To the Editor: Acinetobacter baumannii is an opportunistic pathogen that is a source of nosocomial infections, mostly pneumonia (1). Treatment of infections caused by A. baumannii is becoming a serious clinical concern as this microorganism becomes increasingly resistant to multiple antimicrobial drugs (2). A. baumannii resistance to carbapenems is mostly associated with production of carbapenem-hydrolyzing class D β-lactamases and metallo-β-lactamases (2). New Delhi metallo-β-lactamase 1 (NDM-1) is one of the most recently discovered metallo-β-lactamases among various gram-negative species, including A. baumannii (3). We recently reported the recovery of NDM-1–producing A. baumannii isolates throughout Europe (4). In that study, the genetic background of several strains was identified and corresponded to sequence types (STs) 1, 25 and 85. The ST85 clone was isolated in France from 2 patients previously hospitalized in Algeria (4,5).
The present study was initiated by the recent isolation of 6 more NDM-1–producing A. baumannii linked with North Africa. To determine the extent of spread of this organism from Africa to France, we genetically analyzed 8 other NDM-1-producing A. baumannii isolates collected from different towns in France during 2011–2012. Of these 8 isolates, 6 were from patients previously hospitalized in different cities in Algeria (including Algiers, Setif, Constantine, and Tlemcen), 1 from a patient previously hospitalized in Tunisia, and 1 from a patient previously hospitalized in Egypt. These 8 isolates came from 2 clinical samples (blood cultures and wound) from 6 screening rectal swab samples collected at the time of hospital admission (Technical Appendix). Because the 8 samples were recovered from 5 hospitals, nosocomial acquisition can be ruled out.
The isolates were identified by 16S rRNA gene sequencing. Susceptibility testing was performed by disk diffusion (Sanofi-Diagnostic Pasteur, Marnes-La-Coquette, France) and interpreted according to updated Clinical and Laboratory Standards Institute guidelines (6). The MICs of β-lactams (imipenem, meropenem and doripenem) were determined by the Etest technique (AB bioMérieux, Solna, Sweden) according to the manufacturer’s recommendations. All isolates were resistant to β-lactams, including all carbapenems (MICs >32mg/L). The isolates were also resistant to fluoroquinolones, gentamicin, sulfonamides, and chloramphenicol but susceptible to amikacin, netilmicin, rifampin, tetracycline, and tigecycline according to Clinical and Laboratory Standards Institute guidelines (6) and colistin according to European Committee on Antimicrobial Susceptibility Testing guidelines (www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v1.3_pdf.pdf).
The production of metallo-β-lactamases was suspected by use of a combined disk test, based on the inhibition of the metallo-β-lactamase activity by EDTA as described (4). All isolates were positive for production of metallo-β-lactamases.
For all 8 isolates, PCRs aimed at detecting carbapenemase genes, using primers described elsewhere (7), followed by sequencing, led to identification of the blaNDM-1 gene. The isolates also carried a naturally-occurring blaOXA-51-like gene, namely blaOXA-94 (Technical Appendix). The blaOXA-51-like β-lactamase confers a low level of resistance to carbapenems.
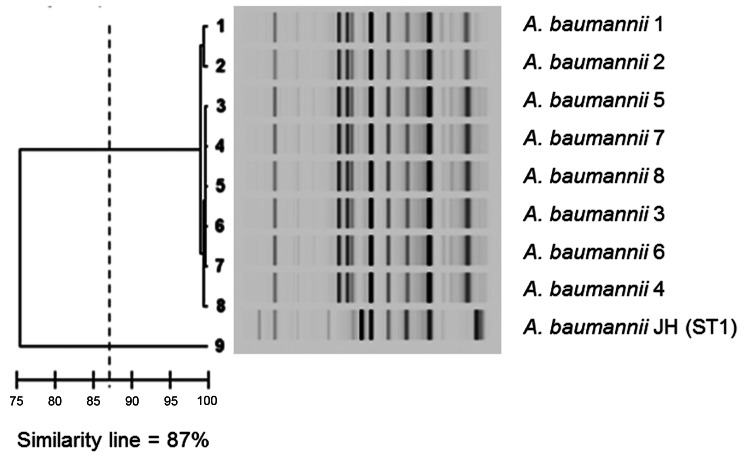
Genotypic comparison was performed by multilocus sequence typing as described (8) and by repetitive extragenic palindromic sequence-based PCR by using the DiversiLab system (bioMérieux, La Balme-les-Grottes, France) according to the manufacturer’s instructions. The genomic pattern of all isolates was identical (Figure). Further multilocus sequence typing indicated that all isolates belonged to ST85. This ST was identified in Greece during a nationwide study that focused on carbapenem resistance in clinical isolates of A. baumannii and identified mainly carbapenem-hydrolyzing carbapenemase OXA-58 (9).
Figure.
Results of Diversilab system (bioMérieux, La Balme-les-Grottes, France) analysis of Acinetobacter baumannii isolates. Similarity line shows the cutoff that separates the different clones.
Recently, we showed that the blaNDM-1 gene was carried by a composite transposon bracketed by 2 copies of ISAba125 in A. baumannii (10). Cloning and sequencing of the genetic context of the blaNDM-1 in the first isolate showed that transposon Tn125 was truncated at its 3′-end extremity by insertion sequence ISAba14, giving rise to a truncated Tn125 (ΔTn125). PCR mapping of all isolates showed that they possessed this truncated isoform of Tn125, which was therefore probably no longer functional.
The identification of several clinical A. baumannii isolates that possessed the blaNDM-1 gene and originated from North Africa, with no obvious link to the Indian subcontinent, strongly suggests that 1 NDM-producing A. baumannii clone is probably widespread in North Africa and that it might now act as a reservoir for NDM-1. This finding might indicate that control of spread of multidrug-resistant A. baumannii would have a primary role in controlling spread of NDM-1.
Clinical features of New Delhi metallo-β-lactamase–producing Acinetobacter baumannii.
Acknowledgments
This work was funded by a grant from the Institut National de la Santé et de la Recherche Médicale.
Footnotes
Suggested citation for this article: Bonnin RA, Cuzon G, Poirel L, Nordmann P. Multidrug-resistant Acinetobacter baumannii clone, France [letter]. Emerg Infect Dis [Internet]. 2013 May [date cited]. http://dx.doi.org/10.3201/eid1905.121618
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life. 2011;63:1061–7. 10.1002/iub.532 [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19:588–95. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 4.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, et al. Dissemination of New Delhi metallo-β-lactamase-1–producing Acinetobacter baumannii in Europe. Clin Microbiol Infect. 2012;18:E362–5. 10.1111/j.1469-0691.2012.03928.x [DOI] [PubMed] [Google Scholar]
- 5.Boulanger A, Naas T, Fortineau N, Figueiredo S, Nordmann P. NDM-1-producing Acinetobacter baumannii from Algeria. Antimicrob Agents Chemother. 2012;56:2214–5. 10.1128/AAC.05653-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. M100–S22. Wayne (PA): The Institute; 2012. [Google Scholar]
- 7.Bonnin RA, Rotimi V, Al Hubail M, Gasiorowski E, Al Sweih N, Nordmann P, et al. Wide dissemination of GES-type carbapenemases in Acinetobacter baumannii in Kuwait. Antimicrob Agents Chemother. 2013;57:183–8. 10.1128/AAC.01384-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE. 2010;5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, Zarrilli R, et al. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10-year study in Greece (2000–09). J Antimicrob Chemother. 2011;66:2767–72. 10.1093/jac/dkr390 [DOI] [PubMed] [Google Scholar]
- 10.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:1087–9. 10.1128/AAC.05620-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical features of New Delhi metallo-β-lactamase–producing Acinetobacter baumannii.