthetic reactions on arbitrary chemical substrates would enable complex syntheses to be carried out bio-catalytically. Computationally engineered enzymes can in principle provide control over every step of a reaction and computation offers the possibility to go directly from a theoretical understanding of reaction mechanism to a working protein catalyst. Recent computational work has created catalysts for simple unimolecular reactions (1–3), and more recently for a bimolecular reaction (4). Work in experimental enzyme engineering has created enzymes for more useful reactions starting from natural enzymes by reworking substrate specificity (5). The Morita-Baylis-Hillman (MBH) reaction is a carbon-carbon bond forming reaction that creates products in which multiple functional groups have been preserved for subsequent synthetic steps (6). No enzyme or catalytic antibody catalyzes the reaction, and existing catalysts are often quite slow for biologically-relevant substrates, usually requiring one or more days for high yields. The MBH is a useful test case for the development of methods to create a protein catalyst for a slow, multi-step reaction for which no catalyst yet exists.
In the MBH reaction a nucleophile catalyzes the formation of a C-C single bond between the alpha position of an alpha, beta-unsaturated carbonyl and an electrophilic carbon (such as an aldehyde). The MBH reaction was first reported by Morita in 1968 (7), later refined to use less expensive catalysts by Baylis and Hillman (8), and has been used widely in inter- and intramolecular forms, e.g. in the syntheses of epopromycin B and salinosporamide A (9, 10). The most common catalysts in synthetic use are DABCO, quinuclidine, and cinchona-derived alkaloids, all of which have a tertiary amine nucleophile, and likely follow the mechanism indicated in Figure 1A (11, 12). More recently, N-methylimidazole alone has been shown to catalyze the reaction at a low level, with acceleration by proline (13). Miller and co-workers have observed MBH catalysis in chloroform/THF by a variety of N-methylhistidine-containing peptides in combination with proline (14). No catalytic antibodies have been made for the reaction, but various proteins, including albumins and a lipase, have a low level of promiscuous activity for the MBH reaction. (15) The Rauhut-Currier (RC) reaction is similar to the MBH, proceeding with nucleophile catalysis where the nucleophile can be a cysteine-like thiol. Evidence from bioorganic catalysis (16) and examination of a natural synthetic pathway (17) strongly suggest that the RC is catalyzed by natural enzymes. An MBH enzyme would provide a conceptual “missing link” between small-molecule and protein catalysis.
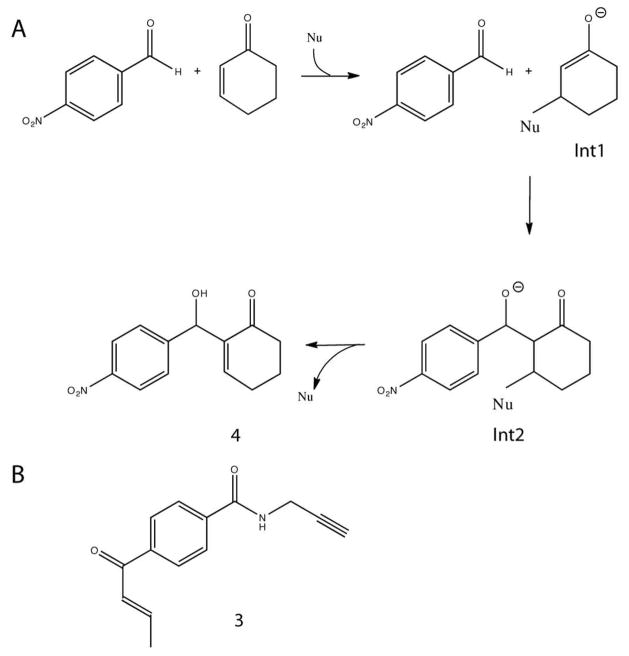
Figure 1.
(A) Morita-Baylis-Hillman reaction between cyclohexenone and 4-nitrobenzaldehyde proceeds via two intermediates Int1 and Int2. Int1 is enolate formed after the nucleophilic (Nu) attack on the C-beta carbon of the enone. Next 4-nitrobenzaldehyde attacks the enolate of Int1 to form Int2. The final product is released after the proton rearrangement and the catalyst is recycled. (B) Enone probe 3 (E)-(4-But-2-enoyl-N-(prop-2-ynyl)benzamide and MBH product 4 2-(Hydroxy(4-nitrophenyl)methyl)cyclohex-2-enone.
Here we focus on the simple model MBH reaction between 2-cyclohexenone (henceforth, “cyclohexenone”) and 4-nitrobenzaldehyde (Figure 1A). The MBH occurs via formation of two distinct catalyst-substrate intermediates and four transition states. We focus our designs on nucleophilic histidines, following the imidazole catalyzed MBH (18) example, and on nucleophilic cysteines, following the example of the RC reaction (19). We used Rosetta scaffold-identification and protein design protocols, including new tools to enable scanning over the entire Protein Data Bank (PDB), to develop candidate MBH protein catalysts. We screened those designs for intermediate labeling in lysate, and carried out a plate assay in lysate for full MBH activity to detect more active variants. Two of the designs modestly accelerate the full MBH reaction. One of the designs binds cyclohexenone in the first step of an MBH reaction, while both show poor binding of the second substrate, limiting overall efficiency. Crystal structures of both designs show nucleophilic residues positioned as designed. The protocol of computational scaffold identification and design, detection of intermediates in lysate, and full MBH reaction activity measurement in lysate should be extendable to develop protein catalysts of the MBH reaction with arbitrary substrates.
Results
We chose the MBH reaction between cyclohexenone and 4-nitrobenzaldehyde (Figure 1A) for design because it is a common test reaction for traditional catalysts and peptide-based catalysts (14). The goal of the design process was to computationally model a protein active site that would bind tightly to the transition state (TS) of the reaction, thereby lowering the activation energy and catalyzing the reaction. We used an “inside-out” computational design protocol with three steps (1, 2): (1) definition of an ideal active site predicted to catalyze the reaction – the catalytic residues and the TS are a “theozyme” (2) dentification of sets of positions in protein structures capable of accommodating the theozyme, and (3) design of the surrounding residues to optimize TS binding and catalytic residue placement. The designed computationally protein sequences were experimentally tested for activity.
Based on the mechanism of the MBH reaction depicted in Figure 1A the ideal active site would contain: (1) a cysteine or histidine nucleophile; (2) two hydrogen bond donors to stabilize Int1; (3) hydrogen bond donors for Int2; (4) a hydrophobic binding site for cyclohexenone; and (5) a hydrophobic, possibly aromatic, binding site for benzaldehyde. This site would contain at least 10 residues with precise geometric placements relative to the substrates. It is extremely unlikely to find an existing protein backbone capable of meeting all of these criteria. We chose to focus on the nucleophile, Int1 stabilization and benzaldehyde stacking. We built a model of the transition state for carbon-carbon bond formation using short MD simulations and QM modeling (20). The theozyme was modeled as a composite transition state – a superposition of the transition state (from QM and MD) and int2, not a single structure. Coordinates for the composite transition state are provided in the SI (Appendix A). For the nucleophile we utilized a cysteine activated by a lysine and histidine backed up by an aspartate or glutamate. For the first enolate stabilization we employed pairs of backbone amide, serine, threonine, histidine, tyrosine, or asparagine/glutamine. Binding of the benzaldehyde was accomplished by stacking interactions from aromatic residues. We also model in a water molecule to provide the hydrogen-bond bridging predicted to stabilize the final deprotonation of Figure 1A (20, 21).
Initial Testing
We tested 48 designs for MBH activity in a pH 7.4 PBS (10 mM phosphate) buffer at room temperature as indicated in Figure 1A using HPLC/MS/MS detection of the product. We found two designs, BH25 and BH32 with activity that was abolished (27-fold and 19-fold for the best variants, respectively) upon mutation of the nucleophilic residue to alanine. BH25 employs a cysteine nucleophile backed up by a lysine residue built into the structure of alanine racemase from G. Stearothermophilus, pdb code 1FTX (deposited to PDB but not published). BH32 has a histidine nucleophile positioned by hydrogen-bonding to a glutamate, built into the structure of haloacid dehalogenase from P. horikoshii, pdb code 1X42 (22). Both nucleophiles have precedence in the MBH literature: The nucleophilic cysteine thiol in the RC reaction (19), and the pi-methylhistidine used in various peptide-catalysts for the MBH (14). Note that the product here is chiral, but this MS assay does not distinguish enantiomers, and the results therefore necessarily focus on overall yield.
Upon confirmation of activity we tested point mutants of BH25 and BH32 based on visual inspection and automated scanning in Rosetta. For BH25 we identified N43Y as the most active mutant, with W164Y, G312M, and Y129F also increasing activity. For BH32 we identified the higher activity variants S124A, S9H, and S91V. After solving the crystal structure (see below), we identified new variants based on discrepancies between design and crystal structure, including the most active point-mutants N14I and S9G.
MS measurement of protein labeling
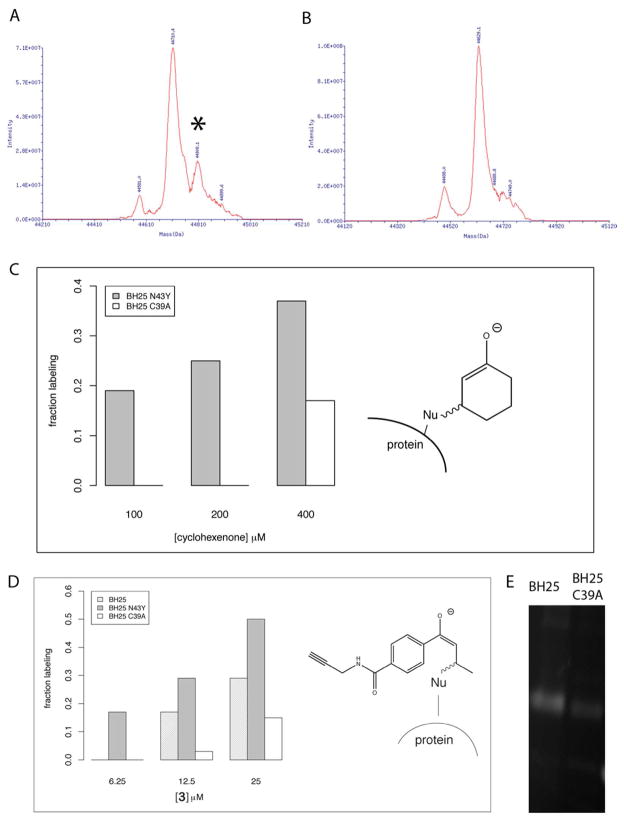
We directly measured the accumulation of Int1 (Figure 1A) by protein MS of samples incubated with cyclohexenone. BH25 and BH32 both are labeled by one cyclohexenone (96 Daltons) at 100 μM while both become multiply labeled at higher concentrations. The labeling of BH25 to form Int1 (Figure 2C, right) is abolished by mutating the nucleophilic cysteine, C39A (Figure 2A, B). BH32 binds cyclohexenone, but deletion of the nucleophilic histidine (which abolishes most activity for the full MBH reaction) does not appreciably decrease cyclohexenone labeling (data not shown). While BH32 is able to catalyze product formation at H23 (and that activity is abolished in H23A), it apparently has other sites that are randomly labeled by cyclohexenone, and the intermediate at H23 cannot be directly observed.
Figure 2.
Cyclohexenone and 3 labeling of BH25 and variants by HPLC/MS and fluorescent in-gel visualization. (A) Cyclohexenone labeling of BH25 N43Y measured by ESI MS and (B) labeling of the cysteine nucleophile knockout BH25 C39A. Both A and B were labeled at 200 μM cyclohexenone and 1 hour. Cyclohexenone at 200 μM labels specifically on the BH25 design (peak position marked by *), but not on the BH25 C39A knockout. (C) Specific labeling is also confirmed for 100 μM of cyclohexenone, but at a higher concentration of 400 μM a non-specific labeling is detectable in protein MS. (D) 3 labels BH25 N43Y specifically already at a very low concentration of 6.25 μM. At 12.5 μM the BH25 is labeled as well, however a small detectable peak arises also for the non-specific 3 labeling for the cysteine to alanine mutant (nucleophile knockout). (E) Specific binding of 3 is also confirmed for BH25 by in lysate “click” chemistry attachment of TAMRA dye (carboxytetramethyl-rhodamine) onto the free alkyne of the substrate. BH25 shows detectable labeling at 200 μM of 3 and 48 μM of TAMRA dye, while the cysteine to alanine mutant is labeled only weakly.
To quantify the affinity of BH25 N43Y for cyclohexenone, the fraction of protein bound was calculated from the raw MS peaks depicted in Figure 2A. Cyclohexenone binding begins at 100 μM, with an approximate dissociation constant (measured as the concentration resulting in 50% labeling) of approximately 500 μM. BH25 N43Y C39A does not bind cyclohexenone below 400 μM (Figure 2C).
Development of a probe for direct labeling and visualization
To detect Int1 formation without MS, thereby enabling a mid-throughput activity screen on lysate, we designed an enone with an alkyne for a “click” cyclization reaction with a fluorescent azide compound. Active BH designs are then visible by fluorescence. We synthesized and tested compound 3 (Figure 1B and protein-bound intermediate depicted in Figure 2D) and found it to bind tightly to the catalytic cysteine (Figure 2D).
Titration of compound 3 on BH25 N43Y measured by protein labeling in HPLC/MS shows a dissociation constant of 25 μM (Figure 2D). This probe labels C39 at approximately 20x lower concentration than cyclohexenone, partly due to enhanced intrinsic reactivity of the non-cyclic enone. The titration of the C39A active-site knockout shows binding only at higher concentrations to other sites (Figure 2D, white columns). For BH25 and BH25 N43Y at concentrations below 20 μM 3 binds only to the designed C39.
To investigate the labeling potential of 3 we incubated it for one hour with pure BH25 protein and BH25 C39A, followed by a “click” cyclization reaction with an azide-substituted fluorescent dye (TAMRA-azide). The mix was visualized on an SDS-PAGE gel. 3 only labeled BH25, but not the knockout (data not shown). We tested the same reaction on an E. coli cell lysate expressing BH25: 3 at a concentration of 200 μM labeled BH25 strongly and knockout very weakly (Figure 2E). We note that this labeling is at significantly higher concentration of 3 than was used in the MS assay showed in Figure 2D, probably due to non-specific interaction of 3 with other compounds in the lysate. This method can discriminate active from inactive variants, but direct MS/MS measurement of product is more quantitative.
Kinetic parameters
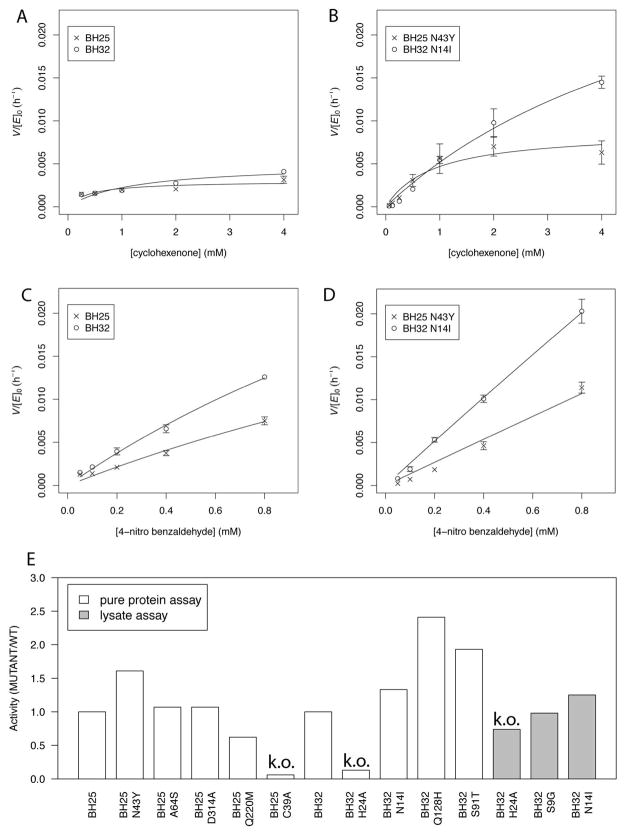
The reaction proceeds with linear increase in product up to 6 hours (Figure S1). To compare the activity of designs to catalytic residue knockout variants (the knockouts do not have detectable product after 6 hours) we measured the amount of product formed after 18 hours by HPLC/MS/MS for a range of substrate concentrations, and extrapolate a product formation rate from the 18 hour time point assuming a linear increase in product as demonstrated in shorter incubations (Figure S1). We fit the rate data with the steady-state Michaelis-Menten equation to estimate KM where appropriate, although we do not observe rate saturation so these numbers are therefore approximate; the KM numbers for cyclohexenone can be cross-checked with the independent direct labeling experiments (Figure 2). For BH25 and BH25 N43Y we measure a KM, cyclohexenone of roughly 600 μM (Figure 3A, B), matching well with the dissociation constant measured by direct MS visualization of Int1 (Figure 2). For BH32 we were not able to observe rate-saturation at high cyclohexenone concentration, and estimate the KM, cyclohexenone at > 4 mM.
Figure 3.
Kinetic characterization of BH25, BH25 N43Y, BH32 and BH32 N14I variants and BH32 lysate assay. (A) BH25 and BH32, and (B) BH25 N43Y and BH32 N14I kinetics for fixed 400 μM concentration of 4-nitrobenzaldehyde with 0.25, 0.5, 1, 2 and 4 mM of cyclohexenone, respectively. The trend is similar for BH25 N43Y while no clear saturation is observed for BH32 and the BH32 N14I variant. (C) and (D) show kinetics for same MBH variants but with a fixed concentration of cyclohexenone at 4 mM while the 4-nitrobenzaldehyde concentration is fixed at 25, 50, 100, 200, 400 and 800 μM, respectively. The low solubility of 4-nitrobenzaldehyde prevents measurements at higher concentrations and no saturation is observed at 800 μM. (E) Pure protein and crude lysate reaction assay of MBH variants. C39A diminishes the BH25 activity about 20 times. BH32 Q128H mutant is the most active variant. BH32 N14I is detectable in lysate and the knockout mutant BH32 H24A is at the lysate background activity. Activities are normalized to BH25 and BH32 activity in both assays.
Titration of 4-nitrobenzaldehyde did not reveal rate saturation for either BH25 or BH32 at concentrations up to 800 μM. While a number of variants have a higher catalytic rate, they do not saturate at high 4-nitrobenzaldehyde concentration (Figure 3D).
Identification of more active variants in lysate
The most active mutants were tested for activity detectable over background in clarified lysate. Background activity was measured using nucleophile knockouts. We found a consistently repeatable level of activity in BH32 N14I approximately 50% over the knockout background (Figure 3E, grey columns). BH25 N43Y was not over the background level in lysate.
We reasoned that more-active mutants of BH25 might be detectable in lysate. We performed saturation mutagenesis at selected positions in the active site of BH32: 10, 14, 19, 22, 42, 45, 46, 91, 95, and 128. We also tested BH25 at positions: 64, 86, 210, 220, 286, 314, and 364. At each position roughly 48 single clones were examined.
A number of point-mutants showed increased activity in lysate for BH32: L10W, Q128H, S91T and the already-identified N14I (data not shown). For BH25 we also identified variants with higher activity in lysate: N43Y (already found), A64S, Q220M and D314A. The lysate assay was able to correctly recapitulate more active variants from pure-protein assays, confirming that we were observing authentic activity signal.
The most active variants from the lysate assay were tested in a pure-protein assay (Figure 3E, white columns). The best BH32 variant (Q128H) was 2.4-fold more active than BH32, and 19-fold more active than the H23A active-site knockout. The most-active BH25 variant (N43Y) is 27-fold more active than the BH25 knockout (which is comparable to buffer-only background rate) and 1.7 fold over the original BH25. We also tested our designs in parallel with BSA samples (Sigma), which shows catalysis over buffer with variation from batch to batch (as observed by Reetz and co-workers (15)). In spite of the variance in BSA catalytic activity, both BH25 and BH32 are significantly more active than the best BSA samples. We also note that in previous work with these substrates in PBS buffer no background reaction was detectable, while our highly sensitive HPLC/MS/MS assay is able to detect the small level of background activity under our assay conditions. The possible contributions of the point mutants to activity are discussed in detail in Supplementary Information.
We tested the reaction with the new variants at high protein concentration to find the maximum obtainable yield in a 28-hour incubation. BH32 N14I at 152 μM gave 24% yield at 38% molar ratio of catalyst to 4-nitrobenzaldehyde and BH25 N43Y at 139 μM gave 9% yield at 35% molar ratio of catalyst.
Comparison of MBH designs to experimental structures
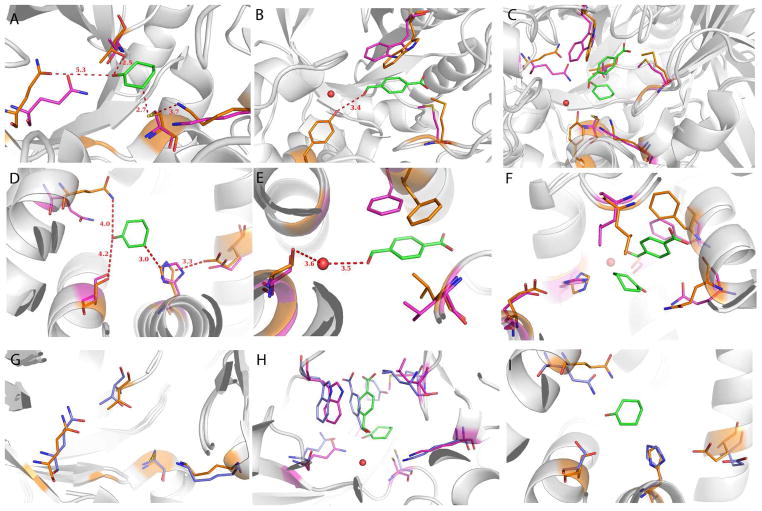
We crystallized and solved the apo crystal structures of BH25 N43Y (pdb code 3UW6) and BH32 (pdb code 3U26). BH25 N43Y crystallizes as a trimer with 2.8 Å resolution. The backbone is largely unchanged from the design or from the wild-type scaffold 1FTX, with 0.82 Å all-atom RMSD to the BH25 design structure (Figure 4A). BH32 crystallizes as a monomer with 1.6 Å resolution. The backbone overlays well on the design and the wild-type scaffold 1X42, with 0.67 Å all-atom RMSD to the BH32 design (Figure 4D). However, backbone changes around the active site are significant, with consequences for sidechain positioning. The largest shifts are at helices 126–132 (1.9 Å deviation), and 87–98 (0.7 Å).
Figure 4.
Superposition of Morita-Baylis-Hillman (MBH) designs BH25 N43Y and BH32 (purple) on the corresponding X-ray structures, 3UW6 and 3U26 (orange). Both are apo structures, but for modeling purposes we superimpose the designed transition state model. (A) Cyclohexenone is positioned well for the nucleophilic attack by BH25 cysteine, (B) while the 4-nitrobenzaldehyde pocket shows some slight deviations from the design. A water molecule is depicted as a red sphere, which is occupied by N43Y mutant. (C) Transition state of the reaction as modeled during the design. (D) BH32 design shows similar binding of the cyclohexenone as BH25 design but with H23 as a nucleophile. Hydrogen-bonding Q128 adopts a different conformation from the designed one as Q219 in BH25 N42Y structure. (E) BH32 4-nitrobenzaldehyde binding pocket rearranges in the similar way as in BH25 N43Y. (F) BH32 with the transition state of the reaction depicting the whole active site. (G) The active site of BH25 after 50ns of MD (blue) superimposed on the crystal structure (orange). Note the good overlay of sidechains, indicating that the MD has correctly predicted both well-designed sidechains (C39 and K285) as well as those that shift from the design (Q219). (H) The active site of BH25 with cyclohexenone and 4-nitrobenzaldehyde substrates after 50 ns (blue) superimposed on the design structure (purple). (I) The active site of BH32 after 50 ns of MD (blue) superimposed on the crystal structure (orange).
In BH25 the nucleophilic cysteine (C39) and backing-up lysine (K285) are in place as designed (Figure 4A). T85 is in position to form a hydrogen bond with the cyclohexenone oxygen. Residues packing around the cyclohexenone form a shallow groove as designed. The Q219 designed for stabilization of the developing enolate negative charge in Int1 is at a backbone position that moves away, and the sidechain moves out of the designed position. The W166 designed to stack on 4-nitrobenzaldehyde to stabilize binding and formation of Int2 is rotated out of position, precluding the exact designed binding configuration (Figure 4B). Y43 is well placed to hydrogen bond with the aldehyde oxygen of Int2 (Figure 4B). Overall, the cyclohexenone binding site and C39 are positioned as designed, while most aspects of catalysis and binding for the later steps of the MBH reaction, except Y43, do not appear as designed (Figure 4C and Table S2).
The situation in BH32 is similar – cyclohexenone packing and the nucleophilic histidine are largely in place as designed, while other aspects of catalysis and binding are not positioned exactly as intended (Figure 4F). Much of the change from the design results from backbone helix movement (Table S3). The catalytic H23 is in the correct design location, but in the structure the ring has turned such that the N-epsilon is not facing the beta-carbon of cyclohexenone, although the electron density does not define the plane of the imidazole well (Figure 4D and S6). The backing-up E46 is positioned as designed to H-bond with the N-delta of H23. Hydrophobic packing around the cyclohexenone leaves a shallow groove into which the substrate fits without clashes. The Q128, designed to interact with the oxyanion of Int1, is not in place, largely due to a shift of the helix 126–132 (Figure 4D). Packing around the 4-nitrobenzaldehyde is largely shifted, especially F132 and L10 (Figure 4E). The S22 positioned to bind the water stabilizing Int2 is in position to bind the designed water molecule, although it has shifted rotamers (Figure 4E).
Comparison of MD simulations to experimental structures
MD simulations offer predictions of the holo structure and a glimpse of alternative states in the apo structure. The end-points from each apo MD simulation (30–50 ns run) overlay well on the crystal structures of BH25 N43Y and BH32 with RMSD values of 1.22 Å and 1.02 Å respectively (Figure 4). The trajectories suggest that the nucleophilic residues maintain their designed positions (Figure 4G, I). In apo simulations H-bond donors designed to stabilize Int1 are engaged in alternative binding patterns for both BH25 and BH32, while C39 remains mostly preorganized (Figure S2) with excursions to interact with D313 (Figure S4).
In the absence of holo structures we turn to MD to predict the binding mode of the substrates and intermediate in BH25. Simulations of BH25 with both substrates predict stacking of 4-nitrobenzaldehyde on W166, suggesting that the binding pocket may be intact and capable of binding the substrate with ligand present (Figure 4H) even though the substrate pocket is remodeled in the apo crystal structure (Figure 4C). The carbonyl group of the enone interacts with T85 and Q219 and the aldehyde forms a face-to-face π-stacking interaction with W166. In simulations of the covalently bound intermediate these designed H-bond donors are engaged in alternative interactions (Figure S3). The negatively charged carbonyl oxygen is predicted to move towards the protonated K285, moving from its intended interaction with Q219.
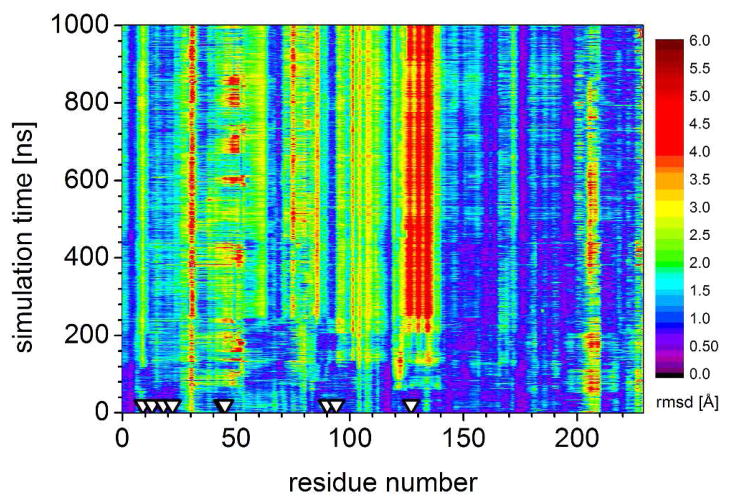
Long timescale MD simulations offer the promise of interrogating large-scale backbone motions in designs. We conducted a long, one microsecond simulation run on the Anton MD-specialized supercomputer. In this simulation the helix 126–132 has the largest deviation of any part of the design, but it ends at a final location 5–6 Å away from the design crystal structure (Figure 5). Portions of the helix 38–56 around the backing-up residue E46 also deviate from the design, with an oscillating helix kink at position 50 that is not observed in the structure.
Figure 5.
Backbone RMSD calculated for each residue at 1 ns steps (after fitting all backbone atoms to the original BH32 structure) during a 1 microsecond Anton run of BH32. RMSD is indicated by color, lower in purple/blue and higher in red. The most significant deviations from the design occur in the helix 126–132. Triangles indicate residues 10, 14, 19, 23, 45, 46, 91, 95, and 128, discussed in the text.
Summary
We have generated protein catalysts for the MBH reaction using a combination of computational design and plate screening by MS in lysate. While the designs are to our knowledge the most active protein catalysts reported for the MBH reaction, the absolute levels of activity are very low. The rate enhancement over the uncatalyzed reaction in buffer (kcat/kuncat) is only 27 for BH25 N43Y and 54 for BH32 Q128H (Figure 3E). These activities are clearly due to the designed active sites because active site knockouts by nucleophile removal reduce the activities of BH25 N43Y and BH32 Q128H by 27-fold and 19-fold, respectively. The low overall activities lead to the question of whether the nucleophiles alone are sufficient to produce the observed levels of catalysis. The many other designs which were solubly expressed at reasonable levels provide a negative control: All had exposed cysteine and histidine residues but none had measurable activity over background. Furthermore, the 6xHis-tag present on all of the proteins tested had no catalytic activity (as discussed below, imidazole catalyzes the reaction, but in basic mixed solvents at significantly higher concentrations).
Since a cysteine or histidine inside a surface pocket on a protein is not sufficient to produce an MBH catalyst with BH25 and BH32 levels of activity, the designed sites are providing more than a nucleophile alone. Why then are the overall levels of activity so low? A partial answer is that in solution water stabilizes the intermediates and facilitates proton transfer, while our crystal structures suggest that intermediate stabilization is not optimal in the designed active site. For example, the catalytic glutamine designed to bind an oxyanion in both designs is not positioned as intended. More generally, the reaction has multiple steps -- Nucleophile attack, C-C bond formation, and product release -- and imperfection in any of these could result in low activity.
How can the design process be improved? The crystal structures reveal the importance of properly buttressing sidechains that contribute to catalysis. To maximize the likelihood of correct sidechain placement for complex multi-step reactions it will be important to more extensively search sequence space to insert second-shell amino acids to stabilize direct catalytic interactions. An alternative is to use sidechains with less rotameric freedom wherever possible. To achieve better specificity in binding, substrates with hydrogen-bonding functional groups and other molecular handles would be desirable at this early stage in enzyme design methodology development. With improvements in force-fields, MD on the microsecond timescale could be used to predict inaccuracies in a design due to large-scale backbone motion, such as the shift of helix 126–132. Here the Anton MD run correctly identifies a problematic shift in this helix, but does not predict the right direction of the helix displacement.
The designed catalysts produce up to 24% yield over 18 h in fully aqueous and neutral buffer conditions compatible with green chemistry or with activity. The simplest catalysts for the MBH, such as imidazole at 500 mM, are able to catalyze the reaction on identical substrates in a 1:1 THF/H2O mixture over 72 h with 52% yield (accelerated to 69% yield over 8 h at pH 11.9; as noted above in water a 6xHis-tag has no activity at a concentration 0.18 mM)(18). The more common tertiary amine MBH catalysts are relatively slow for substrates involved in the synthesis of interesting biological compounds, achieving 70–100% yield in 1–7 days with varying enantioselectivity(9, 10, 23). Peptide catalystswith pi-methyl-histidine are used at 10 mol% and achieve up to 95% yield in 16 h with up to 81% ee in THF/H2O (3:1)(19). Because protein MBH catalysts derived from these designs would be genetically encodable, they could be used in a whole-cell or cell lysate system as part of a larger one-pot synthesis of small molecule compounds or for in vivo labeling of biological systems using chemistry orthogonal to natural enzymes.
Methods
Matching and native matching
We searched for protein backbone PDB structures (“scaffolds”) capable of accommodating the theozyme using the geometric hashing algorithm in the Rosetta Matcher software (24). The Matcher output (a “match”) is a PDB file with the TS in a suitably sized cavity and with the catalytic residues placed on the scaffold in place of the wild-type residues at those positions. We initially searched a small database of 244 monomeric E. coli-expressing, mostly thermostable structures. Later, we modified the matcher algorithm so the nucleophile residue was required to have the same amino acid identity in the “match” as it does in the “native” structure. This version is faster, and allowed us to conduct the scaffold search over the entire PDB.
Enzyme design
For each match we ran the enzyme design protocol of Rosetta (25). The protocol redesigns a sphere of residues close to the ligand (within 8 Angstroms) and repacks (changes sidechain rotamer) residues just outside of the design shell. The catalytic/binding residues from the theozyme are not changed during design. Designable and repackable residues are sampled by Monte Carlo in the torsional space of rotamers derived from the PDB (26). The theozyme stereochemistry was set as described in the Supplemental Methods.
The designed structures were sorted by scaffold and automatically analyzed to find designs with the best overall energy, ligand binding energy, and accuracy of the required theozyme constraints. Final structures were filtered and manually modified as described in the Supplemental Methods. We produced 48 different designs for experimental testing.
Genes were synthesized and proteins purified as described in Supplementary Methods.
HPLC/MS/MS product assay
Designs were incubated at 30 μM with cyclohexenone and 4-nitrobenzaldehyde at the indicated concentrations for 6–18 hours at room temperature (final acetonitrile volume 3%). After the reaction the solutions were filtered (PTFE 0.45 micron hydrophilic filter plates, Millipore), proteins precipitated with 3x volume acetonitrile, and the precipitate filtered (PTFE filter plates). Samples were run over an HPLC C18 column (Thermo scientific, Hypersil GOLD, 1.9 micron particle size) and directly injected for tandem MS/MS analysis (Thermo TSQ Quantum Access, triple stage quadrupole MS using APCI) with a 4-nitrophenol internal standard. Product concentration was quantified by automatic peak integration with a product standard curve. The MBH product 4 (Figure 1A) was synthesized as described in Supplementary Information.
Protein HPLC/MS measurement of direct labeling
For direct labeling, protein samples were incubated at 1 mg mL−1 with the desired concentration of the enone compound (at 2 volume % acetonitrile) for 1–2 hours. Samples were run over an HPLC C4 column (Thermo Scientific, BioBasic-4, 5 micron particle size) and directly injected for MS analysis using ESI.
Fluorophore “Click” labeling in lysate
Direct labeling with the cyclohexenone analog compound 3 (cf. Supplementary Information for synthesis) was detected by fluorescent in-gel visualization after azide-alkyne Huisgen cyclization (“click”) labeling of a TAMRA dye (carboxytetramethylrhodamine) onto the free alkyne of the substrate. Crude lysate of E. coli BL21 cells was incubated with compound 3 for 1 hour at room temperature. After incubation, TAMRA-azide was added, followed by TCEP (tris(2-carboxyethyl)phosphine), the “click” cyclization ligand TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine) and Cu(II) sulfate catalyst, with a one hour “click” reaction incubation. Labeling was stopped by addition of 1 volume SDS gel dye buffer, boiled, and the mixture run on an SDS-PAGE gel. Fluorescent images were recorded on a fluorescent gel imager (Fotodyne Inc., FOTO Analyst FX).
Lysate assay of MBH reaction
Single colonies were inoculated into a 96 well plate with 0.5 mL of TB (with kanamycin) and grown overnight at 37 deg C. A new 96 well plate with 0.48 mL of TB/kanamycin was inoculated with 20 μL of starter culture grown for 3h at 37 degrees and induced with IPTG (Sigma) overnight at 18 deg C. Cells were lysed by freeze-thaw cycling, clarified by centrifugation and supernatant assayed by HPLC/MS/MS.
Structure determination and MD simulations
The procedures of crystallization, structure determination and MD simulations are described in the Supplementary Methods.
Supplementary Material
Acknowledgments
S.B. acknowledges support from The Swedish Research Council (623-2008-497) and Foundation BLANCEFLOR Boncompagni-Ludovisi, née Bildt. L.G.N. acknowledges support from NSF Minority Post-Doctoral Fellowship. D.B. thanks DARPA/DTRA for financial support.. K.N.H. thanks the National Institute of General Medical Sciences, National Institutes of Health (GM075962) for financial support and acknowledges computer time on DESRES’s Anton special purpose supercomputer at the National Resource for Biomedical Supercomputing (NRBSC) at the Pittsburgh Supercomputing Center, and on the UCLA Institute for Digital Research and Education (IDRE) Hoffman 2 and Dawson 2 clusters.
Footnotes
Accession codes
The structure of BH25 N43Y, NESG target OR120, has been deposited in the PDB with code 3UW6. BH32, NESG target OR48, has been deposited in the PDB with accession code 3U26.
Additional methods, figures and tables. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retroaldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–U194. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 3.Richter F, Blomberg R, Khare SD, Kiss G, Kuzin AP, Smith AJT, Gallaher J, Pianowski Z, Helgeson RC, Grjasnow A, Xiao R, Seetharaman J, Su M, Vorobiev S, Lew S, Forouhar F, Kornhaber GJ, Hunt JF, Montelione GT, Tong L, Houk KN, Hilvert D, Baker D. Computational Design of Catalytic Dyads and Oxyanion Holes for Ester Hydrolysis. J Am Chem Soc. 2012;134:16197–16206. doi: 10.1021/ja3037367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, Clair JLS, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational Design of an Enzyme Catalyst for a Stereoselective Bimolecular Diels-Alder Reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 6.Basavaiah D, Reddy BS, Badsara SS. Recent Contributions from the Baylis-Hillman Reaction to Organic Chemistry. Chem Rev (Washington, DC, U S) 2010;110:5447–5674. doi: 10.1021/cr900291g. [DOI] [PubMed] [Google Scholar]
- 7.Morita K, Suzuki Z, Hirose H. A Tertiary Phosphine-Catalyzed Reaction of Acrylic Compounds with Aldehydes. Bull Chem Soc Jpn. 1968;41:2815. [Google Scholar]
- 8.Baylis AB, Hillman MED. German Patent 2155113. Chemical Abstracts. 1972;77:34174. [Google Scholar]
- 9.Iwabuchi Y, Sugihara T, Esumi T, Hatakeyama S. An enantioand stereocontrolled route to epopromycin B via cinchona alkaloid-catalyzed Baylis-Hillman reaction. Tetrahedron Lett. 2001;42:7867–7871. [Google Scholar]
- 10.Reddy LR, Saravanan P, Corey EJ. A simple stereocontrolled synthesis of salinosporamide A. J Am Chem Soc. 2004;126:6230–6231. doi: 10.1021/ja048613p. [DOI] [PubMed] [Google Scholar]
- 11.Hill JS, Isaacs NS. Mechanism of Alpha-Substitution Reactions of Acrylic Derivatives. J Phys Org Chem. 1990;3:285–288. [Google Scholar]
- 12.Santos LS, Pavam CH, Almeida WP, Coelho F, Eberlin MN. Probing the mechanism of the Baylis-Hillman reaction by electrospray ionization mass and tandem mass spectrometry. Angew Chem Int Ed. 2004;43:4330–4333. doi: 10.1002/anie.200460059. [DOI] [PubMed] [Google Scholar]
- 13.Shi M, Jiang JK. An exploration of asymmetric Baylis-Hillman reactions catalyzed by quinidine-derived chiral amines. Tetrahedron- Asymmetry. 2002;13:1941–1947. [Google Scholar]
- 14.Imbriglio JE, Vasbinder MM, Miller SJ. Dual catalyst control in the amino acid-peptide-catalyzed enantioselective Baylis-Hillman reaction. Org Lett. 2003;5:3741–3743. doi: 10.1021/ol035466b. [DOI] [PubMed] [Google Scholar]
- 15.Reetz MT, Mondiere R, Carballeira JD. Enzyme promiscuity: first protein-catalyzed Morita-Baylis-Hillman reaction. Tetrahedron Lett. 2007;48:1679–1681. [Google Scholar]
- 16.Dermenci A, Selig PS, Domaoal RA, Spasov KA, Anderson KS, Miller SJ. Quasi-biomimetic ring contraction promoted by a cysteine-based nucleophile: Total synthesis of Sch-642305, some analogs and their putative anti-HIV activities. Chemical Science. 2011;2:1568–1572. doi: 10.1039/C1SC00221J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HJ, Ruszczycky MW, Choi SH, Liu YN, Liu HW. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo SZ, Wang PG, Cheng JP. Remarkable rate acceleration of imidazole-promoted Baylis-Hillman reaction involving cyclic enones in basic water solution. J Org Chem. 2004;69:555–558. doi: 10.1021/jo035345p. [DOI] [PubMed] [Google Scholar]
- 19.Aroyan CE, Miller SJ. Enantioselective rauhut-currier reactions promoted by protected cysteine. J Am Chem Soc. 2007;129:256–257. doi: 10.1021/ja067139f. [DOI] [PubMed] [Google Scholar]
- 20.Robiette R, Aggarwal VK, Harvey JN. Mechanism of the Morita-Baylis-Hillman reaction: A computational investigation. J Am Chem Soc. 2007;129:15513–15525. doi: 10.1021/ja0717865. [DOI] [PubMed] [Google Scholar]
- 21.Aggarwal VK, Dean DK, Mereu A, Williams R. Rate acceleration of the Baylis-Hillman reaction in polar solvents (water and formamide). Dominant role of hydrogen bonding, not hydrophobic effects, is implicated. J Org Chem. 2002;67:510–514. doi: 10.1021/jo016073y. [DOI] [PubMed] [Google Scholar]
- 22.Arai R, Kukimoto-Niino M, Kuroishi C, Bessho Y, Shirouzu M, Yokoyama S. Crystal structure of the probable haloacid dehalogenase PH0459 from Pyrococcus horikoshii OT3. Protein Sci. 2006;15:373–377. doi: 10.1110/ps.051922406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar V, Das P, Ghosal P, Shaw AK. Syntheses of (−)-gabosine A, (+)-4-epi-gabosine A, (−)-gabosine E, and (+)-4-epi-gabosine E. Tetrahedron. 2011;67:4539–4546. [Google Scholar]
- 24.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De Novo Enzyme Design Using Rosetta3. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunbrack RL, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.