Abstract
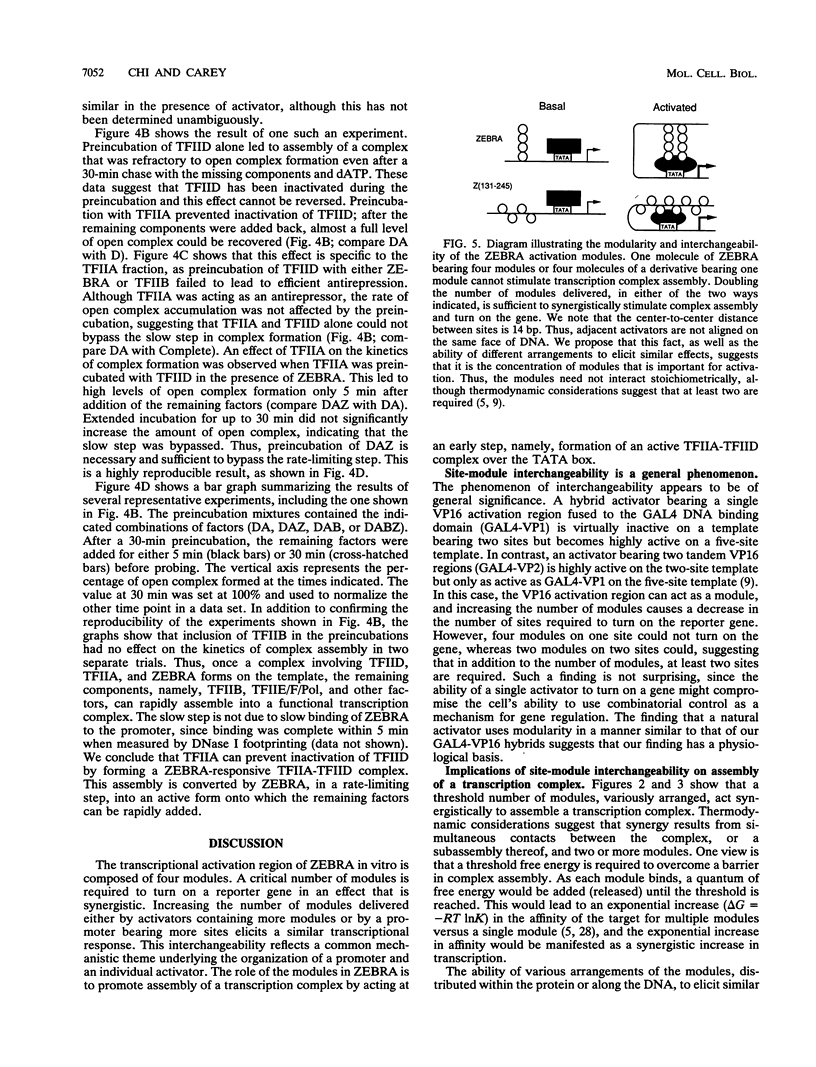
An RNA polymerase II activator often contains several regions that contribute to its potency, an organization ostensibly analogous to the modular architecture of promoters and enhancers. The regulatory significance of this parallel organization has not been systematically explored. We considered this problem by examining the activation domain of the Epstein-Barr virus transactivator ZEBRA. We performed our experiments in vitro so that the activator concentrations, stabilities, and affinities for DNA could be monitored. ZEBRA and various amino-terminal deletion derivatives, expressed in and purified from Escherichia coli, were assayed in a HeLa cell nuclear extract for the ability to activate model reporter templates bearing one, three, five, and seven upstream ZEBRA binding sites. Our data show that ZEBRA contains four modules that contribute to its potency in vitro. The modules operate interchangeably with promoter sites to determine the transcriptional response such that the loss of modules can be compensated for by increasing promoter sites. Potassium permanganate footprinting was used to show that transcriptional stimulation is a consequence of the activator's ability to promote preinitiation complex assembly. Kinetic measurements of transcription complex assembly in a reconstituted system indicate that ZEBRA promotes formation of a subcomplex requiring the TFIIA and TFIID fractions, where TFIIA acts as an antirepressor. We propose a model in which the concentration of DNA-bound activation modules in the vicinity of the gene initiates synergistic transcription complex assembly.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buratowski S., Hahn S., Guarente L., Sharp P. A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989 Feb 24;56(4):549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Carey M., Kolman J., Katz D. A., Gradoville L., Barberis L., Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J Virol. 1992 Aug;66(8):4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Leatherwood J., Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990 Feb 9;247(4943):710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990 May 24;345(6273):361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Carey M. Mechanistic advances in eukaryotic gene activation. Curr Opin Cell Biol. 1991 Jun;3(3):452–460. doi: 10.1016/0955-0674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Dynan W. S. Modularity in promoters and enhancers. Cell. 1989 Jul 14;58(1):1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Emami K. H., Carey M. A synergistic increase in potency of a multimerized VP16 transcriptional activation domain. EMBO J. 1992 Dec;11(13):5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P. J., Rowe D. T., Rooney C. M., Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989 Jan;8(1):127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington E. K., Borras A. M., Lytle J. P., Speck S. H. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J Virol. 1992 Feb;66(2):922–929. doi: 10.1128/jvi.66.2.922-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O., Lu H., Killeen M., Greenblatt J., Burton Z. F., Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T., Balmaceda C. G., Roeder R. G. The cell type-specific octamer transcription factor OTF-2 has two domains required for the activation of transcription. EMBO J. 1990 May;9(5):1635–1643. doi: 10.1002/j.1460-2075.1990.tb08283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Sadowski I., Ptashne M. Mutations that increase the activity of a transcriptional activator in yeast and mammalian cells. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2127–2131. doi: 10.1073/pnas.87.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot J. F., Mikaelian I., Buisson M., Manet E., Joab I., Nicolas J. C., Sergeant A. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 1991 Mar 25;19(6):1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J. RNA polymerase-associated transcription factors. Trends Biochem Sci. 1991 Nov;16(11):408–411. doi: 10.1016/0968-0004(91)90165-r. [DOI] [PubMed] [Google Scholar]
- Hardwick J. M., Tse L., Applegren N., Nicholas J., Veliuona M. A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992 Sep;66(9):5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. Transcriptional activation: enter TFIIB. Trends Biochem Sci. 1991 Sep;16(9):317–318. doi: 10.1016/0968-0004(91)90130-n. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Johnson F. B. Synergism in transcriptional activation: a kinetic view. Genes Dev. 1993 Feb;7(2):173–179. doi: 10.1101/gad.7.2.173. [DOI] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O., Gill G., Chen J. L., Dynlacht B. D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993 Jan 29;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Evans R. M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988 Dec 2;55(5):899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Ingles C. J., Shales M., Cress W. D., Triezenberg S. J., Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991 Jun 13;351(6327):588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- Inostroza J. A., Mermelstein F. H., Ha I., Lane W. S., Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992 Aug 7;70(3):477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991 Oct 18;67(2):365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991 Dec;5(12B):2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Hardwick J. M., Hayward S. D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989 Jul;63(7):3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Carey M. F., Ptashne M., Green M. R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988 Aug 26;54(5):659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991 Mar 8;64(5):971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Lu H., Zawel L., Fisher L., Egly J. M., Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992 Aug 20;358(6388):641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Structural features of the herpes simplex virus alpha gene 4, 0, and 27 promoter-regulatory sequences which confer alpha regulation on chimeric thymidine kinase genes. J Virol. 1982 Dec;44(3):939–949. doi: 10.1128/jvi.44.3.939-949.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M., Roeder R. G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991 Nov 1;67(3):557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Roy A. L., Lieu H. M., Roeder R. G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991 Sep 6;66(5):981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- Miller G. The switch between latency and replication of Epstein-Barr virus. J Infect Dis. 1990 May;161(5):833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Müller-Immerglück M. M., Schaffner W., Matthias P. Transcription factor Oct-2A contains functionally redundant activating domains and works selectively from a promoter but not from a remote enhancer position in non-lymphoid (HeLa) cells. EMBO J. 1990 May;9(5):1625–1634. doi: 10.1002/j.1460-2075.1990.tb08282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Pascal E., Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991 Sep;5(9):1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J. H., Chambon P., Egly J. M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993 Apr 2;260(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schaffner G., Schirm S., Müller-Baden B., Weber F., Schaffner W. Redundancy of information in enhancers as a principle of mammalian transcription control. J Mol Biol. 1988 May 5;201(1):81–90. doi: 10.1016/0022-2836(88)90440-8. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Sundseth R., Hansen U. Activation of RNA polymerase II transcription by the specific DNA-binding protein LSF. Increased rate of binding of the basal promoter factor TFIIB. J Biol Chem. 1992 Apr 15;267(11):7845–7855. [PubMed] [Google Scholar]
- Sutherland J. A., Cook A., Bannister A. J., Kouzarides T. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 1992 Sep;6(9):1810–1819. doi: 10.1101/gad.6.9.1810. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990 Feb 9;60(3):375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Tora L., White J., Brou C., Tasset D., Webster N., Scheer E., Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989 Nov 3;59(3):477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- Wampler S. L., Kadonaga J. T. Functional analysis of Drosophila transcription factor IIB. Genes Dev. 1992 Aug;6(8):1542–1552. doi: 10.1101/gad.6.8.1542. [DOI] [PubMed] [Google Scholar]
- Wang W., Carey M., Gralla J. D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992 Jan 24;255(5043):450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- Wang W., Gralla J. D., Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992 Sep;6(9):1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- White J., Brou C., Wu J., Lutz Y., Moncollin V., Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992 Jun;11(6):2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]





