Abstract
Merozoite surface protein 1 of Plasmodium vivax (PvMSP1), a glycosylphosphatidylinositol-anchored protein (GPI-AP), is a malaria vaccine candidate for P. vivax. The paralog of PvMSP1, named P. vivax merozoite surface protein 1 paralog (PvMSP1P; PlasmoDB PVX_099975), was recently identified and predicted as a GPI-AP. The similarities in genetic structural characteristics between PvMSP1 and PvMSP1P (e.g., size of open reading frames, two epidermal growth factor-like domains, and GPI anchor motif in the C terminus) led us to study this protein. In the present study, different regions of the PvMSP1P protein, demarcated based on the processed forms of PvMSP1, were expressed successfully as recombinant proteins [i.e., 83 (A, B, and C), 30, 38, 42, 33, and 19 fragments]. We studied the naturally acquired immune response against each fragment of recombinant PvMSP1P and the potential ability of each fragment to bind erythrocytes. The N-terminal fragment (83A) and two C-terminal fragments (33 and 19) reacted strongly with sera from P. vivax-infected patients, with 50 to 68% sensitivity and 95 to 96% specificity, respectively. Due to colocalization of PvMSP1P with PvMSP1, we supposed that PvMSP1P plays a similar role as PvMSP1 during erythrocyte invasion. An in vitro cytoadherence assay showed that PvMSP1P, especially the 19-kDa C-terminal region, could bind to erythrocytes. We also found that human sera from populations naturally exposed to vivax malaria and antisera obtained by immunization using the recombinant molecule PvMSP1P-19 inhibited in vitro binding of human erythrocytes to PvMSP1P-19. These results provide further evidence that the PvMSP1P might be an essential parasite adhesion molecule in the P. vivax merozoite and is a potential vaccine candidate against P. vivax.
INTRODUCTION
In the life cycle of Plasmodium vivax, the asexual erythrocytic-stage parasites produce all of the clinical symptoms, disease, and pathology associated with malaria (1). During this period, schizont-stage-infected erythrocytes produce merozoites and invade new erythrocytes. The molecular interactions between the ligand on the merozoite and the receptor on the host erythrocyte membrane are essential during erythrocyte invasion by Plasmodium parasites (2, 3). Erythrocyte invasion by the P. vivax merozoite appears to be dependent on two ligands, the Duffy-binding protein (DBP) and reticulocyte-binding protein (RBP). The P. vivax DBP (PvDBP) binds the Duffy blood group antigen during erythrocyte invasion (4). However, a recent report found that vivax malaria is also observed in Duffy-negative populations (5, 6), suggesting that the P. vivax parasite contains a ligand other than DBP.
Once a protein is confirmed as associated with the surface of the merozoite, it usually becomes a candidate molecule for (i) possible adhesion to the erythrocyte surface and thus a potential ligand for invasion and (ii) potential malaria vaccine candidates. Many proteins expressed during host cell invasion by malaria parasites have emerged as important vaccine candidates because they are involved in the attachment, junction formation, or internalization of merozoites into the host erythrocytes. Several of these are located on the surface or in the apical organelles (body or neck of rhoptries, micronemes, and dense granules) of merozoites (3).
Among the proteins involved in the invasion by merozoite, the glycosylphosphatidylinositol-anchored proteins (GPI-APs) are suggested as potential vaccine candidates because of their localization to apical organelles and the surface; these candidates are predicted to play essential roles during invasion (7). The GPI-APs of P. falciparum have been found to participate in erythrocyte invasion by merozoite. Proteomic analysis of schizont/merozoite proteins found 11 GPI-APs (merozoite surface protein 1 [MSP-1], MSP-2, MSP-4, MSP-5, MSP-10, rhoptry-associated membrane antigen, apical sushi protein, Pf92, Pf38, Pf12, and Pf34). These proteins represent approximately 94% of the GPI-anchored schizont/merozoite proteome and constitute by far the largest validated set of GPI-anchored proteins in Plasmodium falciparum (7). Most of these GPI-APs are important candidates for a blood-stage malaria vaccine (8–10); for example, it is impossible to knock out MSP1 in P. falciparum (11, 12), suggesting that it is essential for invasion and survival of the parasite. Thirty-one putative GPI-APs from the P. vivax genome have been identified, and 30 of them are predicted to be orthologs of GPI-APs in P. falciparum (13). Among them, only eight GPI-APs (PvMSP-1, -4, -5, -8, and -10, Pv12, Pv34, and Pv38) have been identified as possible blood-stage vaccine candidates (14–21), and the rest remain uncharacterized.
As a well-known blood-stage antigen of P. vivax, PvMSP1 localizes to the merozoite surface, and its C terminus holds great promise in the development of a malaria vaccine (22, 23). PvMSP1-19, the C-terminal region of PvMSP1, is a core-binding region for erythrocytes (24). Antibodies against the C terminus of PvMSP1 in P. vivax-immune humans and immunized animals play a major role in the parasite inhibitory response (15, 22, 25). The paralog of pvmsp1, named pvmsp1p (PVX_099975), is highly conserved among worldwide isolates (26). The primary structure of PvMSP1P contains a putative GPI anchor attachment signal and double epidermal growth factor (EGF)-like domains at the C terminus. The predicted molecular mass is about 215 kDa, which is similar to that of PvMSP1 (13). Because most of the GPI-APs with EGF-like domains have been confirmed as playing crucial roles in parasite survival and invasion (11), PvMSP1P may also play an important role during invasion by the merozoite.
To elucidate the function of PvMSP1P, we investigated the humoral immune response in patients against the recombinant PvMSP1P fragments, subcellular localization of PvMSP1P, and the erythrocyte-binding activity of the C terminus of PvMSP1P in comparison with that of PvDBP.
MATERIALS AND METHODS
Human serum and parasite samples.
The sera from the vivax malaria patients were collected from people with symptoms and positive vivax parasitemia by microscopic examination (mean parasitemia, 0.121%; range, 0.027 to 0.630%) at local health centers and clinics in areas where malaria is endemic in Gangwon Province, Republic of Korea (ROK). Their mean age was 28 years (range, 18 to 60 years). The sera of healthy individuals were collected from malaria-naïve people living in areas of nonendemicity in the ROK. The study was approved by the Institutional Review Board at Kangwon National University Hospital. Human sera from 150 patients with vivax malaria and 100 healthy individuals were screened for immunoreactivity using a protein array method (27). We used sera from 50 patients with vivax malaria and 40 malaria-naïve individuals and eight fragments of PvMSP1P for primary humoral immune response screening by protein array. In addition, three fragments of PvMSP1P with higher immune responses from results of primary screening were used for secondary screening with sera of 150 patients with vivax malaria and 100 malaria-naïve individuals by protein array. We also used both pooled sera from 10 vivax patients (ROK+) randomly selected from antibody-positive samples against PvMSP1P-19 and 10 healthy individuals (ROK−). We also made two pooled sera from the ROK+, combining five high antibody titers (high ROK+) and five low antibody titers (low ROK+) against PvMSP1P-19 for measurement of erythrocyte-binding inhibitory activity. Genomic DNA from the parasite was prepared from 200 μl of whole blood from a P. vivax patient in ROK using QIAamp DNA blood minikits (Qiagen, Hilden, Germany), which provided 200-μl aliquots of template DNA.
Expression of recombinant PvMSP1P proteins.
The primers for PvMSP1P fragments were designed based on PvMSP1P of the P. vivax Sal-1 strain sequence (PlasmoDB PVX_099975) and used for amplification of PvMSP1P fragments from genomic DNA of P. vivax isolates from ROK. Eight fragments of PvMSP1P and PvMSP1-19 (19-kDa C-terminal fragment of PvMSP1) (Fig. 1A and B) were expressed using the wheat germ cell-free (WGCF) system (CellFree Sciences, Matsuyama, Japan) (28–30). Briefly, to clone each fragment into the pEU-E01-His-TEV-MCS vector (CellFree Sciences), eight pairs of forward and reverse primers were synthesized and used for each fragment gene amplification (see Table S1 in the supplemental material). These were used to generate the DNA fragments encoding PvMSP1P-83A (amino acid [aa] positions 29 to 278), -83B (aa 229 to 528), -83C (aa 479 to 731), -30 (aa 732 to 1046), -38 (aa 1047 to 1474), -42 (aa 1475 to 1834), -33 (aa 1475 to 1748), and -19 (aa 1749 to 1834) (Fig. 1B). Eight fragments of DNA encoding PvMSP1P-83A, -83B, -83C, -30, -38, -42, -33, and -19 were amplified and cloned into pEU-E01-His-TEV-MCS vector (CellFree Sciences), and the cloned inserts were sequenced using an ABI 3700 genetic analyzer (Genotech, Daejeon, South Korea). Those proteins, including PvDBP region II (PvDBPII) and PvMSP1-19, were also expressed using a WGCF expression system (CellFree Sciences) and purified using Ni-affinity chromatography as described previously (31–33).
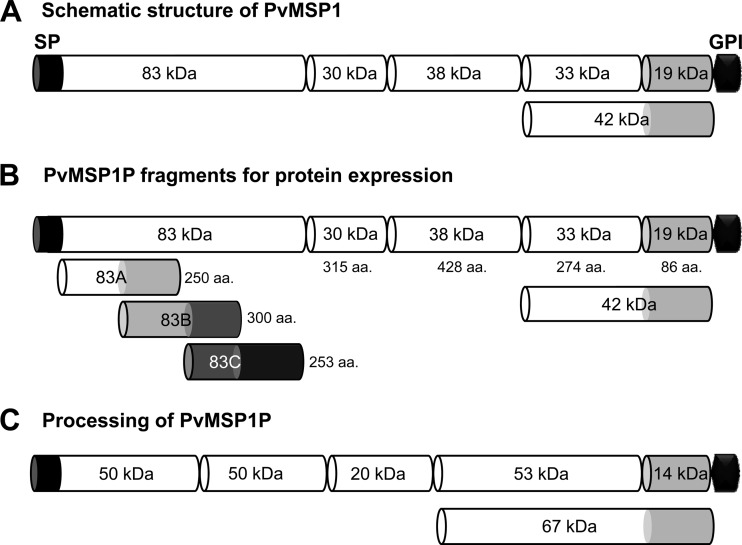
Fig 1.
Schematic diagram of PvMSP1, fragments of PvMSP1P for recombinant protein expression, and processing of native PvMSP1P. (A) Schematic diagram of processing of PvMSP1. (B) Demarcation of PvMSP1P, similar to the processed forms of PvMSP1, for protein expression of 8 fragments of PvMSP1P. For the N-terminal 83-kDa region, three 50-amino-acid overlapping fragments were expressed as 83A, 83B, and 83C. The MSP1P protein comprises 1,856 amino acids, with a calculated molecular mass of 214.5 kDa. The predicted signal peptide (SP; aa positions 1 to 28) and GPI anchor signal (GPI; aa 1835–1854) are shown. Eight fragments of MSP1P (83A [aa 29 to 278], 83B [aa 229 to 528], 83C [aa 479 to 731], 30 [aa 732 to 1046], 38 [aa 1047 to 1474], 42 [aa 1475 to 1834], 33 [aa 1475 to 1748], and 19 [aa 1749 to 1834]) were expressed as recombinant proteins. PvMSP1P-30, -42, -33, and -19 were used to raise specific antisera. (C) Hypothetical processing of PvMSP1P was predicted from Western blot results of the parasite lysate probed with anti-PvMSP1P-42 serum (Fig. 3B). aa, amino acid; kDa, kilodalton; SP, signal peptide; GPI, glycophosphatidylinositol anchor signal.
Production of animal immune sera.
The C termini of PvMSP1P (30, 42, 33, and 19) recombinant proteins were used for raising antibodies. Female BALB/c mice (DaehanBiolink Co., Eumsung, ROK) were used at 5 to 7 weeks of age. Groups of three mice were injected intraperitoneally with 20 μg of MSP1P-30, -42, -33, and -19, or MSP1-19, and phosphate-buffered saline (PBS) with Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO) as described previously (31). Booster injections were given 3 and 6 weeks after the priming using the same amount of antigen with Freund's incomplete adjuvant (Sigma-Aldrich), and mouse sera were collected 2 weeks after the final boost. Meanwhile, to generate antibodies against PvMSP1-19 and PvDBPII by rabbits, one Japanese white rabbit was immunized subcutaneously with 250 μg of purified proteins plus Freund's complete adjuvant, followed by 250 μg with Freund's incomplete adjuvant thereafter. All immunizations were done three times at 3-week intervals. The antisera were collected 2 weeks after the final boost. Two BALB/c mice were also immunized with either PvMSP1-19 or PvDBPII as described above. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Ehime University, and the experiments were conducted according to the Ethical Guidelines for Animal Experiments of Ehime University.
SDS-PAGE and Western blot analysis.
Ten micrograms of each recombinant PvMSP1P protein was prepared in reducing sample buffer, separated by 12% SDS-PAGE, and then stained with Coomassie brilliant blue. P. vivax parasites rich in schizonts were harvested from 10 ml of a patient blood sample (parasitemia > 0.1%) by the Percoll gradient method (20), and the parasite proteins were extracted in SDS-PAGE loading buffer. One tenth of the parasite lysate was loaded in each lane and then separated by 12% SDS-PAGE. For Western blot analysis, the proteins were transferred electrophoretically to polyvinylidene difluoride (PVDF) membranes (Millipore Corp., Bedford, MA) and incubated with blocking buffer (5% nonfat milk in PBS containing 0.2% Tween 20 [PBST]) for 1 h at 37°C. The blots containing recombinant proteins were then incubated for 1 h at 37°C with either anti-penta-His antibody (Qiagen) or each antibody against recombinant PvMSP1P fragment diluted 1:1,000 in PBST. The membranes were washed with PBST and incubated with a 1:2,000 dilution of alkaline phosphatase-conjugated goat anti-mouse IgG (MP Biomedicals, Solon, OH) for 1 h at 37°C. The blots were washed with PBST and developed with 5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium (BCIP-NBT; Sigma-Aldrich). The blots containing parasite proteins were incubated for 1 h at 37°C with either anti-PvMSP1P-42 antisera or anti-PvMSP1-19 antisera diluted 1:1,000 in PBST. After the primary antibody reaction and washing, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (GE Healthcare, Camarillo, CA) diluted 1:20,000 in PBST and visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) on a LAS 4000 mini luminescent image analyzer (GE Healthcare).
Indirect IFA.
Immunofluorescence assays (IFAs) were performed on acetone-fixed parasites as described previously (19). The following primary antibodies (dilutions) were used: rabbit anti-PvMSP1-19 (1:100) and mouse anti-PvMSP1P-42 (1:20). The following secondary antibodies were used: Alexa-488 goat anti-mouse IgG (1:500; Invitrogen), Alexa-568 goat anti-rabbit IgG (1:500; Invitrogen), and DAPI (4′,6′-diamidino-2-phenylindole) for nuclear staining (1:1,000; Invitrogen). The slides were mounted in ProLong Gold antifade reagent (Invitrogen) and visualized under oil immersion in a confocal scanning laser microscope (LSM710; Carl Zeiss MicroImaging, Thornwood, NY) using a Plan-Apochromat 63×/1.4 oil differential interference contrast (DIC) objective lens. Images were captured with Zen software (Carl Zeiss MicroImaging) and prepared for publication with Adobe Photoshop (Adobe Systems, San Jose, CA).
Protein arrays.
Amine-coated slides were prepared as described previously (27, 30). To develop the protein arrays, sera from patients with vivax malaria and malaria-naïve individuals were used to analyze the humoral immune response by well-type amine arrays. A series of double dilutions was used to optimize the coating antigen concentration (3 to 200 ng/μl) of each fragment. The optimized concentrations of the purified recombinant proteins per well of the array were determined as 100 ng/μl for PvMSP1P-19, 50 ng/μl for PvMSP1-19, PvDBPII, PvMSP1P-42, -83B, and -33, 12 ng/μl for PvMSP1P-83C and -30, and 6 ng/μl for PvMSP1P-83A and -38, all in PBST. The spotted slides were then incubated for 1 h at 37°C. Each well was blocked with 1 μl of blocking buffer (5% bovine serum albumin [BSA] in PBST) incubated for 1 h at 37°C. The arrays also contained an area spotted with purified PvMSP1-19 as a positive control and wheat germ lysate without any plasmid vector as a negative control. The chips were then probed with sera from the malaria patients or healthy individuals (1:10 dilution) that had been preabsorbed against wheat germ lysate (1:100 dilution) to block anti-wheat germ antibodies. Alexa Fluor 546-conjugated goat anti-human IgG (10 ng/μl; Invitrogen) in PBST was used as the detection antibody, and the fluorescent signals were scanned in a fluorescence scanner (ScanArray Express, PerkinElmer, Boston, MA) and quantified as described previously (27). The cutoff value was defined as two standard deviations (SDs) above the mean fluorescence intensity of the negative-control samples.
Erythrocyte-binding assay on the surface of transfected COS-7 cells.
The pEGFP-HSVgD1-N1 vector was used to construct COS-7 expression vectors encoding each fragment of PvMSP1P, PvMSP1, and PvDBPII as described previously (33). Here, we prepared the PvMSP1-19 fragment (Fig. 1A) and the PvMSP1P-30, -42, -33, and -19 fragments (Fig. 1B) by PCR amplification with primers listed in Table S1 in the supplemental material. Each amplified gene fragment in 2 μl of the PCR product was ligated with 100 ng of linearized pEGFP-HSVgD-N1 vector using the In-Fusion HD cloning kit (Clontech). These constructs are designated pEGFP-MSP1-19 and pEGFP-MSP1P-30, -42, -33, and -19 and were purified using an Ultrapure plasmid extraction system (Viogene, Taipei, Taiwan).
COS-7 cells were plated into 24-well culture plates and transfected with each of the above pEGFP construct DNAs (100 ng per well) using Lipofectamine (Invitrogen) in serum-free Dulbecco's modified Eagle's medium (incomplete DMEM; Invitrogen). After 42 to 44 h of incubation, transfected COS-7 cells were incubated with 250 μl of diluted antiserum against each fragment of the target protein in incomplete DMEM at 37°C for 1 h. After washing, the transfected cells were incubated for 2 h at 37°C with human erythrocytes (blood group O+, 1% hematocrit in incomplete DMEM). To determine transfection efficiency, green fluorescence cells (green fluorescent protein [GFP] tagged) were observed unfixed on a Flowview FV1000 laser scanning confocal imaging system (Olympus, Tokyo, Japan) at a magnification of ×200 to score. Images were analyzed and manipulated using Adobe Photoshop CS5. Cells were washed three times with PBS to remove nonadherent erythrocytes, and the number of rosettes in 30 microscopic fields was counted in total, 1,500 to 2,000 cells at a magnification of ×200 to score erythrocyte-binding activity. Positive rosettes were defined as adherent erythrocytes covering more than 50% of the COS-7 cell surface (33). Cells were counted unfixed on a light microscope (Olympus, Tokyo, Japan). Images were captured by eXcope software (DIXI Optics, Daejeon, South Korea) and analyzed and manipulated using Adobe Photoshop CS5.
Experiments for each antiserum were performed in triplicate and repeated twice unless otherwise stated. All antisera, including high ROK+ and low ROK+, and its immune mouse serum, were first tested at a 1:10 dilution to assess their inhibitory activity. Serial dilutions for each antiserum were then tested to titrate the erythrocyte-binding inhibitory activity. The binding inhibitory activity for each antiserum dilution was compared with the binding inhibitory activity of negative-control sera (either a 1:10 dilution of PBS immune mouse sera or ROK−). The results are expressed as the relative percent binding activity (negative-control serum, 100% binding activity).
Fluorescence microscopy.
Surface expression of each PvMSP1P fragment on COS-7 cells transfected with target plasmid DNA was detected using a 1:20 dilution of mouse antisera against PvMSP1P fragments and rabbit antiserum against PvDBPII as the primary antibody, and Alexa Fluor 543-conjugated goat anti-mouse or Alexa Fluor 568-conjugated goat anti-rabbit antibodies (Invitrogen) as the secondary antibody. Cells were observed unfixed on a Flowview FV1000 laser scanning confocal imaging system (Olympus, Tokyo, Japan). Images were analyzed and manipulated using Adobe Photoshop CS5.
Statistical analysis.
The data were analyzed using GraphPad Prism (GraphPad Software, San Diego, CA), SigmaPlot (Systat Software Inc., San Jose, CA), and Microsoft Excel 2007. For protein array and erythrocyte-binding assay, Student's t test was used to compare the means of each group. A P value of <0.05 was considered significant. Simple scatter regression was used to create standard curves.
RESULTS
Schematic of the primary structure of PvMSP1P.
Pvmsp1p gene sequence information encoded by PVX_099975 on chromosome 7 retrieved from PlasmoDB revealed that PvMSP1P is a 1,856-amino-acid protein with a predicted molecular mass of 214.5 kDa (Fig. 1B). We have described in our previous report that Pvmsp1p protein is encoded by a single exon gene and comprises a signal peptide (aa 1 to 28), GPI-anchor (aa 1834 to 1854), E/Q-rich region (aa 1157 to 1172), heptapeptide tandem repeat region (aa 905 to 918), and a transmembrane domain (aa 1832 to 1854) (26). The predicted molecular masses of PfMSP-1, PvMSP-1, and PvMSP1P are similar, approximately 200 kDa. The C termini of all three proteins have a GPI anchor signal and two tandem EGF-like domains. Based on this structural similarity, PvMSP1P was divided into six regions: PvMSP1P-83 (without the signal peptide), -30, -38, -42, -33, and -19 (Fig. 1B). PvMSP1P-83 was too large to be expressed as a single recombinant protein and hence divided into three smaller subfragments (83A, 83B, and 83C) with a 50-amino-acid overlap between each subfragment. In total, we designed eight fragments to assess the recombinant protein expression using the WGCF system.
Expression of each fragment of PvMSP1P and PvMSP1-19 and production of immune sera.
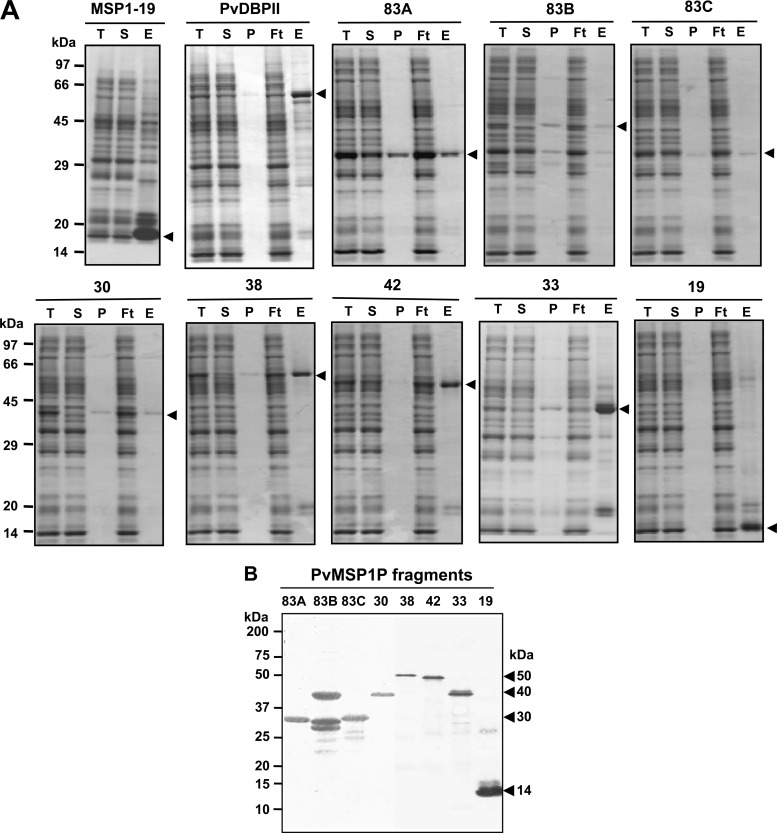
SDS-PAGE showed that all the recombinant proteins were recovered in the supernatant soluble fraction (Fig. 2A). The PvMSP1P-83A, -83B, -83C, -30, -38, -42, -33, and -19 fragments were expressed, as predicted from their theoretical molecular weights, as 30-, 40-, 30-, 40-, 50-, 50-, 40-, and 14-kDa proteins, respectively (Fig. 2B). In the 83B fragment, extra smaller bands were present, and it may be due to the translational arrest (Fig. 2B). The C termini of PvMSP1P (-30, -42, -33, and -19) and PvMSP1-19 were used to immunize mice to produce polyclonal antibodies. The immune serum sample reacted with the respective PvMSP1P fragment proteins in the Western blot (Fig. 3A, arrowheads).
Fig 2.
Recombinant protein expression and purification. (A) PvMSP1-19, PvDBPII, and all PvMSP1P fragments were synthesized using the wheat germ cell-free protein expression system and then purified with a Ni-Sepharose column. The purified MSP1-19 (15 kDa), DBPII (75 kDa), 83A (30 kDa), 83B (40 kDa), 83C (30 kDa), 30 (40 kDa), 38 (50 kDa), 42 (50 kDa), 33 (40 kDa), and 19 (14 kDa) fragments of PvMSP1P existed in soluble elution fractions. Arrowheads indicate specific bands for each recombinant protein. T, total translation mix; S, supernatant; P, pellet; Ft, flowthrough; E, elution. (B) The purified 83A, 83B, 83C, 30, 38, 42, 33, and 19 fragments of PvMSP1P were resolved by SDS-PAGE, transferred to a PVDF membrane, and probed with anti-His-tagged antibody. Arrowheads indicate specific bands for each recombinant protein.
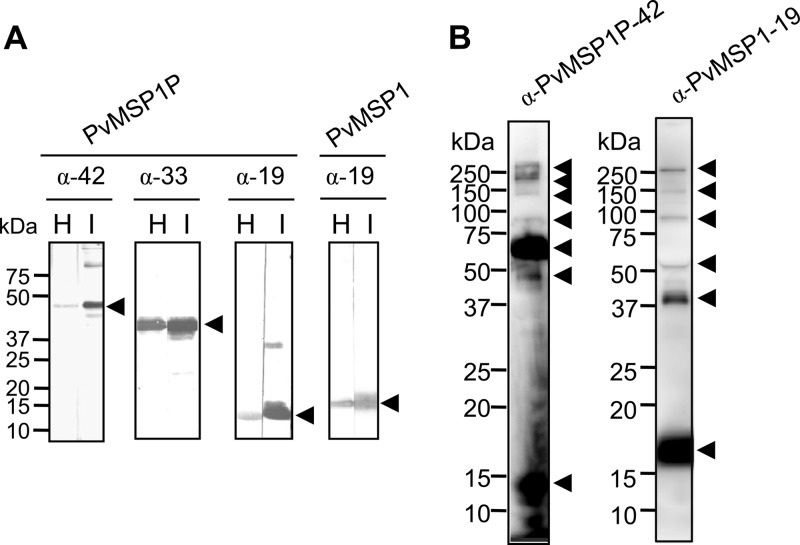
Fig 3.
Western blot analysis of recombinant PvMSP1P C-terminal fragments, PvMSP1-19, and schizont lysates probed with PvMSP1P and PvMSP1 immune sera. (A) Three C-terminal fragments of PvMSP1P (42, 33, and 19 kDa) and PvMSP1-19 were probed with the respective mouse immune sera (I) and with anti-His antibody (H). (B) Western blot analysis. Recognition of the native PvMSP1P and PvMSP1 antigen in the parasite lysate with mouse antisera against PvMSP1P-42 and rabbit antisera against PvMSP1-19, respectively, under reducing conditions. Arrowheads indicate the target bands and putative processed fragments.
Proteolytic processing of PvMSP1P and PvMSP1.
Western blot analysis of the schizont lysate probed with anti-PvMSP1-19 antisera confirmed the previous finding that the full-length PvMSP1 undergoes proteolytic processing to form 19-, 42-, 50-, 80-, 150-, and >200-kDa fragments (Fig. 3B). The blot probed with antibody against PvMSP1P-42 showed that PvMSP1P also undergoes proteolytic processing events to form 14-, 45-, 70-, 80-, 140-, 200-, and >250-kDa fragments. These results confirmed that PvMSP1P is processed, and the processing profile of PvMSP1P is distinct from that of PvMSP1, suggesting that the anti-PvMSP1P-42 antibody did not cross-react with PvMSP1.
High IgG prevalence and titers against the C terminus of PvMSP1P among vivax malaria patients.
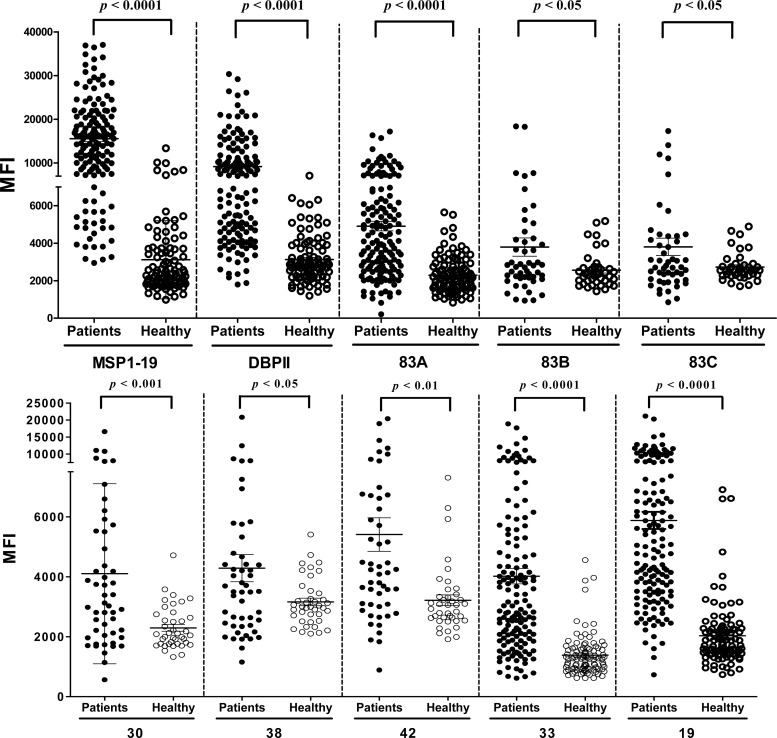
The antigenicity of each fragment of PvMSP1P was evaluated in sera obtained from vivax malaria patients in areas where malaria is endemic and from healthy individuals in areas where malaria is nonendemic in the ROK. The prevalence of total IgG for the N- and C-terminal regions of PvMSP1P was greater than 50% (50%, 60%, and 68% for PvMSP1P-83A, -33, and -19, respectively). The specificities for all eight fragments were more than 90% (Table 1). There were high specificity and significant differences in the total IgG prevalence for the five PvMSP1P fragments (83A, 30, 42, 33, and 19) between vivax patients and healthy individuals (P < 0.05) (Fig. 4). We also analyzed the antigenicity of PvMSP1-19 and PvDBPII using the identical group of sample sera, and our data reconfirmed the high IgG prevalence against PvMSP1-19 and PvDBPII (Table 1 and Fig. 4).
Table 1.
Prevalence, 95% confidence intervals, and mean fluorescence intensity of IgG responses to each fragment of P. vivax MSP1P, MSP1-19, and DBPII in human patients and healthy individual serum samples
| Antigen | No. of patient samples |
95% CIb | MFIc | No. of healthy samples |
95% CI | MFI | P valuee | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Total (%)a | Positive | Negative | Total (%)d | ||||||
| MSP1-19 | 120 | 30 | 150 (80.0) | 67.6–82.5 | 15,573 | 6 | 94 | 100 (94.0) | 90.0–97.8 | 3,122 | P < 0.0001 |
| DBPII | 95 | 55 | 150 (63.3) | 55.7–70.8 | 9,176 | 4 | 96 | 100 (96.0) | 90.1–98.4 | 3,126 | P < 0.0001 |
| 83Af | 75 | 75 | 150 (50.0) | 42.1–57.9 | 6,193 | 5 | 95 | 100 (95.0) | 88.8–97.8 | 2,033 | P < 0.0001 |
| 83Bf | 11 | 39 | 50 (22.0) | 12.8–35.2 | 3,796 | 3 | 37 | 40 (92.5) | 80.1–97.4 | 2,599 | P = 0.03 |
| 83Cf | 12 | 38 | 50 (24.0) | 14.3–37.4 | 3,807 | 3 | 37 | 40 (92.5) | 80.1–97.4 | 2,782 | P = 0.04 |
| 30f | 22 | 28 | 50 (44.0) | 31.1–57.7 | 4,100 | 1 | 39 | 40 (97.5) | 87.1–99.6 | 2,291 | P = 0.0003 |
| 38f | 12 | 38 | 50 (24.0) | 14.3–37.4 | 4,288 | 1 | 39 | 40 (97.5) | 87.1–99.6 | 3,159 | P = 0.03 |
| 42f | 16 | 34 | 50 (32.0) | 20.7–45.8 | 5,408 | 2 | 38 | 40 (95.0) | 83.8–98.6 | 3,216 | P = 0.001 |
| 33f | 90 | 60 | 150 (60.0) | 52.0–67.5 | 4,015 | 4 | 96 | 100 (96.0) | 90.1–98.4 | 1,384 | P < 0.0001 |
| 19f | 102 | 48 | 150 (68.0) | 60.1–74.9 | 5,878 | 4 | 96 | 100 (96.0) | 90.1–98.4 | 2,033 | P < 0.0001 |
Sensitivity: percent positive in patient samples.
CI, confidence interval.
MFI, mean fluorescence intensity.
Specificity: percent negative in healthy samples.
Differences in the total IgG prevalence for each antigen between vivax patients and healthy individuals were calculated with Student's t test. A P value of <0.05 is considered statistically significant.
PvMSP1P fragments used as capture antigens.
Fig 4.
IgG antibody responses to PvMSP1-19, PvDBPII, and eight fragments of PvMSP1P using protein microarrays. Immunoreactivity against each antigen with the sera of malaria patients (patients) and healthy individual samples (healthy) from ROK was determined. There were high specificity and significant differences in the total IgG prevalence for the eight PvMSP1P fragments between vivax patients and healthy individuals (P < 0.05). The P values were calculated using Student's t test. The bar indicates the mean ± standard deviation. MFI, mean fluorescence intensity.
PvMSP1P colocalizes with the merozoite surface marker, PvMSP1.
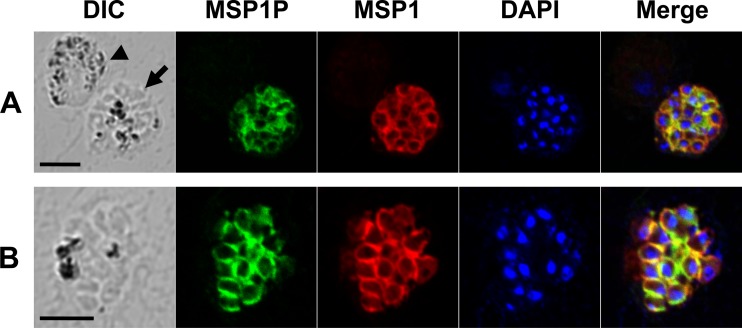
We analyzed the localization of PvMSP1P in merozoite using IFA. Mouse antibody against PvMSP1P showed staining on the surface of the merozoite in mature schizont (Fig. 5A, arrow), but no staining was detected on gametocyte (Fig. 5A, arrowhead). We also stained schizonts with rabbit anti-PvMSP1 antibody indicating the colocalization of both images on the surface of merozoites (Fig. 5A and B). The merozoite surface localization of PvMSP1P explained our finding that PvMSP1P is immunogenic in humans, similar to PvMSP1.
Fig 5.
Subcellular localization of PvMSP1P protein in asexual blood-stage parasites of P. vivax. (A) The acetone-fixed mature schizont (arrow) and a gametocyte (arrowhead) of P. vivax were dually labeled with mouse immune sera against PvMSP1P-42 (green) and rabbit immune sera against PvMSP1-19 (surface marker) (red). Nuclei are visualized with DAPI (blue). (B) Another mature schizont of P. vivax was also probed with the same antibodies as above. Bar represents 5 μm.
Erythrocyte-binding activity of PvMSP1P.
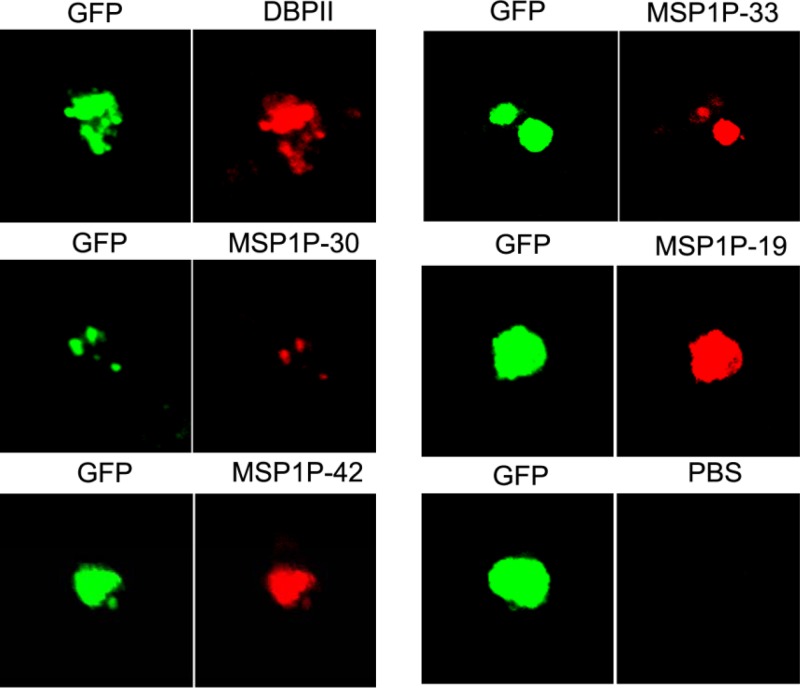
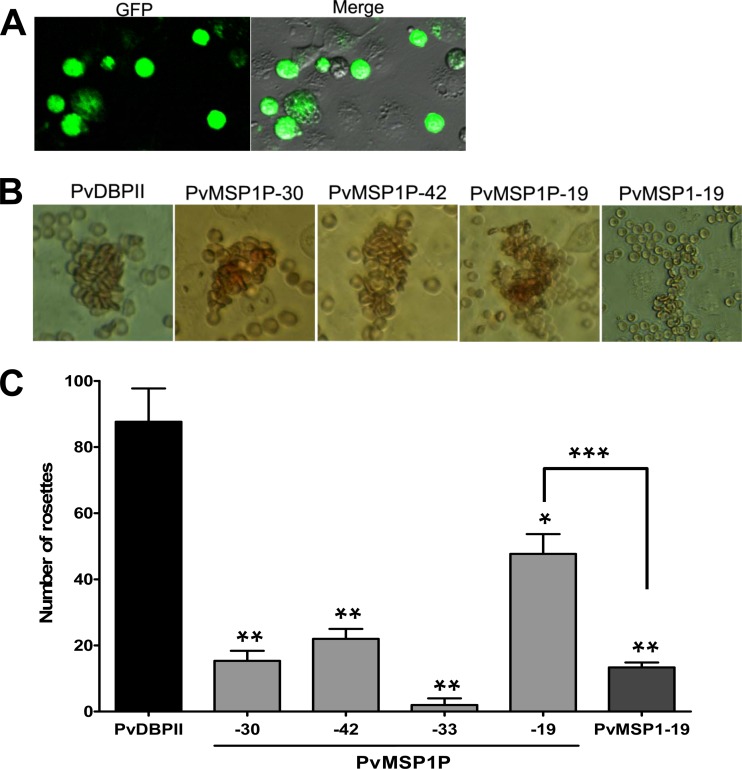
Merozoite surface expression of PvMSP1P suggests an important role during parasite invasion of the erythrocyte. To elucidate the function of PvMSP1P, the erythrocyte-binding activity of recombinant PvMSP1P proteins was evaluated. Successful COS-7 cell transfection was assessed by detecting the expression of the green fluorescent protein (Fig. 6, GFP) and reaction with anti-PvMSP1P-30, -42, -33, and -19 and anti-PvDBPII immune sera under fluorescence microscopy. All of the fragments were expressed on the surface of transfected COS-7 cells labeled with GFP (Fig. 6). In a previous study (34), the transfection efficiency of each fragment was more than 80% by counting green fluorescent cells (Fig. 7A), and the positive rosettes were defined as adherent erythrocytes covering more than 50% of the COS-7 cell surface as shown in Fig. 7B. To confirm whether these PvMSP1P fragments can bind to erythrocytes, we counted the number of rosettes on COS-7 cells transfected with each PvMSP1P fragment (Fig. 7C). PvMSP1P-19 (mean ± SD, 48 ± 6; 6.5 to 11.4%) possessed the strongest rosetting and binding ability compared with the other PvMSP1P regions, although this was lower than that of PvDBPII (88 ± 10; 12.5 to 20.0%). In contrast, PvMSP1P-30 (also a cysteine-rich region), however, produced only a small number of rosettes (15 ± 3; 1.2 to 4.8%). The binding ability of PvMSP1-19, as the known P. vivax erythrocyte-binding protein other than PvDBPII, was also reconfirmed in this study; however, it is significantly lower (13 ± 2; 1.3 to 4.4%) than that of PvMSP1P-19 (Fig. 7C).
Fig 6.
Transfection induced expression of parasite proteins on the surface of COS-7 cells. Expression of PvDBPII and PvMSP1P C-terminal fragments on the surface of COS-7 cells transfected with pEGFP-HSVgD1_PvDBPII and PvMSP1P-30, -42, -33, and -19 plasmid DNA was detected by IFA. Green fluorescent protein (GFP) (green, control), Alexa Fluor 543-conjugated goat anti-mouse antibody (red, MSP1P fragments), or Alexa Fluor 568-conjugated goat anti-rabbit antibody (red, DBPII) was used and visualized by confocal microscopy. Mouse antiserum against PBS was used as a negative control.
Fig 7.
Binding of PvMSP1P fragments expressed on COS-7 cells to erythrocytes. (A) The transfection efficiency of each plasmid construct was calculated by counting green fluorescent cells (GFP) and COS-7 cells in bright field image (Merge). Average transfection efficiency was more than 80%. (B) Erythrocyte-binding rosettes formed on the surfaces of COS-7 cells expressing PvDBPII or different fragments of PvMSP1P or PvMSP1-19 were visualized under light microscopy. (C) The number of rosettes formed by the COS-7 cells transfected with genes coding for either PvDBPII or different fragments of PvMSP1P or PvMSP1-19. Detection of the transfection efficiency into COS-7 cells of all constructs by counting green signal cells within 30 microscope fields (×200). A positive result was defined as more than half the surface of the transfected cells covered with attached erythrocytes, and the total number of COS-7 cells per coverslip was recorded. Data are shown as the mean number of rosettes of three independent experiments, and the error bar represents ± standard deviation. Statistical differences between PvDBPII and the other proteins are indicated with a single asterisk (P < 0.05) and double asterisks (P < 0.001). Statistical difference between PvMSP1P-19 and PvMSP1-19 is shown with triple asterisks (P < 0.001).
Blocking activity of antibodies on erythrocyte binding of PvMSP1P.
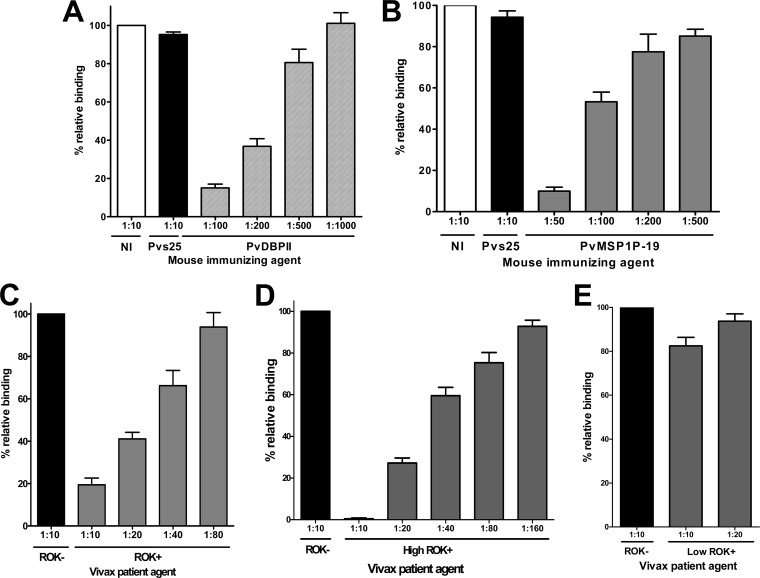
An in vitro binding assay was used to determine whether the antibody response correlated with an inhibitory effect of human sera against PvMSP1P-19 cytoadherence activity. A 1:10 dilution of mouse serum against PBS as nonimmunized control (Fig. 8, NI) and mouse serum against Pvs25, the ookinete surface protein (Fig. 8, Pvs25) (35), were used as negative controls. To ensure the propriety and dependability of our assay, we included PvDBPII as a positive control and did an erythrocyte-binding inhibition assay using PvDBPII-transfected COS-7 cells and serially diluted anti-PvDBPII sera (Fig. 8A). The inhibitory effect was measured by counting the number of rosettes in the presence of increasing concentrations of test antibody. The difference in the inhibitory efficacies of these antisera may reflect the difference in their antibody titers against the binding epitope(s). The mouse anti-PvMSP1P-19 antibody had a significant dose-dependent inhibitory effect (P < 0.05) on the in vitro binding of human erythrocytes to the transfected COS-7 cells expressing the PvMSP1P-19 (Fig. 8B). To investigate whether the human immune sera also contain the blocking antibody against PvMSP1P-19, we tested the effects of human immune sera on the erythrocyte-binding inhibitory activity. ROK+ blocked the binding of PvMSP1P-19 to human erythrocytes (Fig. 8C). Binding activity of ROK+ at a 1:10 dilution of the sera was reduced to approximately 20%, and this inhibitory activity was still significant even at the 1:40 dilution; however, the binding activity was increased to 93.8% at a 1:80 dilution (Fig. 8C). We then selected the high responders (high ROK+) and low responders (low ROK+) from the ROK+ samples. The high ROK+ showed significantly stronger blocking activity even at a higher dilution (1:80) (Fig. 8D) than those from ROK+ at 1:40 dilution (Fig. 8C). In contrast, the low ROK+ showed no blocking ability even at the highest serum concentration (1:10) (Fig. 8E). ROK− were included in the above-described experiments as negative controls.
Fig 8.
Inhibition of erythrocyte binding to P. vivax MSP1P-19 expressed on COS-7 cells by naturally acquired antibodies and mouse anti-PvMSP1P-19 antibody. (A) COS-7 cells were transfected with pEGFP-HSVgD1_DBPII plasmid DNA expressing a GFP-DBPII fusion protein. Cells were subsequently incubated with anti-PvDBPII immune mouse sera at various dilutions before the addition of human erythrocytes. Binding was scored by counting the number of rosettes bound to COS-7 cells in 30 microscope fields (×200). Mouse antiserum against PBS as nonimmunized control (NI) and anti-Pvs25 (no erythrocyte-binding activity) immune mouse sera diluted 1:10 were included as negative controls (Pvs25). (B) Inhibition of erythrocyte binding to PvMSP1P-19 expressed on COS-7 cells by PvMSP1P immune mouse sera. Cells were then incubated with PvMSP1P immune mouse sera at various dilutions before the addition of human erythrocytes. (C) Inhibition of erythrocyte binding to PvMSP1P-19 expressed on COS-7 cells by vivax immune human sera. Cells were incubated with various dilutions of pooled sera from vivax malaria patients of ROK (ROK+) before the addition of human erythrocytes. Erythrocyte-binding activity was scored by counting the number of rosettes in 30 fields at a magnification of ×200. The percent binding was determined relative to a 1:10 dilution of pooled sera from areas of nonendemicity of ROK (ROK−) as a negative control. Cells were incubated with various dilutions of the high-responding (high ROK+) (D) and low-responding (low ROK+) (E) vivax malaria sera before the addition of human erythrocytes. Error bars represent ± standard deviations.
DISCUSSION
Availability of the comparative genomics of Plasmodium has provided the impetus to understand the complex processes of the malaria parasite life cycle, such as merozoite invasion, and thus find new vaccine candidate antigens and drug targets. In this study, we have characterized PvMSP1P, a paralog of PvMSP1.
To date, in P. vivax, molecular structure, as well as protein processing during invasion in blood stage, was well studied only in PvMSP1 (36). The commonality between PvMSP1 and PvMSP1P in their primary structural features, and their stage-specific expression profile, led us to speculate that PvMSP1P might be a member of the PvMSPs. However, PvMSP1P has a limited number of polymorphisms in comparison with PvMSPs and is highly conserved, including the C terminus that contains the EGF-like domains (26), suggesting that the function of PvMSP1P-19 is important during parasite invasion of erythrocytes. Although PvMSP1 and PvMSP1P are expressed on the surface of merozoite, transcriptome analysis of blood-stage vivax parasites (PlasmoDB) shows that PvMSP1 is highly upregulated in the schizont-stage parasite; however, expression of PvMSP1P is only moderately upregulated, suggesting that the role of PvMSP1P may be different from that of PvMSP1 (37). In addition to this, the detection of processed fragments (∼14 and 70 kDa) of PvMSP1P in the parasite lysate (Fig. 3B) demonstrates that the PvMSP1P processing pattern is similar but not identical to that of its paralog PvMSP1 (Fig. 1C). However, further mass spectrometric studies are required to validate whether the fragments detected are indeed processed species of native PvMSP1P.
PvMSPs with GPI anchors (PvMSP1, PvMSP5, and PvMSP8) are immunogenic in humans in areas where malaria is endemic (14, 15, 18, 27, 32). Acquired immune responses to PvMSP1 suggest that antibodies and cellular immune responses to PvMSP1 are strongly induced during vivax malaria (15, 38). Previous reports confirmed that the C terminus of PvMSP1 is more immunogenic than the N terminus of PvMSP1 (39). Our analysis of human antibody responses against a number of P. vivax blood-stage antigens using protein array indicated that PvMSP1-19 had the highest immunogenicity among vivax patients (27). It also indicated that a high titer and prevalence of the total IgG response to both the N terminus (PvMSP1P-83A) and C terminus (PvMSP1P-33 and -19) to PvMSP1P were detected. Importantly, similar to the immune response against PvMSP1, the C terminus of PvMSP1P had significantly higher immunogenicity than did the N terminus (P < 0.01) (Fig. 4 and Table 1). Taken together, these findings suggest that, similar to PvMSP1-19, the C-terminal regions of PvMSP1P, containing EGF-like domains, are the major target of immune response in humans and hence they may play crucial roles during invasion.
Formation of tight junction and invasion of human erythrocytes by P. vivax require the interaction of PvDBP with the Duffy antigen on the erythrocyte (4, 40, 41). Recent reports in Madagascar showed that genetically diverse isolates of P. vivax can cause both asymptomatic and symptomatic vivax malaria and show frequent transmission even in the Duffy-negative population (5, 6), suggesting that P. vivax, similar to P. knowlesi (42, 43) and P. falciparum (3), can use an alternative pathway(s) to invade erythrocytes. Therefore, it will be interesting to identify molecules involved in alternative pathways in P. vivax invasion and also explain the mechanism of P. vivax invasion into Duffy-negative erythrocytes. In our experiments, the erythrocyte-binding activity of PvMSP1P, as shown in Fig. 7 and 8, suggests that PvMSP1P is a novel erythrocyte-binding ligand of P. vivax, suggesting that PvMSP1P might be one of the molecules that help invasion of P. vivax through the alternative pathway. Studies of P. falciparum MSPs show that they may play a role in the ligand-receptor interaction during the initial attachment of the merozoite (44, 45). Similar results in P. vivax have also been reported in a study showing that the C terminus of PvMSP1 binds strongly to the erythrocyte surface (24). Our IFA data have demonstrated for the first time that PvMSP1P is expressed on the surface of P. vivax merozoites. Intriguingly, it shows that the localization of PvMSP1P and PvMSP1 overlaps completely (Fig. 5), suggesting that PvMSP1P might play a role similar to PvMSP1 during merozoite invasion. Our Western blotting, done with antibody against the C terminus of PvMSP1P, has demonstrated that native PvMSP1P is processed and that the dominant species remaining in the parasite were about 70 and 14 kDa from the C terminus (Fig. 3B) and our erythrocyte-binding assays have demonstrated that erythrocyte-binding activity of PvMSP1P was found to be highly associated with the PvMSP1P-19 (14 kDa). Taken together, these data suggest that the erythrocyte-binding domain is in the C terminus of the native PvMSP1P. Further studies are required to find whether the erythrocyte-binding activity of PvMSP1P-19 is responsible for the initial attachment of the merozoite like PvMSP1 or for the junction formation like PvDBP. Overall, our studies have found that PvMSP1P has erythrocyte-binding ability, and it may represent the fourth novel ligand in the identified P. vivax merozoite, after PvDBP, PvRBP, and PvMSP1, which are responsible for erythrocyte invasion by the vivax parasite.
To determine whether the antibodies induced in humans can inhibit the function of PvMSP1P, we did an inhibition assay to measure erythrocyte binding on COS-7 cells expressing PvMSP1P-19 on their surface in the presence of human sera. The mouse anti-PvMSP1P-19 serum had significant inhibitory effects on erythrocyte binding at a 1:100 or lower (i.e., <1:100) dilution (Fig. 8B). The failure of ROK+ (Fig. 8C) to inhibit erythrocyte adhesion completely even at the highest concentration (1:10 dilution) of vivax patient sera suggests that either a lower avidity or lower titer of the anti-PvMSP1P-19 antibodies was induced in vivax patients than in immune mice. However, in human sera obtained only from the higher-responding samples (Fig. 8D, high ROK+), the inhibitory activity increased significantly and almost complete inhibition was observed at a 1:10 dilution (Fig. 8D). Taken together, these data suggest that P. vivax infection indeed successfully induces a significant level of blocking antibodies against the erythrocyte-binding epitope of PvMSP1P-19.
In summary, these data suggest that PvMSP1P is a novel surface antigen of P. vivax merozoite that binds to erythrocytes and it is immunogenic in humans; hence, it represents a novel vaccine candidate against P. vivax. PvMSP1P might play an important role in the merozoite invasion process and further studies on the precise molecular characterization of PvMSP1P in comparison with those of PvMSP1 will help us characterize a novel mechanism of P. vivax invasion into erythrocyte and develop a novel control measure against vivax malaria.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to John H. Adams for pEGFP-HSVgD1_PvDBPII_Sal 1 plasmid DNA and Kang-Seung Lee in K.-S. Ha's laboratory for technical assistance. We thank Thangavelu U. Arumugam for the critical reading of the manuscript. We also thank Kana Kato, Sachi Yawata, and Yuiko Ogasawara for their technical assistance.
This work was supported by a National Research Foundation of Korea Grant funded by the Korean Government (2009-0075103) and the Mid-Career Researcher Program through an NRF grant funded by the MEST (2011-0016401). This research was also supported in part by MEXT KAKENHI (23117008), JSPS KAKENHI (23406007), and a grant from the Ministry of Health, Labor, and Welfare (H21-Chikyukibo-ippan-005), Japan.
Footnotes
Published ahead of print 4 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01117-12.
REFERENCES
- 1. Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA. 2009. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect. Dis. 9:555–566 [DOI] [PubMed] [Google Scholar]
- 2. Gaur D, Mayer DC, Miller LH. 2004. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int. J. Parasitol. 34:1413–1429 [DOI] [PubMed] [Google Scholar]
- 3. Cowman AF, Crabb BS. 2006. Invasion of red blood cells by malaria parasites. Cell 124:755–766 [DOI] [PubMed] [Google Scholar]
- 4. Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295:302–304 [DOI] [PubMed] [Google Scholar]
- 5. Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod JF, Domarle O, Colin Y, Bertrand O, Picot J, King CL, Grimberg BT, Mercereau-Puijalon O, Zimmerman PA. 2010. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl. Acad. Sci. U. S. A. 107:5967–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, do Rosario VE, Benito A, Berzosa P, Arez AP. 2011. Duffy negative antigen is no longer a barrier to Plasmodium vivax–molecular evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl. Trop. Dis. 5:e1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilson PR, Nebl T, Vukcevic D, Moritz RL, Sargeant T, Speed TP, Schofield L, Crabb BS. 2006. Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics 5:1286–1299 [DOI] [PubMed] [Google Scholar]
- 8. Wang L, Mohandas N, Thomas A, Coppel RL. 2003. Detection of detergent-resistant membranes in asexual blood-stage parasites of Plasmodium falciparum. Mol. Biochem. Parasitol. 130:149–153 [DOI] [PubMed] [Google Scholar]
- 9. Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, McConville MJ, Schofield L, Hodder AN, Yates JR, III, Crabb BS. 2005. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J. Biol. Chem. 280:40169–40176 [DOI] [PubMed] [Google Scholar]
- 10. Kooij TW, Janse CJ, Waters AP. 2006. Plasmodium post-genomics: better the bug you know? Nat. Rev. Microbiol. 4:344–357 [DOI] [PubMed] [Google Scholar]
- 11. O'Donnell RA, Saul A, Cowman AF, Crabb BS. 2000. Functional conservation of the malaria vaccine antigen MSP-119 across distantly related Plasmodium species. Nat. Med. 6:91–95 [DOI] [PubMed] [Google Scholar]
- 12. Sanders PR, Kats LM, Drew DR, O'Donnell RA, O'Neill M, Maier AG, Coppel RL, Crabb BS. 2006. A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infect. Immun. 74:4330–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woodberry T, Minigo G, Piera KA, Hanley JC, de Silva HD, Salwati E, Kenangalem E, Tjitra E, Coppel RL, Price RN, Anstey NM, Plebanski M. 2008. Antibodies to Plasmodium falciparum and Plasmodium vivax merozoite surface protein 5 in Indonesia: species-specific and cross-reactive responses. J. Infect. Dis. 198:134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valderrama-Aguirre A, Quintero G, Gomez A, Castellanos A, Perez Y, Mendez F, Arevalo-Herrera M, Herrera S. 2005. Antigenicity, immunogenicity, and protective efficacy of Plasmodium vivax MSP1 PV200l: a potential malaria vaccine subunit. Am. J. Trop. Med. Hyg. 73:16–24 [DOI] [PubMed] [Google Scholar]
- 16. Martinez P, Suarez CF, Gomez A, Cardenas PP, Guerrero JE, Patarroyo MA. 2005. High level of conservation in Plasmodium vivax merozoite surface protein 4 (PvMSP4). Infect. Genet. Evol. 5:354–361 [DOI] [PubMed] [Google Scholar]
- 17. Garzon-Ospina D, Romero-Murillo L, Tobon LF, Patarroyo MA. 2011. Low genetic polymorphism of merozoite surface proteins 7 and 10 in Colombian Plasmodium vivax isolates. Infect. Genet. Evol. 11:528–531 [DOI] [PubMed] [Google Scholar]
- 18. Perez-Leal O, Sierra AY, Barrero CA, Moncada C, Martinez P, Cortes J, Lopez Y, Torres E, Salazar LM, Patarroyo MA. 2004. Plasmodium vivax merozoite surface protein 8 cloning, expression, and characterisation. Biochem. Biophys. Res. Commun. 324:1393–1399 [DOI] [PubMed] [Google Scholar]
- 19. Li J, Ito D, Chen JH, Lu F, Cheng Y, Wang B, Ha KS, Cao J, Torii M, Sattabongkot J, Tsuboi T, Han ET. 2012. Pv12, a 6-Cys antigen of Plasmodium vivax, is localized to the merozoite rhoptry. Parasitol. Int. 426:1–6 [DOI] [PubMed] [Google Scholar]
- 20. Mongui A, Angel DI, Gallego G, Reyes C, Martinez P, Guhl F, Patarroyo MA. 2009. Characterization and antigenicity of the promising vaccine candidate Plasmodium vivax 34 kDa rhoptry antigen (Pv34). Vaccine 28:415–421 [DOI] [PubMed] [Google Scholar]
- 21. Mongui A, Angel DI, Guzman C, Vanegas M, Patarroyo MA. 2008. Characterisation of the Plasmodium vivax Pv38 antigen. Biochem. Biophys. Res. Commun. 376:326–330 [DOI] [PubMed] [Google Scholar]
- 22. Perera KL, Handunnetti SM, Holm I, Longacre S, Mendis K. 1998. Baculovirus merozoite surface protein 1 C-terminal recombinant antigens are highly protective in a natural primate model for human Plasmodium vivax malaria. Infect. Immun. 66:1500–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Polley SD, McRobert L, Sutherland CJ. 2004. Vaccination for vivax malaria: targeting the invaders. Trends Parasitol. 20:99–102 [DOI] [PubMed] [Google Scholar]
- 24. Han HJ, Park SG, Kim SH, Hwang SY, Han J, Traicoff J, Kho WG, Chung JY. 2004. Epidermal growth factor-like motifs 1 and 2 of Plasmodium vivax merozoite surface protein 1 are critical domains in erythrocyte invasion. Biochem. Biophys. Res. Commun. 320:563–570 [DOI] [PubMed] [Google Scholar]
- 25. Dutta S, Ware LA, Barbosa A, Ockenhouse CF, Lanar DE. 2001. Purification, characterization, and immunogenicity of a disulfide cross-linked Plasmodium vivax vaccine candidate antigen, merozoite surface protein 1, expressed in Escherichia coli. Infect. Immun. 69:5464–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Kaneko O, Sattabongkot J, Chen JH, Lu F, Chai JY, Takeo S, Tsuboi T, Ayala FJ, Chen Y, Lim CS, Han ET. 2010. Genetic polymorphism of Plasmodium vivax msp1p, a paralog of merozoite surface protein 1, from worldwide isolates. Am. J. Trop. Med. Hyg. 84:292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen JH, Jung JW, Wang Y, Ha KS, Lu F, Lim CS, Takeo S, Tsuboi T, Han ET. 2010. Immunoproteomics profiling of blood stage Plasmodium vivax infection by high-throughput screening assays. J. Proteome Res. 9:6479–6489 [DOI] [PubMed] [Google Scholar]
- 28. Rui E, Fernandez-Becerra C, Takeo S, Sanz S, Lacerda MV, Tsuboi T, del Portillo HA. 2011. Plasmodium vivax: comparison of immunogenicity among proteins expressed in the cell-free systems of Escherichia coli and wheat germ by suspension array assays. Malar. J. 10:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsuboi T, Takeo S, Iriko H, Jin L, Tsuchimochi M, Matsuda S, Han ET, Otsuki H, Kaneko O, Sattabongkot J, Udomsangpetch R, Sawasaki T, Torii M, Endo Y. 2008. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect. Immun. 76:1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsuboi T, Takeo S, Sawasaki T, Torii M, Endo Y. 2010. An efficient approach to the production of vaccines against the malaria parasite. Methods Mol. Biol. 607:73–83 [DOI] [PubMed] [Google Scholar]
- 31. Arumugam TU, Takeo S, Yamasaki T, Thonkukiatkul A, Miura K, Otsuki H, Zhou H, Long CA, Sattabongkot J, Thompson J, Wilson DW, Beeson JG, Healer J, Crabb BS, Cowman AF, Torii M, Tsuboi T. 2011. Discovery of GAMA, a Plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen. Infect. Immun. 79:4523–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen JH, Wang Y, Ha KS, Lu F, Suh IB, Lim CS, Park JH, Takeo S, Tsuboi T, Han ET. 2011. Measurement of naturally acquired humoral immune responses against the C-terminal region of the Plasmodium vivax MSP1 protein using protein arrays. Parasitol. Res. 109:1259–1266 [DOI] [PubMed] [Google Scholar]
- 33. Michon P, Fraser T, Adams JH. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect. Immun. 68:3164–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayer DC, Cofie J, Jiang L, Hartl DL, Tracy E, Kabat J, Mendoza LH, Miller LH. 2009. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc. Natl. Acad. Sci. U. S. A. 106:5348–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arakawa T, Tsuboi T, Kishimoto A, Sattabongkot J, Suwanabun N, Rungruang T, Matsumoto Y, Tsuji N, Hisaeda H, Stowers A, Shimabukuro I, Sato Y, Torii M. 2003. Serum antibodies induced by intranasal immunization of mice with Plasmodium vivax Pvs25 co-administered with cholera toxin completely block parasite transmission to mosquitoes. Vaccine 21:3143–3148 [DOI] [PubMed] [Google Scholar]
- 36. Babon JJ, Morgan WD, Kelly G, Eccleston JF, Feeney J, Holder AA. 2007. Structural studies on Plasmodium vivax merozoite surface protein-1. Mol. Biochem. Parasitol. 153:31–40 [DOI] [PubMed] [Google Scholar]
- 37. Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NP, White NJ, Carlton JM, Preiser PR. 2008. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 105:16290–16295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soares IS, Levitus G, Souza JM, Del Portillo HA, Rodrigues MM. 1997. Acquired immune responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1 in individuals exposed to malaria. Infect. Immun. 65:1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandez-Becerra C, Sanz S, Brucet M, Stanisic DI, Alves FP, Camargo EP, Alonso PL, Mueller I, del Portillo HA. 2010. Naturally-acquired humoral immune responses against the N- and C-termini of the Plasmodium vivax MSP1 protein in endemic regions of Brazil and Papua New Guinea using a multiplex assay. Malar. J. 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ceravolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, Braga EM, McHenry AM, Adams JH, Brito CF, Carvalho LH. 2009. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin. Exp. Immunol. 156:502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, Sattabongkot J, Cui L, Bockarie M, Chitnis C, Adams J, Zimmerman PA, King CL. 2007. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 4:e337 doi:10.1371/journal.pmed.0040337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chitnis CE, Miller LH. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chitnis CE, Sharma A. 2008. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol. 24:29–34 [DOI] [PubMed] [Google Scholar]
- 44. Goel VK, Li X, Chen H, Liu SC, Chishti AH, Oh SS. 2003. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 100:5164–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Chen H, Oo TH, Daly TM, Bergman LW, Liu SC, Chishti AH, Oh SS. 2004. A co-ligand complex anchors Plasmodium falciparum merozoites to the erythrocyte invasion receptor band 3. J. Biol. Chem. 279:5765–5771 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.