Abstract
Atlantis II and Discovery are two hydrothermal and hypersaline deep-sea pools in the Red Sea rift that are characterized by strong thermohalo-stratification and temperatures steadily peaking near the bottom. We conducted comprehensive vertical profiling of the microbial populations in both pools and highlighted the influential environmental factors. Pyrosequencing of the 16S rRNA genes revealed shifts in community structures vis-à-vis depth. High diversity and low abundance were features of the deepest convective layers despite the low cell density. Surprisingly, the brine interfaces had significantly higher cell counts than the overlying deep-sea water, yet they were lowest in diversity. Vertical stratification of the bacterial populations was apparent as we moved from the Alphaproteobacteria-dominated deep sea to the Planctomycetaceae- or Deferribacteres-dominated interfaces to the Gammaproteobacteria-dominated brine layers. Archaeal marine group I was dominant in the deep-sea water and interfaces, while several euryarchaeotic groups increased in the brine. Across sites, microbial phylotypes and abundances varied substantially in the brine interface of Discovery compared with Atlantis II, despite the near-identical populations in the overlying deep-sea waters. The lowest convective layers harbored interestingly similar microbial communities, even though temperature and heavy metal concentrations were very different. Multivariate analysis indicated that temperature and salinity were the major influences shaping the communities. The harsh conditions and the low-abundance phylotypes could explain the observed correlation in the brine pools.
INTRODUCTION
The Red Sea, a part of the Great Rift Valley, was formed when the Arabian plate split from the African plate. Due to the high surface temperature, excess evaporation, and scarcity of rainfall coupled with a lack of a major source of fresh water influx, the Red Sea is one of the hottest and saltiest seawater bodies in the world. Over the past 40 years, oceanographers have discovered 25 brine-filled deeps in the rift valley of the Red Sea (1–6). These deeps are characterized by their anoxic, hypersaline, hyperthermal, and metalliferous conditions at the bottom, representing some of the most unusual and extreme environments on earth (4, 7–9).
The deep-sea brine pools Atlantis II and Discovery, which are located in the middle axial rift zone of the Red Sea, represent a rare type of extreme environment with both hypersaline and hydrothermal geochemical characteristics. Since their discovery in 1965, detailed geological and geochemical investigations have been carried out on these two deeps (1). The Atlantis II Deep is a hydrothermally active brine pool with a temperature that has continuously increased from the earliest record of 44.8°C (1, 10) to the current recorded temperature of 68°C. This increase has been attributed to the influx of hot brine supplied by a geyser spring at the bottom of the Atlantis II pool (11), which divides the brine into two stratified anoxic layers with substantially different temperatures (12): the lower convective layer and the upper convective layer. Subsequent surveys revealed the formation of two new convective layers within the previously reported upper layer. The new layers were characterized by near-constant temperatures and salinities, bringing the total number of convective layers to four (13). Besides the high temperature and salinity, the Atlantis II brine pool is also characterized by its high metal concentration, with trace elements such as iron, zinc, and copper present in concentrations 1,000 times higher than in normal seawater. The high temperature, pressure (due to depths reaching over 2,100 m), and metal concentrations make the Atlantis II brine pool a typical hydrothermal system (14). More interestingly, this vertical stratification of water temperature and salinity as well as metal contents in the pool increases drastically with water depth. In contrast, the neighboring Discovery Deep, located 7.4 km southwest of the Atlantis II Deep, is a typical brine basin, with a nearly constant temperature of 45°C reported between 1965 and 1998. There has been no reported increase since (15). The temperature stability in Discovery indicates the absence of any substantial hydrothermal input from the substrata (7, 16, 17).
Previous studies revealed a great diversity of bacteria and archaea in hypersaline and hydrothermal environments (18, 19), many of which were novel organisms or were previously unknown to exist in such environments (20–22). Studies using 16S rRNA gene sequencing and cultivation methods have shown that novel groups of Archaea and Bacteria thrive in the Kebrit Deep in the Red Sea (23, 24). Although detailed geological and geochemical investigations have been carried out at the Atlantis II and Discovery Deeps during the last 40 years, information about the microbial communities of these deeps is scarce. To date, only four recent publications have investigated the microbial diversity and composition in the water column above the two deeps and in the sediment (25–28). Thus, we adopted a comprehensive approach using 16S rRNA tag pyrosequencing and multivariate statistics to characterize the bacterial and archaeal communities of these two deeps in order to answer the following fundamental questions. (i) How do the vertical stratification of physical and chemical environmental conditions shape the microbial community in these deep-sea brine pools? (ii) How do the microbial communities respond to the contrasting environmental conditions (the rapidly changing environmental conditions in the Atlantis II Deep versus the relatively stable environmental conditions in the Discovery Deep)?
MATERIALS AND METHODS
Sample collection.
Water samples were collected in April 2010 from the Atlantis II Deep (21°20.97′ N, 38°04.65′ E) and Discovery Deep (21°16.82′ N, 38°2.95′ E) during the Aegaeo expedition in the Red Sea. A rosette water sampler equipped with 10-liter Niskin bottles and a SIMRAD EK500 scientific echo sounder system that monitors depth were dispatched at each location. A Seabird conductivity-temperature-depth (CTD) device was deployed down to the deep water to measure pressure, temperature, and salinity, but for the brine pools, a high-range CTD device was deployed instead. At the Discovery Deep, water samples were collected from the brine-overlying deep water at 1,800 m (DIS-NDW), the brine-water interface at 2,028 m (DIS-BWI), and the lower convective layer of the brine pool at 2,114 m (DIS-LCL). At the Atlantis II Deep, in addition to these three layers (ATL-NDW at 1,800 m, ATL-BWI at 1,998 m, and ATL-LCL at 2,139 m), water samples were collected from the two upper convective layers at 2,022 and 2,049 m (ATL-UCL1 and ATL-UCL2, respectively) (Fig. 1; Table 1). The depth was estimated as a function of pressure (see equation 25 in reference 29). Each water sample collected (10 liters per specified depth at each location) was immediately filtered through a 1.6-μm-pore-size glass fiber filter (GF/A; diameter, 125 mm; Whatman, Maidstone, United Kingdom) to remove suspended particles and eukaryotes and then filtered through six 0.22-μm-pore-size polycarbonate membranes (diameter, 45 mm; Millipore, Bedford, MA) to capture microbial cells. The polycarbonate membranes were then frozen at −80°C in 0.8 ml of extraction buffer (40 mM EDTA, 0.75 M sucrose, 0.5 M NaCl, 50 mM Tris; pH 8) and transported back to the Hong Kong University of Science and Technology on dry ice for DNA extraction.
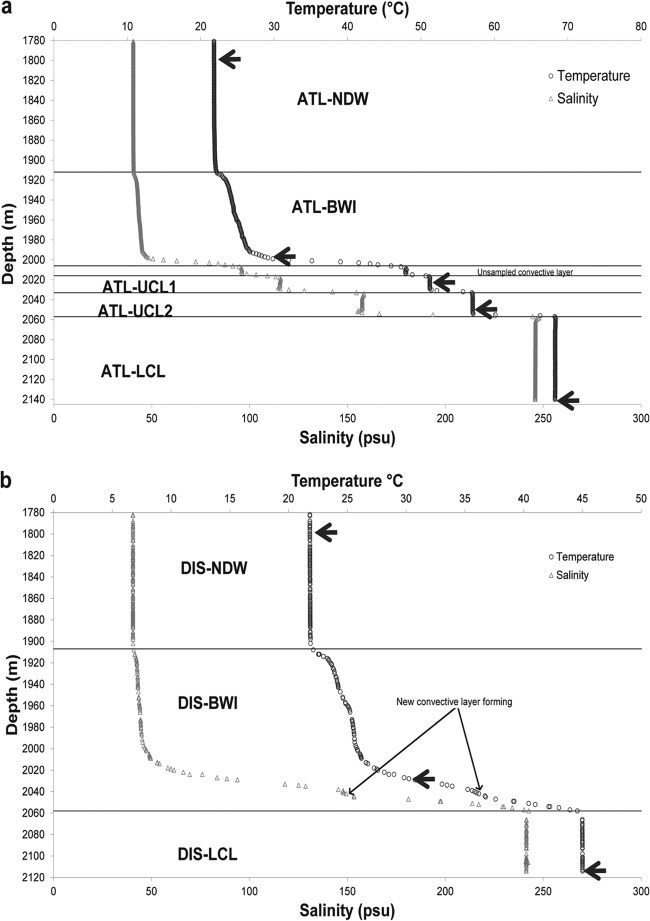
Fig 1.
Temperature and salinity profiles of Atlantis II (a) and Discovery (b) sampling sites. Values were obtained with a high-range CTD device. For Atlantis II, four layers of stable temperature and salinity were clearly observed and labeled, while for Discovery, a single homogeneous convective layer was observed. An upper convective layer seems to be forming above the lower convective layer of Discovery Deep. Sampling depths are indicated with the arrows on the temperature lines.
Table 1.
Uppermost limits of convective layers and transition zones in the Atlantis II and Discovery Deepsa
| Atlantis II |
Discovery |
||
|---|---|---|---|
| Layer | Upper limit (m) | Layer | Upper limit (m) |
| ATL-BWI | 1,902 | DIS-BWI | 1,902 |
| ATL-UCLb | 2,006 | ||
| ATL-UCL1 | 2,015 | ||
| ATL-UCL2 | 2,034 | ||
| ATL-LCL | 2,057 | DIS-LCL | 2,057 |
Determined by the temperature and salinity profiles generated by the high-range CTD device.
This is the uppermost convective layer in Atlantis II and was not sampled because of its thinness.
Measurement of environmental parameters and determination of microbial cell count.
Environmental parameters, including temperature, salinity, nutrient concentrations (NO3−, PO43−, Si, and SO42−), and metal contents (Cu, Fe, and Mn) were determined on board by the CTD device and a HACH DR/890 portable colorimeter according to the manufacturer's protocol (Hach, Loveland, CO). The total organic carbon (TOC) and total inorganic carbon (TIC) were analyzed using a TOC analyzer in the laboratory (TOC-VCPH/TNM-I; Shimadzu, Kyoto, Japan), and NH4+ and NO2− were also analyzed in the laboratory (Skalar 4000 analyzer; Skalar, Breda, The Netherlands).
For the total microbial cell counts, a 10-ml aliquot of each sample (3 replicates each) was immediately fixed with formaldehyde at a final concentration of 4% upon collection. The fixed samples were then filtered through black polycarbonate membranes (0.2-μm pore size; 25-mm diameter; Isopore; Millipore, Bedford, MA) and stained with 4,6-diamidino-2-phenylindole for 15 min in the dark. Stained cells were visualized by using an Olympus epifluorescence microscope (FSX100, Hamburg, Germany) with excitation/emission filters of 365/420 nm and counted on 10 randomly selected fields.
DNA extraction, PCR amplification, and 16S rRNA gene tag pyrosequencing.
The total DNA was extracted and purified using an SDS-based method described by Qian et al. (25). A set of primers was designed by adding 6 nucleotide barcodes (see Table S1 in the supplemental material) to the universal forward primer U789F (5′-TAGATACCCSSGTAGTCC-3′) and the reverse primer U1068R (5′-CTGACGRCRGCCATGC-3′) for amplification of Bacteria and Archaea (30, 31). Each 100-μl reaction mixture contained 4 units of Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), 1× Pfu reaction buffer, 0.2 mM concentrations of deoxynucleoside triphosphate (dNTPs), a 0.1 μM concentration of each primer, and 20 ng of total DNA. PCR amplification was conducted in a thermocycler (Bio-Rad, Hercules, CA) under the following conditions: initial denaturation at 94°C for 5 min; 25 cycles of 94°C for 50 s, 54°C for 50 s, and 72°C for 50 s; and a final extension at 72°C for 6 min. PCR amplicons were purified using a TaKaRa agarose gel DNA purification kit (TaKaRa, Dalian, China) and quantified with a NanoDrop ND-1000 device (Nanodrop, DE). Amplicons of every sample were mixed and subjected to pyrosequencing on the 454 FLX Titanium platform (Roche, Basel, Switzerland) at the Chinese National Human Genome Centre in Shanghai, China.
Classification and statistical analyses.
A downstream bioinformatics analysis was performed using QIIME 1.4.0 (32). All reads had to satisfy the following criteria: (i) no ambiguous bases, (ii) a minimum length of 150 bp, (iii) no homopolymers of 6 bp and above, and (iv) application of a quality window of 50 bp with an average flowgram score of 25. Demultiplexing was done using the barcodes, and then a second round of quality control was carried out using Denoiser (33). Operational taxonomic units (OTUs) were assigned to qualified reads at 3% dissimilarity using Mothur's average-neighbor algorithm (34). Representatives from each OTU were selected based on abundance for subsequent analysis. Representative OTUs were de novo aligned using MUSCLE (35), and a phylogenetic tree was produced using FastTree (36). Chimeric reads were identified and excluded using ChimeraSlayer (37) after alignment with PyNAST (38) against Silva108 as a reference set (39). Species diversity, richness, and rarefaction curves were computed at 97% similarity as part of QIIME's alpha diversity pipeline. Four metrics were calculated, including observed species, Chao1 (a species richness index), Shannon index (a diversity index that takes into account abundance and evenness), and Simpson index (a diversity index that measures the probability that two randomly selected individuals selected from the sample will belong to the same group). We also calculated single and double OTUs to quantify the “rare biosphere” in the various samples. Jackknife-supported beta diversity analysis was conducted using both the weighted and the unweighted UniFrac metrics after the samples had been rarefied at the smallest library and then visualized using the unweighted pair group method with arithmetic mean (UPGMA) and principal coordinate analysis (PCoA). A step size of 100 was used, with 100 repetitions at each step. Taxonomy assignment was conducted using the RDP classifier, version 2.2 (40), against Silva108 with a bootstrap confidence level of 80%. The number of reads assigned to different genera were converted into percentages, which served as the input for Cluster3 (41). A divergence greater than 0.5% was used to filter out genera with small differences among the samples. The remaining genera were further normalized and centered by mean. Complete linkage method with a metric of correlation (uncentered) was used to generate a hierarchical cluster and a heatmap by using Java Treeview (42).
Relationship between microbial communities and environment.
The correlations between microbial communities and environmental factors were analyzed with ordination methods using the software Canoco for Windows, v4.5 (43). For both constrained and unconstrained ordination methods, the percentage abundance data of bacterial groups (at the genus level) in each library were used as the species input, and the environmental variables were normalized by log transformation and served as the environmental input. Canoco's automatic forward selection and Monte Carlo permutations were used to build the optimal models of microbe-environment relationship (44).
Sequence data accession number.
The pyrosequencing reads were deposited in the NCBI Sequence Read Archive database under accession number SRA052277.
RESULTS
Geochemical characterization of the sampling sites.
Temperature and salinity profiling of the Atlantis II Deep and the Discovery Deep revealed a vertical stratification along both water columns (Fig. 1; Table 2). Temperature and salinity in the normal deep water, defined as the water column between a depth of 1,780 m and the brine interface (the transition layer), were the same at both sampling stations. The interface region between the Red Sea deep water and the Atlantis II and Discovery brine basins was divided into an upper zone and a lower zone. The former is defined from the top by the 1,900-m southern contour and from the bottom by the 1,990-m sill that divides the Atlantis II Deep from the Discovery Deep (45, 46). This upper zone was outlined by a similar gradual elevation in temperature and salinity above both basins (Fig. 1). In contrast, both basins' lower zones showed an accentuated increase in temperature and salinity (Fig. 1). This lower zone extended from the 1,990-m sill to ∼ 2,006 m in the Atlantis II Deep and to ∼2,056 m in the Discovery Deep. Below the interface, a total of four convective layers were observed in the Atlantis II brine pool, while the Discovery pool constituted a single homogeneous body of brine (Fig. 1). All layers were sampled as described in Materials and Methods, except the uppermost layer in the Atlantis II Deep, due to its narrow range.
Table 2.
Geochemical and microbial profiling of Atlantis II and Discovery brine and overlying deep seawater
| Sample ID | Depth (m) | Concn of: |
Microbial cell density (104/ml) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fe (mg/liter) | Mn (mg/liter) | Cu (μg/liter) | Si (mg/liter) | SO4−2 (mg/liter) | PO4−3 (mg/liter) | NO3− (mg/liter) | NO2− (μM) | NH4+ (mg/liter) | TOC (mg/liter) | TIC (mg/liter) | |||
| DIS-NDW | 1,800 | <0.1 | <0.1 | 7.73 ± 0.15 | 1.03 ± 0.15 | 2,700 ± 100 | 2.32 ± 0.13 | 1.00 ± 0.10 | 0.055 | 0.003 | 1.32 ± 0.73 | 28.6 ± 0.14 | 4.5 ± 1.91 |
| DIS-BWI | 2,028 | <0.1 | 9.53 ± 0.83 | <0.1 | 10.13 ± 0.15 | 2,400 ± 264 | <0.1 | 1.40 ± 0.30 | 0.37 | 1.13 | 3.32 ± 0.47 | 18.7 ± 0.02 | 54.4 ± 15.45 |
| DIS-LCL | 2,114 | 0.72 ± 0.13 | 4.93 ± 2.37 | 2.20 ± 0.20 | 7.83 ± 1.44 | 667 ± 58 | 1.29 ± 0.07 | 2.80 ± 0.10 | 0.94 | 10.91 | 2.10 ± 0.39 | 3.0 ± 0.02 | 1.9 ± 1.18 |
| ATL-NDW | 1,800 | <0.1 | <0.1 | 5.03 ± 0.15 | 0.77 ± 0.06 | 2,866 ± 57 | 0.18 ± 0.02 | 7.47 ± 0.31 | 0.009 | 0.009 | 3.31 ± 0.22 | 29.8 ± 0.04 | 5.0 ± 1.00 |
| ATL-BWI | 1,998 | <0.1 | <0.1 | 1.43 ± 0.66 | 3.07 ± 0.06 | 2,933 ± 602 | <0.1 | 1.13 ± 0.59 | 0.01 | 0.01 | 5.51 ± 0.38 | 26.4 ± 0.13 | 18.2 ± 2.23 |
| ATL-UCL1 | 2,022 | <0.1 | 49.00 ± 2.00 | 2.4 ± 0.44 | 20.50 ± 1.75 | 1,900 ± 173 | 0.28 ± 0.07 | 3.27 ± 2.47 | 0.1 | 0.38 | 4.36 ± 0.25 | 25.3 ± 1.38 | 28.3 ± 10.41 |
| ATL-UCL2 | 2,049 | 0.23 ± 0.02 | 94.67 ± 5.13 | 3.20 ± 2.77 | 25.77 ± 1.58 | 1,733 ± 58 | 0.61 ± 0.28 | 1.20 ± 0.36 | 0.39 | 1.95 | 1.87 ± 0.01 | 22.5 ± 0.01 | 3.33 ± 2.08 |
| ATL-LCL | 2,139 | 83.00 ± 10.44 | 141.00 ± 39.00 | 18.37 ± 4.50 | 69.33 ± 4.05 | 467 ± 57 | 1.11 ± 0.29 | 4.80 ± 0.17 | 2.38 | 10.62 | NDa | 15.3 ± 0.87 | 0.83 ± 0.29 |
Not detected.
Table 2 summarizes the chemical parameters measured. The Atlantis II Deep was richer than the Discovery Deep in heavy metals (Fe, Mn, Si, and Cu), and concentrations increased with depth. Nutrient concentrations, on the other hand, varied along the depth and across sites. In both brines, NO2−, NH4+, NO3− and PO43− concentrations increased with depth, while SO42−, TIC (which were present noticeably in the normal deep water), and TOC concentrations decreased with depth. Intriguingly, TOC in ATL-LCL was at depletion level, while TIC was relatively high. In contrast, DIS-LCL showed a different TOC/TIC profile, suggesting the presence of an active carbon mineralization process in the Atlantis II Deep and not in the Discovery Deep.
Diversity and species richness of microbial communities.
The microbial cell density varied markedly between different layers (Table 2) and correlated negatively with Fe, PO4, and Cl and positively with TOC (Spearman's rank correlation; P > 0.05; see Table S2 in the supplemental material). Cell density was the highest in both transition layers and ATL-UCL1, up to 12-fold higher than that in the normal deep water (Table 2). Meanwhile, the lower layers harbored the lowest cell counts (Table 2).
Using 454 pyrosequencing targeting the 16S rRNA gene-V6 tag sequences, more than 190,000 qualified reads (18,867 Archaea, 162,870 Bacteria, and the remaining 10,600 unclassified; Table 3), with an average read length of 280 bp, were obtained from the Atlantis II Deep and Discovery Deep. Bacteria were the major constituents of the microbial communities in both deeps, with the percentage ranging from 72% in ATL-BWI to 96.1% in ATL-LCL. The proportion of reads classified as Archaea varied between the different layers within and between the sampling locations, with the highest proportion being in ATL-BWI (23.4%) and the lowest in DIS-BWI (1.3%). No correlation was drawn between the relative proportion of Bacteria or Archaea with depth (both Spearman and Pearson correlations, P > 0.05).
Table 3.
Qualified reads, OTU counts, and alpha diversity estimates of microbial populationsa
| Sample ID |
Bacteria |
Archaea |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of qualified reads | OTU count | No. of observed species | Chao1 index | Shannon index | Simpson indexb | No. of qualified reads | OTU count | No. of observed species | Chao1 index | Shannon index | Simpson indexb | |
| DIS-NDW | 22,309 | 2,031 | 2,024.000 | 2,759.10 | 7.68 | 0.971 | 3,757 | 102 | 101.900 | 125.906 | 3.500 | 0.828 |
| DIS-BWI | 23,515 | 1,828 | 1,827.500 | 2,673.23 | 6.67 | 0.957 | 357 | 126 | 72.400 | 95.796 | 4.718 | 0.911 |
| DIS-LCL | 19,545 | 2,297 | 2,295.500 | 3,080.16 | 8.53 | 0.989 | 1,359 | 110 | 109.900 | 219.150 | 5.083 | 0.937 |
| ATL-NDW | 18,787 | 1,816 | 1,808.300 | 2,643.19 | 7.93 | 0.981 | 3,299 | 97 | 97.000 | 171.000 | 3.654 | 0.853 |
| ATL-BWI | 22,578 | 2,117 | 2,116.700 | 3,095.92 | 7.93 | 0.985 | 7,356 | 130 | 130.000 | 164.731 | 3.269 | 0.798 |
| ATL-UCL1 | 18,925 | 1,959 | 1,953.500 | 2,664.68 | 8.33 | 0.990 | 955 | 93 | 92.900 | 146.564 | 5.065 | 0.944 |
| ATL-UCL2 | 16,737 | 2,169 | 2,166.800 | 2,994.93 | 8.40 | 0.987 | 1,145 | 101 | 100.700 | 151.184 | 4.862 | 0.936 |
| ATL-LCL | 20,474 | 2,299 | 2,298.700 | 2,934.74 | 8.38 | 0.985 | 639 | 71 | 70.000 | 109.985 | 4.648 | 0.932 |
Samples were rarefied at the smallest libraries, and results were averaged over 100 repetitions. In Atlantis II, all metrics more or less agree that bacteria and archaea have higher diversity in transition zones and the brine layers than in normal deep water. In Discovery, the LCL was consistently the highest in diversity for bacteria and archaea, followed by NDW and then BWI (with some differences between species richness and diversity indices). The number of unclassified reads was 10,600.
The Simpson index used here is the Gini-Simpson index.
Bacterial rarefaction curves plateau or nearly plateau for all the alpha diversity metrics used: observed species, Chao1, Shannon index, and Simpson index (see Fig. S1a to d in the supplemental material). Overall, the curves reveal higher diversity in the deeper layers of the brine pools. Conversely, rarefaction curves for the archaeal amplicons suggest that more sequencing effort is required for most layers in both sites (see Fig. S1e to h). Likewise, though, the deepest layers harbor higher archaeal diversity than the brine interface or the overlaying deep-sea water. It is interesting that the samples collected from the brine layers were the highest in diversity according to both Shannon and Simpson indices for both bacterial and archaeal reads, indicating more evenness in the brines. The agreement between Shannon and Simpson indices and the Chao1 and observed-species metrics was further confirmed using Pearson's correlation (see Table S3 in the supplemental material).
Bacterial community structure revealed by 16S rRNA gene pyrosequencing.
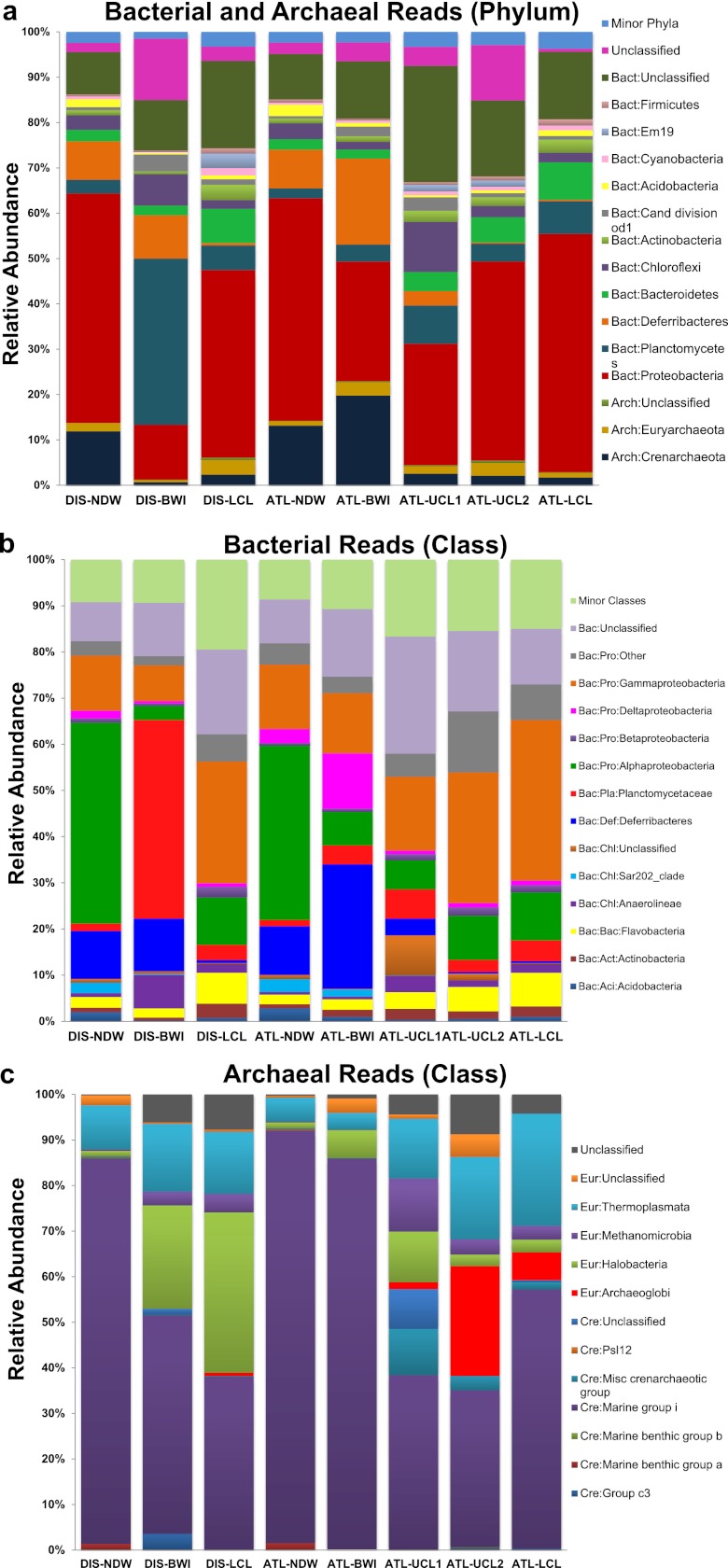
A high percentage of reads from the convective layers of both sites could not be assigned to any phylum, but 43 bacterial phyla were detected in the samples, of which four were ubiquitous in all layers in both locations (Fig. 2a). Proteobacteria was the most abundant phylum in all but DIS-BWI. Relative abundance estimations of the underlying classes reveal a changing relationship with depth. Alphaproteobacteria, dominated by the SAR11 clade, was the main class in deep-sea water, while Gammaproteobacteria, dominated by Alteromonadales and Oceanospirillales, was predominant in the lower convective layers (Fig. 2b).
Fig 2.
Taxonomic classification of the pyrosequencing reads using the RDP classifier at 80% confidence level. (a) Classification at the phylum level for total reads. (b) Class-level classification for bacterial reads. (c) Class-level classification for archaeal reads. Bact, Bacteria; Arch, Archaea; Pro, Proteobacteria; Pla, Planctomycetes; Def, Deferribacteres; Chl, Chloroflexi; Bac, Bacteroidetes; Act, Actinobacteria; Aci, Acidobacteria; Eur, Euryarchaeota; Cre, Crenarchaeota.
The SAR406 clade (phylum Deferribacteres) was abundant in the deep-sea water and brine interface samples but diminished in abundance almost entirely in the brine pools (see Fig. S2 in the supplemental material). On the other hand, the number of Bacteroidetes increased with depth (Fig. 2a). Careful inspection of over 53 affiliated minor genera indicated a situation where the bacteria were ubiquitous, not a dramatic depth-related hike (see Fig. S2, although minor groups are not shown). Two groups with high read numbers from the phylum Chloroflexi showed different vertical distributions. While one group associated with an uncultured member of the Anaerolineales was present in all of the sampled layers (prominent in DIS-BWI and ATL-UCL1 samples from the same depth); the SAR202 clade was exclusive to the deep-sea water and brine interface layers. Amplicons classified as “Candidatus Scalindua” and an unclassified “Candidatus Brocadiales” group (phylum Planctomycetes) were exclusively present in the two brine interfacial samples and ATL-UCL1 (see Fig. S2), albeit at a level 20 times higher in DIS-BWI. Other less abundant groups, such as W4 (order “Candidatus Brocadiales”), were exclusive to ATL-UCL1, while groups such as the PIR4 lineage, Planctomyces, and Rhodopirellula favored the convective layers of the brine pools (see Fig. S2).
Archaeal community structure as revealed by 16S rRNA gene pyrosequencing.
Archaea accounted for nearly 10% of the total read number. The ATL-BWI contained the highest archaeal proportion, followed by the deep-sea water samples, while DIS-BWI and all convective layers contained relatively low archaeal counts. Crenarchaeota and Euryarchaeota were the only two phyla found in both sites. Marine group I (MGI), which was the major group in Crenarchaeota, was abundant in the NDW and BWI samples but was dramatically lower in the brine pools (Fig. 2c). Diversity in the phylum Euryarchaeota was high because several minor groups were present, including “Candidatus Parvarchaeum,” the deep sea euryarchaeotic group (DSEG), deep sea hydrothermal vent group 6, Halobacteriales, marine group II, and marine group III. Thus, the euryarchaeotic groups appeared to favor the brine pool samples. Despite the low count, vertical stratification was clear in both sites (see Fig. S3 in the supplemental material).
Similarities between microbial communities: β diversity.
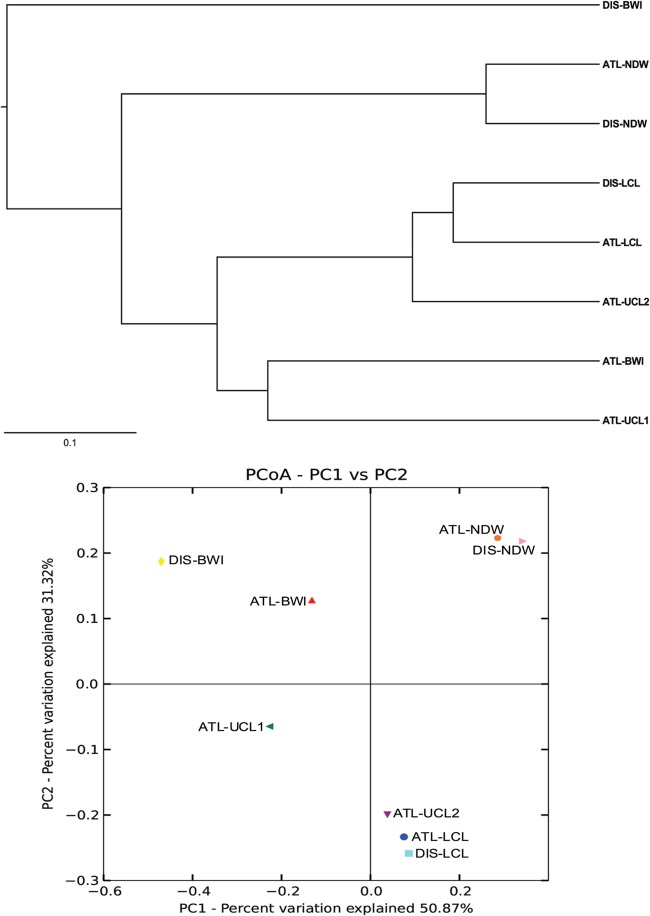
Overall, a clear separation between the deep-sea samples and the brine pool samples was evident, as demonstrated by the weighted-UniFrac UPGMA and PCoA (Fig. 3). The DIS-BWI sample is a notable exception that appears to be unique. Interestingly, within the brine subtree, the lowest convective layers show the most resemblance irrespective of the site (Fig. 3). ATL-UCL1 and ATL-BWI were clustered together, albeit with a large distance. Unweighted UniFrac, UPGMA, and PCoA show a clustering that is largely consistent with that obtained with weighted UniFrac but with longer nodes (see Fig. S4a and b in the supplemental material).
Fig 3.
Jackknife-supported UPGMA tree showing clusters of microbial communities based on weighted UniFrac with 100% support at all nodes (top), and the PCoA plot (bottom).
Comparison to previous work.
Previous work analyzed the metagenomes of samples collected in 2008 from a comparable depth (>2,100 m) in both brine pools (26). Extracted 16S rRNA gene fragments were taxonomically assigned, revealing abundant bacterial taxa shared between the brine pools. Some genera, such as Cupriavidus and Ralstonia, were abundant in both pools, while other genera, such as Acinetobacter, Mycobacterium, and Alkanindiges, were more abundant in Atlantis II than Discovery. No statistical analysis was provided to demonstrate the similarities or differences between the metagenomes.
In this study, we re-examined the metagenomes in a taxonomical framework and conducted a brief temporal comparison. Data were uploaded to the MG-RAST server for analysis, and 16S rRNA genes were downloaded and statistically analyzed using the STAMP package as described in Materials and Methods. We used ABP-Meta for the Atlantis II 2008 metagenome and DBP-Meta for the Discovery equivalent.
16S rRNA gene abundance profiles analyzed using principal component analysis separated the 2008 samples from all of the 2010 samples (see Fig. S5 in the supplemental material). Samples from 2008 share more phylotypes than samples collected 2 years later from similar depths (see Fig. S6 in the supplemental material). In fact, Cupriavidus and Ralstonia, which were abundant in the 2008 metagenomes, are absent in the 2010 samples (see Fig. S7 in the supplemental material). The 2008 and 2010 data correlate better with each other based on the year of sampling than based on the sampling location. Archaea were abundant in the Discovery 2008 metagenome (nearly 45%) but entirely absent in the Atlantis II counterpart (26). In the samples in the present study, Archaea were present in all layers at both sites in various proportions.
Relationship between microbial community and the environment: a statistical perspective.
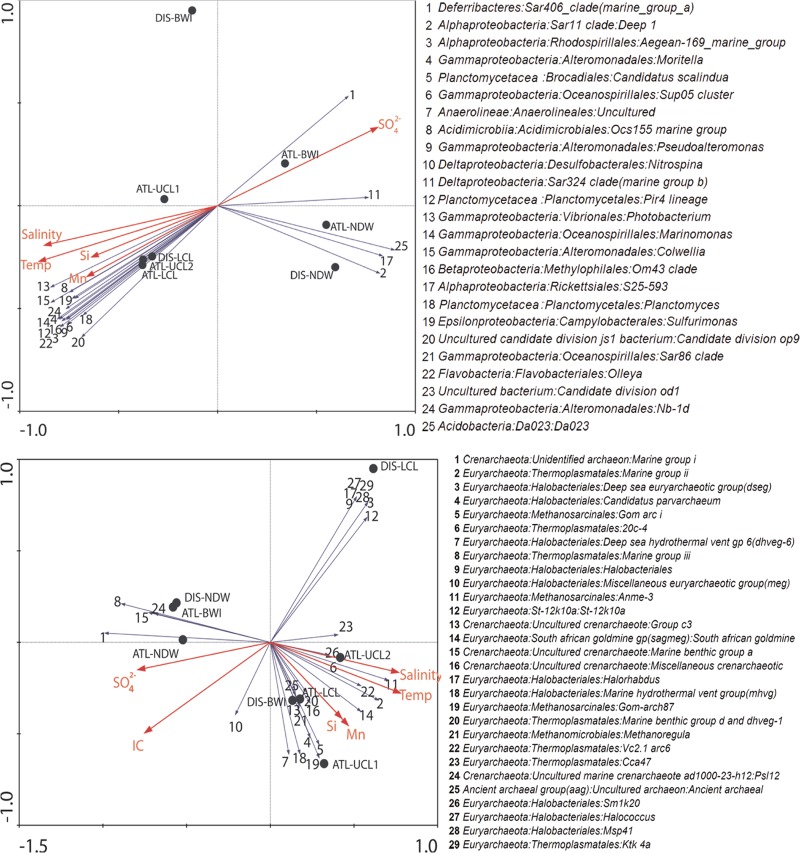
Detrended correspondence analysis (DCA) was initially performed to determine the nature of the underlying bacterial and archaeal genera response and the appropriate statistical model to examine the influence of the environmental conditions on the microbial community. The length of the longest estimated gradient indicated that a linear model is more suitable for the present data (for both bacteria and archaea). For the bacterial data, a combination of temperature, salinity, and SO42−, Si, and Mn concentrations produced the highest correlation with the first and second axes (see Table S4 in the supplemental material). Subsequently, redundancy analysis (RDA) was restricted to these environmental parameters.
Bacteria that thrive in the overlying deep-sea water, such as SAR406, SAR11, SAR423, S25-593 (Rickettsiales), and DA023 (Acidobacteria), showed a highly negative relationship with temperature and a positive correlation with SO42− concentration (Fig. 4). Conversely, several bacteria, including the Aegean 169 marine group, Moritella, “Candidatus Scalindua,” the SUP05 cluster, Nitrospina, Marinomonas, Planctomyces, and Sulfurimonas, showed a preference for the convective layers, and their presence correlated positively with temperature, salinity, and Si and Mn concentration (Fig. 4).
Fig 4.
RDA ordination plots showing relationship between statistically significant environmental factors with bacterial (top) and archaeal (bottom) communities. The numbers correspond to the genera in the keys at right and are ranked according to abundance. Environmental parameters were selected in a parsimonious manner using a DCA preassessment analysis whereby parameters are added in order of highest variation explained to saturation. Bacterial and archaeal genera were selected against an abundance threshold of 0.01% in at least one sample.
RDA analysis of the archaeal data revealed a similar set of environmental variables that statistically influenced the archaeal community. Colinearity between environmental variables was evident, further complicating efforts to identify a factor that truly influences archaeal composition in the brine pools. The first axis separated temperature, salinity, and Si and Mn concentrations from SO42− and TIC concentrations. Three main archaeal clusters were observed: (i) a group including MGI and MGIII correlated negatively with temperature but positively with SO42− and was prominent in the NDW samples and ATL-BWI; (ii) the second group, including DSEG, an unclassified Halobacteriales, Halorhabdus, and Halococcus, was dominant in DIS-LCL and correlated negatively with TIC but showed no correlation with other factors; and (iii) the remaining majority, including MGII, “Candidatus Parvarchaeum,” GOM arc I, and others, correlated with temperature, salinity, and Mn and Si concentrations (Fig. 4).
DISCUSSION
Physicochemical and geographical settings in Atlantis II and Discovery Deeps.
This study investigated and compared the microbial communities in the geographically adjacent Atlantis II and Discovery brine pools in the Red Sea by using 16S rRNA gene tag pyrosequencing technology. Our results shed some light on possible interactions between microbial community structures and environmental conditions in the following respects.
First, the hydrographic profiles show that little or no change has occurred in either brine pool over the last 15 years (7, 13, 15, 17). The highest temperatures recorded during the 2010 Aegaeo expedition were 68.3°C in ATL-LCL and 45°C in DIS-LCL. These temperatures are similar to those measured during the 2008 Oceanus expedition (26, 46). No new thermal stratification was observed in Atlantis II beyond the already-reported layers (47). In Discovery, a thin region within the DIS-BWI lower zone (2,038 to 2,044 m) showed a slower increase in temperature and salinity than the surrounding regions (Fig. 1b). This suggests that a new convective layer could be forming slowly. This new convective layer would be the equivalent of and at the same depth as ATL-UCL2, which is located immediately above ATL-LCL. Overall, the relatively stable conditions in both the Atlantis II and Discovery brine pools over the last 2 decades (from 1992 to 2010) may have allowed the microbial community to adapt and flourish.
Second, the upper boundary of ATL-LCL is at the same depth as that of DIS-LCL (∼2,057 m). This implies that the processes that formed the convective layers in one brine pool are somehow related to the other's, perhaps due to a historical physical connection (48). However, ATL-LCL was rich in iron, manganese, copper, and silica, while DIS-LCL had barely detectable levels of iron and low concentrations of manganese and silica. These significant differences are counterintuitive if we are to assume the existence of a connection between the lower zones of the brines.
Overall, over the last 15 years temperature and salinity have remained relatively stable in the stratified layers at both sites. Starting from the surface to the deep sea above the brine pools, the temperature profile is within the normal range (25), in which temperatures gradually drop to zero as depth increases. Here, we have unusual bodies of brines that reverse the trend, so that temperature increases with depth. The concentrations of heavy metals and nutrients varied from one layer to another and across sites. We therefore hypothesized that the microbial communities should exhibit (i) substantial differences within layers of the same site, (ii) significant similarities between equivalent layers across sites at the top of the brine, and (iii) dissimilarities between the lower layers of the brine pools.
Diversity, composition, and similarity of microbial communities.
Alpha diversity estimations revealed a higher diversity in the lower convective layers of the brine pools where cell count is lower (Table 2) than in the deep-sea or the transition zone samples (Table 3). In the brine pools, temperature (up to 68°C), salinity (up to 248 practical salinity units [PSU]), and pressure (20 MPa) are high, and survival pressure is in favor of halo- and thermophilic (49, 50) and perhaps even piezophilic or pressure-tolerant microbes (51–53). It is therefore rather surprising to observe a higher diversity here.
We observed an increase in the single- and double-OTU diversity along the depth (see Fig. S8 in the supplemental material). These low-abundance members of the “rare biosphere” explain the higher diversity in the lower convective layers of the brine pools (54). Conflicting conclusions about the source of these low-abundance populations have been reported in the literature. Some argue for a cosmopolitan distribution due to downward dispersion of highly abundant groups (55, 56), while others argue for a biogeographical distribution governed by physical boundaries and environmental gradients (57). In the extreme and oligotrophic conditions of the brine pools, microbes perhaps tune in to survival mode by activating stress responses and detoxification and repair mechanisms. Consequently, no single genus is able to outcompete and dominate successfully (58–61), hence the higher diversity and absence of highly abundant groups.
In normal deep water, where conditions are far less extreme, highly abundant groups did emerge as expected. The highest microbial count was observed in DIS-BWI (Table 2), but the estimated α-diversity was the lowest (Table 3). This is an indication of the presence of highly abundant or even specialized groups, such as Planctomycetes, thriving under favorable conditions. Gammaproteobacteria and several minor classes, including Flavobacteria, dominated the microbial communities in the convective layers (Fig. 2b).
Moritella is a genus of psychropiezophilic bacteria (62–64) that attained its highest relative abundance of associated reads in the lowest convective layers. To our knowledge, this is the first time this genus has been found under hydrothermal conditions, warranting further work to identify the members of this genus.
The SUP05 group of Gammaproteobacteria, which also had a higher relative abundance in the brine convective layers, are sulfur-oxidizing chemolithoautotrophic bacteria that have been found in hydrothermal vents and minimum-oxygen zones (65). A recent study suggested that SUP05 bacteria may depend heavily on H2 oxidation as an energy source (66). Furthermore, the metagenome of this cluster harbors a versatile repertoire of genes mediating autotrophic carbon assimilation, sulfur oxidation, and nitrate respiration (67, 68). This highlights the genomic plasticity of these bacteria and their ability to deploy versatile metabolic machinery to survive and adapt to the extreme conditions in the brine pools (66).
We put forward two alternative explanations to rationalize the low number of reads assigned to Archaea. (i) The primers used in the PCR amplification were a source of bias. The problem of primer bias is discussed below. (ii) The low archaeal count was in fact a true indicator of low archaeal presence in the presented samples.
Beta diversity analysis revealed a strong resemblance of the microbial communities from the normal deep water samples in both sites (Fig. 3). This is likely a result of the near-identical environmental conditions in the deep-sea water in both sites, due to water circulation between the two sites.
The type of abundant Bacteria and Archaea at the interface of the two deeps differed dramatically, despite the similarities observed in the deep-sea water above. The Discovery basin is 7.4 km southwest of the Atlantis II SW basin. The two brines are separated by a sill at a depth of around 1,990 m (45, 46). Our BWI samples were collected roughly 8 and 38 m below the sill from the Atlantis II Deep and the Discovery Deep, respectively. The observed temperature and salinity gradient within both interfaces may cause large shifts in the microbial community within short distances. It is therefore a challenge to compare the microbial community across sites on equal grounds. The microbial community in DIS-BWI bore the least resemblance to all other samples studied. Planctomycetes were uniquely abundant in this layer (Fig. 2a). Overall, the transition zone was characterized by steep changes in environmental conditions, which might have contributed to the drastic changes in the microbial communities observed within short distances.
In contrast, the microbial communities in the lower convective layers of both sites were surprisingly similar, given the striking differences in heavy metal contents and temperature between ATL-LCL and DIS-LCL. An existing physical connection is unlikely, unless fluid exchange between the brine pools could somehow allow only microbial passage and not that of heavy metals. The absence of dominant genera in both layers from both sites suggests that microbes are in survival mode in this anoxic, nutrient-limited environment. Another possible explanation is that microbial communities in the lower convective layers of both brine pools share an origin (when both brine pools were connected in the past) and have somehow been preserved via unknown mechanisms. Additionally, it is possible that the source of the microbial community in either of the brine pools is a bottom-up migration of microorganisms from the sediment into the brine water (27). The subsequent lateral and vertical distribution was probably governed by turbulent convective mixing in the uniform layers (reference 46 and references therein) and through vent injections across layers (69).
ATL-UCL2 is separated from ATL-LCL by an ∼5-m transition zone that was characterized by a sharp increase in temperature (57 to 68°C) and salinity (157 to 244 PSU), but our sampling depths from the two layers were 90 m apart. Furthermore, ATL-LCL showed the highest levels of heavy metals, which were significantly different from those in ATL-UCL2. The largely similar microbial communities in both brine layers under such drastic different environmental settings were unexpected and may be due to slow vertical diffusion or vent injection (69) or simply due to the fact that these two layers were once one body of brine that has been segmented into different layers by double diffusion (47). A common pool of microbes would have been separated in the formation of the brine layers.
The microbial community in ATL-UCL1 was different from the other convective layers, as demonstrated by weighted and unweighted UniFrac (Fig. 4). Indeed, a closer inspection revealed a transitional-type community where some phyla increased in abundance and others dropped, compared to ATL-BWI and ATL-UCL2. The transitional-type community in ATL-UCL1 may be attributed to the intermediacy of this layer in terms of environmental parameters between ATL-BWI and ATL-UCL2 and the ability of certain microbes to survive within a range of conditions.
Comparison to previous work.
Wang et al. (26) captured a snapshot of the microbial communities in the lowest layers of the brine pools. Those authors correlated functional data predicted from the metagenomes to the environment therein. They also provided a brief taxonomical overview of the microbial community, albeit one based on a limited number of extracted 16S rRNA gene fragments. When the present data were compared with the previous data, some differences were observed. We attribute these differences to the following possible reasons. (i) The depth in the 2008 sampling exercise could not be accurately determined. This uncertainty should not affect the conclusions made in the previous work. But for our purposes, sampling depth is an important factor, because we compared the vertical distribution of prokaryotes. (ii) The sampling locations from 2008 are 350 to 390 m away from the 2010 locations. Because of sampling drift, resampling the same spots was technically not feasible. (iii) Whole-genome amplification (WGA) was used by Wang et al. (26) to obtain sufficient quantities of DNA for pyrosequencing. WGA has been shown to introduce bias in metagenomic analysis, leading to erroneous abundance-based conclusions (70, 71). The extent of the bias cannot be predicted currently. (iv) Primer-based PCR that targets 16S rRNA genes is a source of bias in the present study. Here, the primers were assessed in silico using TestPrime 1.0 (http://www.arb-silva.de/search/testprime) against nonredundant Silva114 (coverage: Archaea, 88.1%, and Bacteria, 89.5%) and using PProspecter (72) against 97%-clustered representatives of the Greengenes 12.10 data set (coverage: Archaea, 94%, and Bacteria, 92%). Both tools estimated good coverage of both Archaea and Bacteria. Indeed, the in silico coverage of the primer pair used here was 92.7% (of 192 genes) and 79.5% (of 224 genes) for the Cupriavidus and Ralstonia (the abundant genera in the 2008 metagenomes) rRNA genes in the Silva114 database. Additionally, Archaea was detected in all of the current Atlantis II samples, as detailed in Results. (v) If we ignore bias, errors, and sampling and technical differences, the apparent shift in the communities was probably either the result of temporal shifts in the microbial populations or a manifestation of niche-specific communities. Regardless of the above, we aim to demonstrate the microbial stratification in these unique deep-sea brine pools.
Relationship of the microbial communities with the environment.
Given the variation in the environmental factors with depth, we hypothesized a strong causal relationship between the environment and harbored microbes. Temperature and salinity are the distinctive features of these brine pools and are expected to exert a strong selective pressure. Results from the multivariate analysis techniques (RDA analysis) for both Bacteria and Archaea highlight a clear separation of microbes inhabiting the normal deep water from those in the brine pools (Fig. 4). Temperature and salinity were identified as the major environmental factors affecting the microbial community in the brine layers where Bacteria such as the Aegean 169 marine group, Moritella, “Candidatus Scalindua,” the SUP05 cluster, Nitrospina, Marinomonas, Planctomyces, and Sulfurimonas were abundant. Indeed, these genera are extremophiles adapted to extreme and harsh environmental conditions. For instance, “Candidatus Scalindua” and other related anammox bacteria were previously found in oil fields with temperatures ranging from 55 to 75°C (73). The psychropiezophile Moritella profunda was found in regions of high pressure and cold temperatures (74), but a recent study demonstrated that the enzyme dihydrofolate reductase in Moritella profunda has subtle structural changes that allow it to remain active under high pressure and temperatures up to 80°C (75).
On the other hand, in the deep-sea water, SO42− concentration explained a significant proportion of the microbial variation, as it correlated positively with abundant genera. However, SO42− was highly negatively correlated with temperature and salinity (Fig. 4). Hence, the lower temperature and salinity could be the real factors promoting growth of bacterial and archaeal groups such as SAR406 and SAR11 Deep1, SAR423, S25-593, DA023, MGI, and MGIII in the normal deep water.
The decrease of sulfate concentrations in the lower convective layers may be partially explained by sulfate reduction. We found in our data that some of the OTUs were assigned to sulfate-reducing bacteria (SRB) from the orders Desulfarculales, Desulfobacterales, Desulfovibrionales, and Desulfuromonadales. In fact, a sulfate-reducing Desulfovibrio strain was successfully isolated from the brine-seawater interface (76). Furthermore, the detection of an anaerobic methanotrophic archaeal group in the lower brine layers also supports the possible existence of syntrophic sulfate reduction. Despite the preferential presence of the aforementioned SRB in the lower layers of the brine pool, their abundance was low.
We believe that sulfate was chiefly depleted as a result of geochemical processes. The mixing of calcium-rich hydrothermal fluids with sulfate-dissolved brine water generates anhydrite (CaSO4) that then precipitates to the bottom of the pool (77–79). An alternative scenario could be diffusion coupled with the salinity gradient of sulfate from the lower layers up to the upper layers of the brine and the deep-sea water (80, 81).
Previous work demonstrated that the SAR11 clade is by far the most abundant group in surface waters and the mesopelagic zone (82). Indeed, this group was the most abundant within Alphaproteobacteria in the deep-sea water (at depths over 1,800 m) in both sites. SAR11 is believed to play an important role in the mineralization of dissolved organic carbon (83, 84). Notably, the abundance of SAR11 in the deep water samples coincided with the depletion of TOC and an increased concentration of TIC with reported low O2 concentrations (Table 2) (7), yet their abundance greatly diminished at the interfaces and in the main body of the brine pools, where conditions are suboxic and anoxic, respectively (7, 46). Phosphorus is also an important limiting nutrient in the oceans (85–89). Phosphate uptake in oligotrophic zones has been attributed to SAR11 and the cyanobacterium Prochlorococcus (90). The higher concentration of phosphate in the Discovery Deep water may explain the higher abundance of SAR11 in those samples.
Unfortunately, O2 concentrations were not measured at the time of sampling, and so we cannot confidently comment on implications for the microbial community. However, previous studies did measure O2 levels (7, 46). The conditions in the water column changed from oxic in the deep-sea water to suboxic at the interface to anoxic in the brine layers, respectively. Therefore, the low presence of the SAR11 bacteria (aerobic) in the brine layers and interfaces, as opposed to their significant presence in the deep-sea water, could be as a result of anoxia in the brine pools. Also, the marine group A bacteria (SAR406 clade) were particularly abundant at the brine interface and in the deep-sea samples, especially in ATL-BWI (oxic/suboxic), but not in the brine convective layers (anoxic). This suggests that these bacteria are microaerophilic. On the other hand, the SUP05 bacteria were detected in all layers, but their concentrations nearly doubled as depth increased. SUP05 from hydrothermal vent plumes in the Guaymas Basin were shown to possess genes that enable it to use oxygen in oxic and microoxic conditions (66), which could mean that these bacteria are facultative. Overall, O2 levels in the water column may well be a significant factor in causing stratification. However, we could not confidently make this conclusion, as measurements were not available.
Nitrogen cycle.
The archaeal community in the deep-sea water of both sites was dominated primarily by the MGI which is recognized as an abundant archaeal group in meso- and bathypelagic zones (91). It is an active ammonia oxidizer (92) that can survive under various environmental and nutritional conditions (93). In ATL-BWI, MGI doubled in terms of relative abundance (double to triple in terms of read count). The concentration of ammonia in the normal deep water of both sites was very low (<0.009 mg/liter) in comparison with that of the brine waters. Similarly, a low ammonium concentration was observed in ATL-BWI (0.01 mg liter−1), where MGI was highly abundant, whereas ammonium concentration increased in DIS-BWI (1.13 mg liter−1) and MGI abundance was very low. The good negative correlation between ammonium concentration and MGI abundance suggests possible ammonium consumption by the microbes therein via oxidation (94, 95). This is in good agreement with the model that argues that archaea play a more critical role in nitrification and the marine nitrogen cycle than previously believed (95–98).
Anammox is an important reaction in the nitrogen cycle whereby ammonium is oxidized into dinitrogen gas in the absence of oxygen with nitrite as the electron acceptor (99, 100). The extent and interplay of ammonium and nitrate biotic depletion and possible renewal through diffusion from the brines' higher concentrations are unclear, given that the ammonium concentration in DIS-BWI was nearly equal to that of nitrate (1.13 mg liter−1 and 1.40 ± 0.30 mg liter−1, respectively). Untangling the complex substrate-intermediate-product relationship with the responsible microbes is rather challenging. Based on microbial abundance and nutrient concentration measurements, we suspect that “Candidatus Scalindua” competed well in DIS-BWI, reflected by its considerable presence with the near absence of the crenarchaeotic MGI, whereas in ATL-BWI, “Candidatus Scalindua” was outcompeted by the abundant MGI. However, we cannot confidently say that the differences in abundance of these two groups were a result of competition for resources under suitable environmental conditions, because the ammonium concentration in DIS-BWI was higher than that in ATL-BWI.
Concluding remarks.
This study investigated the variation in the microbial community in the Atlantis II and Discovery brine pools relative to their highly stratified environments. We found that the types and abundance levels of prokaryotes in the Red Sea deep water above the brine pools were very different from those within the layers of the brines and the intermediate interface regions. Alphaproteobacterial groups such as SAR11 and the crenarchaeotic MGI were the dominant microorganisms in the normal deep-sea water, whereas gammaproteobacterial and euryarchaeotic groups were more abundant in the brine lower layers. The members of these distinctive communities probably adapted and evolved due to the influence of the highly stratified environment in which they live. Ordination analysis indicates that temperature and salinity were the most likely influential factors affecting the microbes. These are the same factors that cause the stratification in the brine pools.
We also compared the results from both sites to investigate the interactions and relationships between the brine pools and the possible coevolution of the microorganisms therein. The communities in the deep-sea water shared the highest resemblance, followed by the communities in the lowest convective layers. Water flow between the sites and similar environmental conditions in the deep-sea water may explain the first instance; the large differences in temperature and heavy metals in the lower regions of the brine pools made for the opposite case in the second instance. We attribute these similarities to the common origin of these microbes predating the brine pools and the bottom-up and/or the top-down migration of microbes. The migrating low-abundance microbes may become abundant in the new environment. The harsh environment might also have played a role in suppressing competition and sustaining the observed diversity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yue Him Wong, Pok Yui Lai, and Feras Lafi from HKUST for their tremendous efforts in collecting the samples and taking the environmental measurements, and we are grateful to Raymond Schmitt from Woods Hole Institute of Oceanography for the high-range CTD data. We also thank Soumaya Belkharchouche for the English editing.
This study was supported by the National Basic Research Program of China (973 Program, no. 2012CB417304) and an award (SA-C0040/UK-C0016) from the King Abdullah University of Science and Technology to P. Y. Qian.
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00254-13.
REFERENCES
- 1. Swallow JC, Crease J. 1965. Hot salty water at the bottom of the Red Sea. Nature 205: 165– 166 [Google Scholar]
- 2. Craig H. 1966. Isotopic composition and origin of the Red Sea and Salton Sea geothermal brines. Science 154: 1544– 1547 [DOI] [PubMed] [Google Scholar]
- 3. Degens ET, Ross DA. ( ed). 1969. Hot brines and recent heavy metal deposits in the Red Sea. A geochemical and geophysical account. Springer-Verlag, New York, NY [Google Scholar]
- 4. Bäcker H, Schoell M. 1972. New deeps with brines and metalliferous sediments in the Red Sea. Nat. Phys. Sci. 240: 153– 158 [Google Scholar]
- 5. Pautot G, Guennoc P, Coutelle A, Lyberis N. 1984. Discovery of a large brine deep in the northern Red Sea. Nature 310: 133– 136 [Google Scholar]
- 6. Cochran JR, Martinez F, Steckler MS, Hobart MA. 1986. Conrad Deep: a new northern Red Sea deep. Origin and implications for continental rifting. Earth Planet. Sci. Lett. 78: 18– 32 [Google Scholar]
- 7. Schmidt M, Botz R, Faber E, Schmitt M, Poggenburg J, Garbe-Schönberg D, Stoffers P. 2003. High-resolution methane profiles across anoxic brine-seawater boundaries in the Atlantis-II, Discovery, and Kebrit Deeps (Red Sea). Chem. Geol. 200: 359– 375 [Google Scholar]
- 8. Reysenbach AL, Cady SL. 2001. Microbiology of ancient and modern hydrothermal systems. Trends Microbiol. 9: 79– 86 [DOI] [PubMed] [Google Scholar]
- 9. Gurvich EG. 2006. Metalliferous sediments of the world ocean: fundamental theory of deep-sea hydrothermal sedimentation. Springer-Verlag, New York, NY [Google Scholar]
- 10. Ramboz C, Danis M. 1990. Superheating in the Red Sea? The heat-mass balance of the Atlantis II Deep revisited. Earth Planet. Sci. Lett. 97: 190– 210 [Google Scholar]
- 11. Oudin E, Thisse Y, Ramboz C. 1984. Fluid inclusion and mineralogical evidence for high-temperature saline hydrothermal circulation in the Red Sea metalliferous sediments: preliminary results. Mar. Mining 5: 3– 31 [Google Scholar]
- 12. Miller AR, Densmore CD, Degens ET, Hathaway JC, Manheim FT, McFarlin PF, Pocklington R, Jokela A. 1966. Hot brines and recent iron deposits in deeps of the Red Sea. Geochim. Cosmochim. Acta 341: 351– 359 [Google Scholar]
- 13. Blanc G, Anschutz P. 1995. New stratification in the hydrothermal brine system of the Atlantis II Deep, Red Sea. Geology 23: 543– 546 [Google Scholar]
- 14. Winckler G, Kipfer R, Aeschbach-Hertig W, Botz R, Schmidt M, Schuler S, Bayer R. 2000. Sub sea floor boiling of Red Sea brines: new indication from noble gas data. Geochim. Cosmochim. Acta 64: 1567– 1575 [Google Scholar]
- 15. Hartmann M, Scholten JC, Stoffers P. 1998. Hydrographic structure of brine-filled deeps in the Red Sea: correction of Atlantis II Deep temperatures. Mar. Geol. 144: 331– 332 [Google Scholar]
- 16. Danielsson LG, Dyrssen D, Granéli A. 1980. Chemical investigations of Atlantis II and Discovery brines in the Red Sea. Geochim. Cosmochim. Acta 44: 2051– 2065 [Google Scholar]
- 17. Hartmann M, Scholten JC, Stoffers P, Wehner F. 1998. Hydrographic structure of brine-filled deeps in the Red Sea—new results from the Shaban, Kebrit, Atlantis II, and Discovery Deep. Mar. Geol. 144: 311– 330 [Google Scholar]
- 18. Javor B. 1989. Hypersaline environments: microbiology and biogeochemistry. Springer-Verlag, New York, NY [Google Scholar]
- 19. Horikoshi K, Grant WD. 1998. Extremophiles: microbial life in extreme environments. Wiley-Liss, New York, NY [Google Scholar]
- 20. Fiala G, Woese CR, Langworthy TA, Stetter KO. 1990. Flexistipes sinusarabici, a novel genus and species of eubacteria occurring in the Atlantis II Deep brines of the Red Sea. Arch. Microbiol. 154: 120– 126 [Google Scholar]
- 21. Huber R, Burggraf S, Mayer T, Barns SM, Rossnagel P, Stetter KO. 1995. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature 376: 57– 58 [DOI] [PubMed] [Google Scholar]
- 22. Ollivier B, Caumette P, Garcia JL, Mah RA. 1994. Anaerobic bacteria from hypersaline environments. Microbiol. Rev. 58: 27– 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eder W, Ludwig W, Huber R. 1999. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch. Microbiol. 172: 213– 218 [DOI] [PubMed] [Google Scholar]
- 24. Eder W, Jahnke LL, Schmidt M, Huber R. 2001. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 67: 3077– 3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qian PY, Wang Y, Lee OO, Lau SCK, Yang J, Lafi FF, Al-Suwailem A, Wong TYH. 2011. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J. 5: 507– 518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Yang J, Lee OO, Dash S, Lau SCK, Al-Suwailem A, Wong TYH, Danchin A, Qian PY. 2011. Hydrothermally generated aromatic compounds are consumed by bacteria colonizing in Atlantis II Deep of the Red Sea. ISME J. 5: 1652– 1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y, Yang JK, Lee OO, Li TG, Al-Suwailem A, Danchin A, Qian PY. 2011. Bacterial niche-specific genome expansion is coupled with highly frequent gene disruptions in deep-sea sediments. PLoS One 6: e29149 doi:10.1371/journal.pone.0029149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siam R, Mustafa GA, Sharaf H, Moustafa A, Ramadan AR, Antunes A, Bajic VB, Stingl U, Marsis NGR, Coolen MJL, Sogin M, Ferreira AJS, Dorry HE. 2012. Unique prokaryotic consortia in geochemically distinct sediments from Red Sea Atlantis II and Discovery Deep brine pools. PLoS One 7:e42872 doi:10.1371/journal.pone.0042872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fofonoff NP, Millard RC., Jr 1983. Algorithms for computation of fundamental properties of seawater. UNESCO Techn. Pap. Mar. Sci. 44:53 [Google Scholar]
- 30. Wang Y, Qian PY. 2009. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 4:e7401 doi:10.1371/journal.pone.0007401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, Qian PY. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 5:650– 664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pẽa AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7: 335– 336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7: 668– 669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 7537– 7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792– 1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Price MN, Dehal PS, Arkin AP. 2009. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26: 1641– 1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21: 494– 504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266– 267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188– 7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261– 5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453– 1454 [DOI] [PubMed] [Google Scholar]
- 42. Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246– 3248 [DOI] [PubMed] [Google Scholar]
- 43. ter Braak CJF. 2002. Program CANOCO version 4.5. Biometrics—quantitative methods in the life and earth sciences. Plant Research International, Wageningen University and Research Centre, Wageningen, the Netherlands [Google Scholar]
- 44. Lepš J, Šmilauer P. 2003. Multivariate analysis of ecological data using Canoco. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 45. Anschutz P, Blanc G. 1996. Heat and salt fluxes in the Atlantis II Deep (Red Sea). Earth Planet. Sci. Lett. 142: 147– 159 [Google Scholar]
- 46. Swift SA, Bower AS, Schmitt RW. 2012. Vertical, horizontal, and temporal changes in temperature in the Atlantis II and Discovery hot brine pools, Red Sea. Deep-Sea Res. Part I 64:118–128 [Google Scholar]
- 47. Anschutz P, Blanc G. 1995. Geochemical dynamics of the Atlantis II Deep (Red Sea): silica behavior. Mar. Geol. 128: 25– 36 [Google Scholar]
- 48. Turner JS. 1969. A physical interpretation of the observations of hot brine layers in the Red Sea, p 164–173 Degens ET, Ross DA. ( ed), Hot brines and recent heavy metal deposits in the Red Sea: a geochemical and geophysical account. Springer-Verlag, New York, NY [Google Scholar]
- 49. Ferrer M, Werner J, Chernikova TN, Bargiela R, Fernández L, La Cono V, Waldmann J, Teeling H, Golyshina OV, Glöckner FO, Yakimov MM, Golyshin PN. 2012. Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: a metagenomic study. Environ. Microbiol. 14: 268– 281 [DOI] [PubMed] [Google Scholar]
- 50. Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, Zhang CL. 2007. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ. Microbiol. 9: 2603– 2621 [DOI] [PubMed] [Google Scholar]
- 51. Meganathan R, Marquis RE. 1973. Loss of bacterial motility under pressure. Nature 246: 525– 527 [DOI] [PubMed] [Google Scholar]
- 52. Zobell CE, Cobet AB. 1962. Growth, reproduction, and death rates of Escherichia coli at increased hydrostatic pressures. J. Bacteriol. 84: 1228– 1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bartlett DH. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595: 367– 381 [DOI] [PubMed] [Google Scholar]
- 54. Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103: 12115– 12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459: 193– 199 [DOI] [PubMed] [Google Scholar]
- 56. Sjöstedt J, Koch-Schmidt P, Pontarp M, Canbäck B, Tunlid A, Lundberg P, Hagström Å, Riemann L. 2012. Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl. Environ. Microbiol. 78: 1361– 1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. 2009. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. U. S. A. 106: 22427– 22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK, Wu D, Robinson JM, Khouri HM, Eisen JA, Cary SC. 2009. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 5: e1000362 doi:10.1371/journal.pgen.1000362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boroujerdi AFB, Jones SS, Bearden DW. 2012. NMR analysis of metabolic responses to extreme conditions of the temperature-dependent coral pathogen Vibrio coralliilyticus. Lett. Appl. Microbiol. 54: 209– 216 [DOI] [PubMed] [Google Scholar]
- 60. Belnap CP, Pan C, Verberkmoes NC, Power ME, Samatova NF, Carver RL, Hettich RL, Banfield JF. 2010. Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 4: 520– 530 [DOI] [PubMed] [Google Scholar]
- 61. Morozova D, Wagner D. 2007. Stress response of methanogenic archaea from Siberian permafrost compared with methanogens from nonpermafrost habitats. FEMS Microbiol. Ecol. 61: 16– 25 [DOI] [PubMed] [Google Scholar]
- 62. Xu Y, Nogi Y, Kato C, Liang Z, Rüger HJ, De Kegel D, Glansdorff N. 2003. Moritella profunda sp. nov. and Moritella abyssi sp. nov., two psychropiezophilic organisms isolated from deep Atlantic sediments. Int. J. Syst. Evol. Microbiol. 53: 533– 538 [DOI] [PubMed] [Google Scholar]
- 63. Fang J, Zhang L, Bazylinski DA. 2010. Deep-sea piezosphere and piezophiles: geomicrobiology and biogeochemistry. Trends Microbiol. 18: 413– 422 [DOI] [PubMed] [Google Scholar]
- 64. Fang J, Kato C. 2010. Deep-sea piezophilic bacteria: geomicrobiology and biotechnology, p 47–77 In Jain SK, Khan AA, Rai MK. (ed), Geomicrobiology. Science Publishers, Enfield, NH [Google Scholar]
- 65. Sunamura M, Higashi Y, Miyako C, Ishibashi JI, Maruyama A. 2004. Two Bacteria phylotypes are predominant in the Suiyo Seamount hydrothermal plume. Appl. Environ. Microbiol. 70: 1190– 1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anantharaman K, Breier JA, Sheik CS, Dick GJ. 2013. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc. Natl. Acad. Sci. U. S. A. 110: 330– 335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, Tortell PD, Hallam SJ. 2009. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578– 582 [DOI] [PubMed] [Google Scholar]
- 68. Stewart FJ. 2011. Dissimilatory sulfur cycling in oxygen minimum zones: an emerging metagenomics perspective. Biochem. Soc. Trans. 39: 1859– 1863 [DOI] [PubMed] [Google Scholar]
- 69. Anschutz P, Turner JS, Blanc G. 1998. The development of layering, fluxes through double-diffusive interfaces, and location of hydrothermal sources of brines in the Atlantis II Deep: Red Sea. J. Geophys. Res. C Oceans 103: 27809– 27819 [Google Scholar]
- 70. Abbai NS, Govender A, Shaik R, Pillay B. 2012. Pyrosequence analysis of unamplified and whole genome amplified DNA from hydrocarbon-contaminated groundwater. Mol. Biotechnol. 50: 39– 48 [DOI] [PubMed] [Google Scholar]
- 71. Pinard R, de Winter A, Sarkis GJ, Gerstein MB, Tartaro KR, Plant RN, Egholm M, Rothberg JM, Leamon JH. 2006. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 7: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. 2011. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27:1159– 1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li H, Chen S, Mu BZ, Gu JD. 2010. Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb. Ecol. 60: 771– 783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Evans RM, Behiry EM, Tey LH, Guo J, Loveridge EJ, Allemann RK. 2010. Catalysis by dihydrofolate reductase from the psychropiezophile Moritella profunda. Chembiochem 11: 2010– 2017 [DOI] [PubMed] [Google Scholar]
- 75. Ohmae E, Murakami C, Tate SI, Gekko K, Hata K, Akasaka K, Kato C. 2012. Pressure dependence of activity and stability of dihydrofolate reductases of the deep-sea bacterium Moritella profunda and Escherichia coli. Biochim. Biophys. Acta 1824: 511– 519 [DOI] [PubMed] [Google Scholar]
- 76. Trüper HG. 1969. Bacterial sulfate reduction in the Red Sea hot brines, p 263–271 In Degens ET, Ross DA. (ed), Hot brines and recent heavy metal deposits in the Red Sea: a geochemical and geophysical account. Springer-Verlag, New York, NY [Google Scholar]
- 77. Anschutz P, Blanc G, Monnin C, Boulègue J. 2000. Geochemical dynamics of the Atlantis II Deep (Red Sea): II. Composition of metalliferous sediment pore waters. Geochim. Cosmochim. Acta 64: 3995– 4006 [Google Scholar]
- 78. Hartmann M, Lohmann L. 1968. Untersuchungen an der heißen Salzlauge und am Sediment des Atlantis-II-Tiefs im Roten Meer, p 13–20 In “Meteor” Forschungsergebnisse: Reihe C. Geologie und Geophysik. Gebrüder Borntraeger, Berlin, Germany [Google Scholar]
- 79. Zhabina NN, Sokolov VS. 1982. Sulphur compounds in the sediments of the Atlantis-II Deep, Red Sea. Mar. Geol. 50: 129– 141 [Google Scholar]
- 80. Monnin C, Ramboz C. 1996. The anhydrite saturation index of the ponded brines and sediment pore waters of the Red Sea deeps. Chem. Geol. 127: 141– 159 [Google Scholar]
- 81. Lasaga AC. 1981. Influence of diffusion coupling on diagenetic concentration profiles. Am. J. Sci. 281: 553– 575 [Google Scholar]
- 82. Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806– 810 [DOI] [PubMed] [Google Scholar]
- 83. Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE, Landry ZC, Giovannoni SJ. 2011. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6: e23973 doi:10.1371/journal.pone.0023973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Laghdass M, Catala P, Caparros J, Oriol L, Lebaron P, Obernosterer I. 2012. High contribution of SAR11 to microbial activity in the northwest Mediterranean Sea. Microb. Ecol. 63:324– 333 [DOI] [PubMed] [Google Scholar]
- 85. Zehr JP, Kudela RM. 2011. Nitrogen cycle of the open ocean: from genes to ecosystems. Annu. Rev. Mar. Sci. 3: 197– 225 [DOI] [PubMed] [Google Scholar]
- 86. Smith SV. 1984. Phosphorus versus nitrogen limitation in the marine environment. Limnol. Oceanogr. 29: 1149– 1160 [Google Scholar]
- 87. Downing JA. 1997. Marine nitrogen: phosphorus stoichiometry and the global N:P cycle. Biogeochemistry 37: 237– 252 [Google Scholar]
- 88. Tyrrell T. 1999. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 400: 525– 531 [Google Scholar]
- 89. Sãudo-Wilhelmy SA, Kustka AB, Gobler CJ, Hutchins DA, Yang M, Lwiza K, Burns J, Capone DG, Raven JA, Carpenter EJ. 2001. Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature 411: 66– 69 [DOI] [PubMed] [Google Scholar]
- 90. Zubkov MV, Mary I, Woodward EMS, Warwick PE, Fuchs BM, Scanlan DJ, Burkill PH. 2007. Microbial control of phosphate in the nutrient-depleted North Atlantic subtropical gyre. Environ. Microbiol. 9: 2079– 2089 [DOI] [PubMed] [Google Scholar]
- 91. Karner MB, DeLong EF, Karl DM. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507– 510 [DOI] [PubMed] [Google Scholar]
- 92. Santoro AE, Casciotti KL, Francis CA. 2010. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12: 1989– 2006 [DOI] [PubMed] [Google Scholar]
- 93. Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. 2009. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33: 855– 869 [DOI] [PubMed] [Google Scholar]
- 94. Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543– 546 [DOI] [PubMed] [Google Scholar]
- 95. Bouskill NJ, Eveillard D, Chien D, Jayakumar A, Ward BB. 2012. Environmental factors determining ammonia-oxidizing organism distribution and diversity in marine environments. Environ. Microbiol. 14: 714– 729 [DOI] [PubMed] [Google Scholar]
- 96. Martens-Habbena W, Berube PM, Urakawa H, De La Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976– 979 [DOI] [PubMed] [Google Scholar]
- 97. Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C. 2012. Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ. Microbiol. 14: 909– 923 [DOI] [PubMed] [Google Scholar]
- 98. Zhang LM, Hu HW, Shen JP, He JZ. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6: 1032– 1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kuenen JG. 2008. Anammox bacteria: from discovery to application. Nat. Rev. Microbiol. 6: 320– 326 [DOI] [PubMed] [Google Scholar]
- 100. Mulder A, van de Graaf AA, Robertson LA, Kuenen JG. 1995. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 16: 177– 183 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.