Abstract
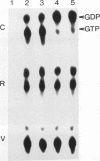
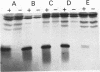
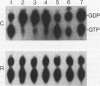
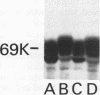
The GTPase activity of p21ras is stimulated by GTPase-activating proteins (GAPs) such as p120GAP and the product of the neurofibromatosis 1 gene, which may negatively regulate p21 function. GAPs are also proposed effectors of ras. We have sought activating substitutions in c-H-ras in the region encoding the effector domain, on the rationale that such mutations would dissociate effector function from negative regulation by GAP. One such activating mutation, Pro-34-->Arg, encodes protein that is substantially bound to GTP in vivo. In vitro, this protein is not stimulated by GAPs, and its binding to p120GAP is grossly impaired. The results support the idea that the p21 structural requirements for effector function and GAP interaction are quite different and suggest that some molecule(s) other than p120GAP serves as the ras effector. In contrast to the results obtained with p120GAP, the Pro-34-->Arg p21 species is effectively coupled to the raf-1 product, as judged from electrophoretic mobility shifts of the Raf-1 phosphoprotein.
Full text
PDF



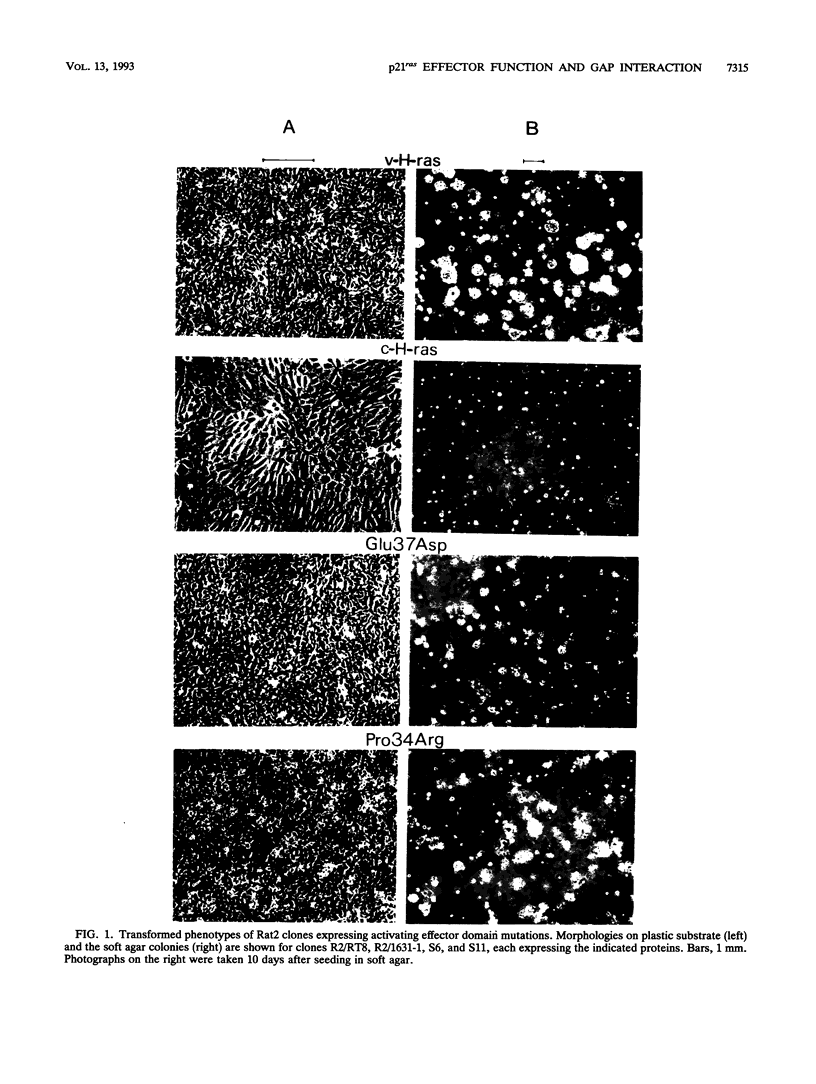

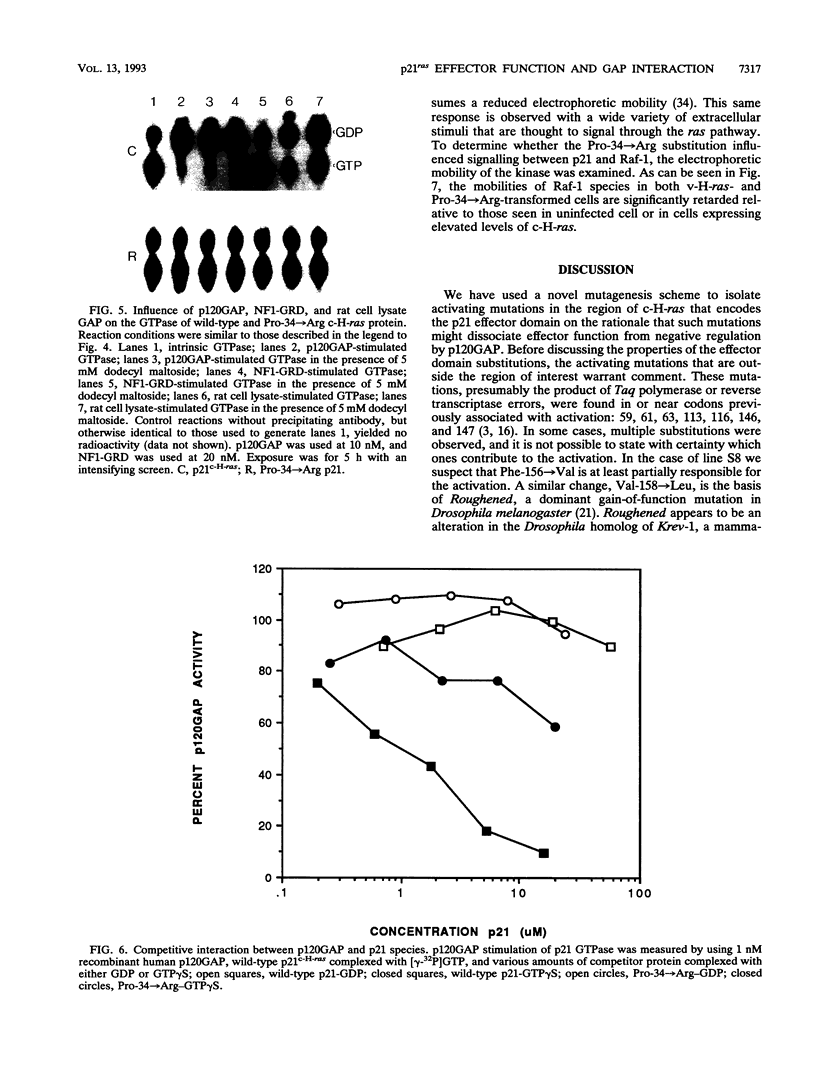



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H., Lowy D. R., Willumsen B. M., Der C. J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988 Apr 22;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Basu T. N., Gutmann D. H., Fletcher J. A., Glover T. W., Collins F. S., Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992 Apr 23;356(6371):713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- Bollag G., McCormick F. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature. 1991 Jun 13;351(6327):576–579. doi: 10.1038/351576a0. [DOI] [PubMed] [Google Scholar]
- Bos J. L. ras oncogenes in human cancer: a review. Cancer Res. 1989 Sep 1;49(17):4682–4689. [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Calés C., Hancock J. F., Marshall C. J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988 Apr 7;332(6164):548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- DeClue J. E., Papageorge A. G., Fletcher J. A., Diehl S. R., Ratner N., Vass W. C., Lowy D. R. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992 Apr 17;69(2):265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- DeClue J. E., Stone J. C., Blanchard R. A., Papageorge A. G., Martin P., Zhang K., Lowy D. R. A ras effector domain mutant which is temperature sensitive for cellular transformation: interactions with GTPase-activating protein and NF-1. Mol Cell Biol. 1991 Jun;11(6):3132–3138. doi: 10.1128/mcb.11.6.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez I., Marshall M. S., Gibbs J. B., García de Herreros A., Cornet M. E., Graziani G., Diaz-Meco M. T., Johansen T., McCormick F., Moscat J. Role of GTPase activating protein in mitogenic signalling through phosphatidylcholine-hydrolysing phospholipase C. EMBO J. 1991 Nov;10(11):3215–3220. doi: 10.1002/j.1460-2075.1991.tb04884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Moran M., McCormick F., Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990 Jan 25;343(6256):377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- Farnsworth C. L., Marshall M. S., Gibbs J. B., Stacey D. W., Feig L. A. Preferential inhibition of the oncogenic form of RasH by mutations in the GAP binding/"effector" domain. Cell. 1991 Feb 8;64(3):625–633. doi: 10.1016/0092-8674(91)90246-u. [DOI] [PubMed] [Google Scholar]
- Feig L. A., Cooper G. M. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988 Jun;8(6):2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth M. E., Davis L. J., Fleurdelys B., Scolnick E. M. Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras gene family. J Virol. 1982 Jul;43(1):294–304. doi: 10.1128/jvi.43.1.294-304.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S., Scolnick E. M., Dixon R. A., Vogel U. S. Modulation of guanine nucleotides bound to Ras in NIH3T3 cells by oncogenes, growth factors, and the GTPase activating protein (GAP). J Biol Chem. 1990 Nov 25;265(33):20437–20442. [PubMed] [Google Scholar]
- Gideon P., John J., Frech M., Lautwein A., Clark R., Scheffler J. E., Wittinghofer A. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol Cell Biol. 1992 May;12(5):2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan I. K., Carthew R. W., Rubin G. M. The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell. 1991 Nov 15;67(4):717–722. doi: 10.1016/0092-8674(91)90066-8. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Nordeen S. K., Vogt K., Edgell M. H. A complete library of point substitution mutations in the glucocorticoid response element of mouse mammary tumor virus. Proc Natl Acad Sci U S A. 1986 Feb;83(3):710–714. doi: 10.1073/pnas.83.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. R., Morrison D. K., Wong G., McCormick F., Williams L. T. PDGF beta-receptor stimulates tyrosine phosphorylation of GAP and association of GAP with a signaling complex. Cell. 1990 Apr 6;61(1):125–133. doi: 10.1016/0092-8674(90)90220-9. [DOI] [PubMed] [Google Scholar]
- Kazlauskas A., Ellis C., Pawson T., Cooper J. A. Binding of GAP to activated PDGF receptors. Science. 1990 Mar 30;247(4950):1578–1581. doi: 10.1126/science.2157284. [DOI] [PubMed] [Google Scholar]
- Kitayama H., Sugimoto Y., Matsuzaki T., Ikawa Y., Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989 Jan 13;56(1):77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- Lacal J. C., Santos E., Notario V., Barbacid M., Yamazaki S., Kung H., Seamans C., McAndrew S., Crowl R. Expression of normal and transforming H-ras genes in Escherichia coli and purification of their encoded p21 proteins. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5305–5309. doi: 10.1073/pnas.81.17.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bollag G., Clark R., Stevens J., Conroy L., Fults D., Ward K., Friedman E., Samowitz W., Robertson M. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell. 1992 Apr 17;69(2):275–281. doi: 10.1016/0092-8674(92)90408-5. [DOI] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Marshall M. S., Davis L. J., Keys R. D., Mosser S. D., Hill W. S., Scolnick E. M., Gibbs J. B. Identification of amino acid residues required for Ras p21 target activation. Mol Cell Biol. 1991 Aug;11(8):3997–4004. doi: 10.1128/mcb.11.8.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Hettich L. A. Characterization of Ras effector mutant interactions with the NF1-GAP related domain. Oncogene. 1993 Feb;8(2):425–431. [PubMed] [Google Scholar]
- Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., Cawthon R. M. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990 Nov 16;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Moodie S. A., Willumsen B. M., Weber M. J., Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993 Jun 11;260(5114):1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- Morgenstern J. P., Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990 Jun 25;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. K., Kaplan D. R., Rapp U., Roberts T. M. Signal transduction from membrane to cytoplasm: growth factors and membrane-bound oncogene products increase Raf-1 phosphorylation and associated protein kinase activity. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8855–8859. doi: 10.1073/pnas.85.23.8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989 Sep 21;341(6239):209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- Pawson T., Gish G. D. SH2 and SH3 domains: from structure to function. Cell. 1992 Oct 30;71(3):359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- Schaber M. D., Garsky V. M., Boylan D., Hill W. S., Scolnick E. M., Marshall M. S., Sigal I. S., Gibbs J. B. Ras interaction with the GTPase-activating protein (GAP). Proteins. 1989;6(3):306–315. doi: 10.1002/prot.340060313. [DOI] [PubMed] [Google Scholar]
- Schlichting I., Almo S. C., Rapp G., Wilson K., Petratos K., Lentfer A., Wittinghofer A., Kabsch W., Pai E. F., Petsko G. A. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature. 1990 May 24;345(6273):309–315. doi: 10.1038/345309a0. [DOI] [PubMed] [Google Scholar]
- Schweighoffer F., Barlat I., Chevallier-Multon M. C., Tocque B. Implication of GAP in Ras-dependent transactivation of a polyoma enhancer sequence. Science. 1992 May 8;256(5058):825–827. doi: 10.1126/science.1317056. [DOI] [PubMed] [Google Scholar]
- Settleman J., Narasimhan V., Foster L. C., Weinberg R. A. Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell. 1992 May 1;69(3):539–549. doi: 10.1016/0092-8674(92)90454-k. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. C., Blanchard R. A. Genetic definition of ras effector elements. Mol Cell Biol. 1991 Dec;11(12):6158–6165. doi: 10.1128/mcb.11.12.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. C., Vass W. C., Willumsen B. M., Lowy D. R. p21-ras effector domain mutants constructed by "cassette" mutagenesis. Mol Cell Biol. 1988 Aug;8(8):3565–3569. doi: 10.1128/mcb.8.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Lin B. K., Wood D. R., Tamanoi F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):468–472. doi: 10.1073/pnas.88.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Wood D. R., Lin B. K., Khalil M., Tamanoi F., Cannon J. F. A dominant activating mutation in the effector region of RAS abolishes IRA2 sensitivity. Mol Cell Biol. 1992 Feb;12(2):631–637. doi: 10.1128/mcb.12.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. M., DeMarco M., D'Arcangelo G., Halegoua S., Brugge J. S. Ras is essential for nerve growth factor- and phorbol ester-induced tyrosine phosphorylation of MAP kinases. Cell. 1992 Mar 20;68(6):1031–1040. doi: 10.1016/0092-8674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Topp W. C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981 Aug;113(1):408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., Gibbs J. B. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988 Sep 1;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Papageorge A. G., Kung H. F., Bekesi E., Robins T., Johnsen M., Vass W. C., Lowy D. R. Mutational analysis of a ras catalytic domain. Mol Cell Biol. 1986 Jul;6(7):2646–2654. doi: 10.1128/mcb.6.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G., Müller O., Clark R., Conroy L., Moran M. F., Polakis P., McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992 May 1;69(3):551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- Wood K. W., Sarnecki C., Roberts T. M., Blenis J. ras mediates nerve growth factor receptor modulation of three signal-transducing protein kinases: MAP kinase, Raf-1, and RSK. Cell. 1992 Mar 20;68(6):1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Yatani A., Okabe K., Polakis P., Halenbeck R., McCormick F., Brown A. M. ras p21 and GAP inhibit coupling of muscarinic receptors to atrial K+ channels. Cell. 1990 Jun 1;61(5):769–776. doi: 10.1016/0092-8674(90)90187-j. [DOI] [PubMed] [Google Scholar]
- Zhang K., DeClue J. E., Vass W. C., Papageorge A. G., McCormick F., Lowy D. R. Suppression of c-ras transformation by GTPase-activating protein. Nature. 1990 Aug 23;346(6286):754–756. doi: 10.1038/346754a0. [DOI] [PubMed] [Google Scholar]