Abstract
The mountain pine beetle, Dendroctonus ponderosae, is a subcortical herbivore native to western North America that can kill healthy conifers by overcoming host tree defenses, which consist largely of high terpene concentrations. The mechanisms by which these beetles contend with toxic compounds are not well understood. Here, we explore a component of the hypothesis that beetle-associated bacterial symbionts contribute to the ability of D. ponderosae to overcome tree defenses by assisting with terpene detoxification. Such symbionts may facilitate host tree transitions during range expansions currently being driven by climate change. For example, this insect has recently breached the historical geophysical barrier of the Canadian Rocky Mountains, providing access to näive tree hosts and unprecedented connectivity to eastern forests. We use culture-independent techniques to describe the bacterial community associated with D. ponderosae beetles and their galleries from their historical host, Pinus contorta, and their more recent host, hybrid P. contorta-Pinus banksiana. We show that these communities are enriched with genes involved in terpene degradation compared with other plant biomass-processing microbial communities. These pine beetle microbial communities are dominated by members of the genera Pseudomonas, Rahnella, Serratia, and Burkholderia, and the majority of genes involved in terpene degradation belong to these genera. Our work provides the first metagenome of bacterial communities associated with a bark beetle and is consistent with a potential microbial contribution to detoxification of tree defenses needed to survive the subcortical environment.
INTRODUCTION
In addition to the well-established roles of pairwise symbioses, the importance of multipartite associations and microbial communities to multicellular organisms is becoming increasingly recognized (1–6). Despite this, our understanding of how symbiotic relationships contribute to large-scale processes, such as ecosystem dynamics and the structure and functioning of biomes, remains poorly understood. Insects serve as a particularly useful model for exploring these cross-scale interactions because of their widespread and diverse associations with microbiota, roles in driving ecosystem processes, influences on human socioeconomic values, rapid evolutionary adaptations, and shifting responses to anthropogenic forces, such as climate change, species invasions, and habitat alteration.
Bark beetles (Curculionidae: Scolytinae) have the ability to overcome host tree defenses and thus colonize and kill healthy conifers. Several species undergo intermittent landscape-scale outbreaks, which appear to be increasing in frequency, magnitude, and interspecific confluence as a result of a warming climate and habitat conversions. For example, bark beetles caused substantial mortality across 47 million acres of conifers in western North America from 1997 to 2007, and the ongoing mountain pine beetle (Dendroctonus ponderosae Hopkins) outbreak is predicted to deplete 1 trillion cubic meters of pine in British Columbia, Canada, alone by 2014 (7).
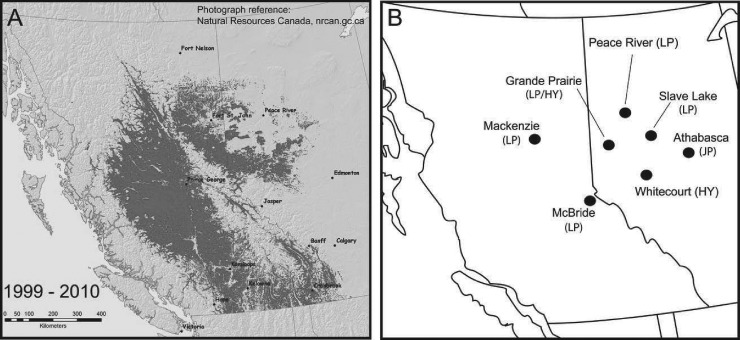
The historical range of D. ponderosae extends from northern Mexico to southern British Columbia and inland to western North Dakota in the United States and the Rocky Mountains in Canada (8). Its preferred host is lodgepole pine, Pinus contorta (Douglas ex. Loud.), which occurs throughout this range. As conditions have warmed, D. ponderosae has expanded to higher latitudes (9–11) and breached the geophysical barrier of the Canadian Rocky Mountains (12) (Fig. 1). It has now colonized the hybrid crossings of P. contorta and jack pine, Pinus banksiana Lamb., and also the pure P. banksiana trees in this region (13, 14). These trees are contiguous with P. banksiana forests throughout all of southern Canada east of the Rocky Mountains, connecting with eastern white pine, Pinus strobus L., and red pine, Pinus resinosa Sol. Ex Aiton, further to the east (15). These beetles therefore potentially threaten pines across North America.
Fig 1.
Outbreak and spread of D. ponderosae in western Canada from 1999 to 2010 (A) and sampling locations of beetles and galleries from lodgepole (LP) and hybrid (HY) pines (B). (The map in panel A is a copy of an official work published by Natural Resources Canada but has not been produced in affiliation with, or with the endorsement of, the Government of Canada.)
An important feature of bark beetles is their reliance on symbiotic microbes, which likely assist with host colonization and utilization. Symbiotic relationships with fungi have been studied for several decades (4, 16, 17) and are known to contribute to larval nutrition. Additionally, some bacterial symbionts have been implicated in defense against antagonistic fungi (18–20) and in nutrient acquisition (21). Moreover, beetle-associated microbes have demonstrated community structures that reflect both host beetle and geographic zones (22).
Both bacterium-beetle and bacterium-fungus-beetle relationships are mediated by host tree chemistry (18, 23). Pines synthesize terpenoids that are toxic at high concentrations to a broad range of insects, including bark beetles and their symbiotic fungi (24). These compounds are present in constitutive resin and phloem and are synthesized and translocated in response to the early stages of beetle-microbe attack. Monoterpenes can kill or repel bark beetles, and both monoterpene and diterpene acids can inhibit fungal symbionts (25–27). Terpenoid-based defenses of healthy trees confront beetles with a significant barrier during periods when their populations are low, and thus colonization is restricted to highly stressed hosts. However, several species, including D. ponderosae, can exhaust tree defenses through pheromone-mediated mass attack when population densities are high (7, 28).
An important question emerging from the current climate-driven range expansion of these beetles is how they will perform in new tree species and whether they will persist in a relatively endemic state or engage in self-driving outbreaks (29–31). Here, we focus on the potential role that D. ponderosae-associated bacterial communities may play during the colonization of both lodgepole and hybrid pines. We first describe how we obtained and examined samples of beetles, galleries, and trees to broadly identify similarities and differences between the communities associated with these environments using denaturing gradient gel electrophoresis (DGGE). Then, using a community metagenomic approach, we provide a detailed analysis of the bacterial communities associated with D. ponderosae and their galleries in both native and hybrid tree hosts. Finally, we explore the hypothesis that bacteria play a role in the detoxification of host tree defenses by specifically analyzing genes involved in terpene degradation encoded by these bacteria.
MATERIALS AND METHODS
Sample collection.
We obtained samples of D. ponderosae adults, their galleries, and unattacked tree phloem from four sites in 2010 across a gradient ranging from historical to naïve tree species of P. contorta (Mackenzie, British Columbia, Canada, and Grande Prairie, Peace River, and Slave Lake, Alberta, Canada), hybrid P. contorta-P. banksiana (Whitecourt, Alberta), and P. banksiana (Athabasca, Alberta) (Table 1; Fig. 1). For DGGE analysis, 291 beetle, gallery, and unattacked tree samples were obtained from multiple sites in Alberta and British Columbia, and for the community metagenomes, sampling consisted of approximately 300 adult beetles and 150 galleries from 20 trees from pines located at each of two sites in July of 2009. One site was located near McBride, British Columbia, in a P. contorta stand, and the other was near Grande Prairie, Alberta, in a mixed P. contorta and hybrid P. contorta-P. banksiana stand (32) (Table 1; Fig. 1). At the time of sampling, D. ponderosae beetles were not observed colonizing P. banksiana.
Table 1.
Site location and sample sizes of the collection of D. ponderosae, galleries, and phloem of unattacked trees
| Site location(s) | Host | Sample type | Sample size | Method of community analysis |
|---|---|---|---|---|
| Mackenzie, British Columbia | P. contorta | D. ponderosae | 26 | DGGE |
| Gallery | 26 | |||
| Phloem | 7 | |||
| McBride, British Columbia | P. contorta | D. ponderosae | 300 | Community metagenomics |
| Gallery | 150 | |||
| Grand Prairie, Peace River, and Slave Lake, Alberta | P. contorta | D. ponderosae | 52 | DGGE |
| Gallery | 15 | |||
| Phloem | 8 | |||
| Whitecourt, Alberta | P. contorta-P. banksiana hybrid | D. ponderosae | 41 | DGGE |
| Gallery | 13 | |||
| Phloem | 24 | |||
| Grand Prairie, Alberta | P. contorta-P. banksiana hybrid | D. ponderosae | 300 | Community metagenomics |
| Gallery | 150 | |||
| Athabasca, Alberta | P. banksiana | D. ponderosae | 53 | DGGE |
| Gallery | 8 | |||
| Phloem | 18 |
For both analyses, D. ponderosae adults and 0.5- by 2-cm sections of phloem that included egg galleries and frass were removed from recently attacked trees and placed on ice before being transported to the laboratory at the University of Wisconsin—Madison. Phloem tissue from trees with no visible signs of bark beetle attack or disease was also sampled for DGGE analysis, with a maximum of three beetles and galleries and two phloem samples from a single tree and no samples from the same gallery (see the supplemental material). Samples for the community metagenomes were collected in the same way and pooled after collection for a total of four samples representing beetles and galleries from P. contorta and hybrid P. contorta-P. banksiana stands.
Community metagenome sequencing and analysis.
Total bacterial genomic DNA was extracted from all four pooled samples. Eukaryotic material was removed through a series of washes, as previously described (33). Briefly, materials were ground with a mortar and pestle and washed in 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) containing 0.1% Tween. Each sample was shaken for 15 min at 150 rpm and centrifuged for 5 min at 500 rpm. This resulted in a 3-layer mixture containing (from the top) bacterial cells, fungal material, and insect material. The bacterial layer was removed and subjected to this process two more times. The final bacterial suspensions were centrifuged for 30 min at 4,100 rpm, resuspended in 1× PBS containing 0.1% Tween and filtered through a 100-μm-pore filter. Total DNA was then extracted using a Qiagen DNeasy plant maxikit (Qiagen Sciences, Germantown, MD).
Community metagenomes were subsequently generated from the extracted DNA using Roche 454 Titanium pyrosequencing (34). These reads were assembled using Newbler v.2.1 with default parameters, and phylogenetic binning of all contigs and singleton reads of the resulting assemblies was performed using BLASTN (35) and PhymmBL (36). All contigs and singletons were first compared to a reference data set containing all completely sequenced bacterial and archaeal genomes available in NCBI as of 1 January 2012. All contigs having BLASTN hits with E values of <1e−10 were classified according to their best hit, while all other contigs were subsequently classified using PhymmBL (36). Contigs with no BLASTN matches that were classified by PhymmBL with confidence scores of <50 were considered unclassified. Proteins were predicted from these metagenomes using the IMG-ER pipeline (37).
Predicted proteins putatively involved in terpene degradation were identified from each metagenome using either the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server (KAAS) (38) or BLASTP (35) against a custom data set of proteins belonging to the dit diterpene acid degradation pathway. Specifically, genes encoding enzymes putatively involved in monoterpene degradation, as defined in the KEGG limonene and pinene degradation pathway (ko00903), were annotated and retained. For the diterpene acid degradation pathway, predicted proteins in the metagenomes were compared to the proteins comprising the dit gene cluster, as described for Pseudomonas abietaniphila BKME-9 (39), using BLASTP (E value of 1e−5). To identify if terpene-degrading genes in the beetle-associated communities were enriched compared to other plant biomass-degrading communities, we performed the same KAAS- and BLASTP-based annotations on publicly available metagenomes associated with the termite hindgut (40), wallaby foregut (41), cow rumen (42), panda gut (43), leaf-cutter ant fungus garden (44), switchgrass compost (45), and poplar biomass bioreactor (46). Using the raw number of proteins annotated in the KEGG limonene and pinene degradation pathway and the dit operon, and the total number of proteins encoded in these metagenomes, we performed Fisher's exact test to identify potential enrichment of these genes in the beetle-associated metagenomes (P < 0.05). Fisher's exact test takes into account the total number of proteins in the data sets being compared and is therefore appropriate for identification of relative changes in coding potential. The BLASTN- and PhymmBL-based phylogenetic binning results were then used to identify the taxonomic origin of the contigs and singletons encoding these proteins.
Nucleotide sequence accession numbers.
Raw pyrosequencing data for the community metagenomes have been deposited in the National Center for Biotechnological Information's (NCBI) short read archive under accession no. SRA4088237, SRA4088238, SRA4088239, and SRA4088241. Assembled community metagenomes are on the Joint Genome Institute's Integrated Microbial Genomes/Microbiomes (IMG/M) database (47) under project identification (ID) no. 2032320008, 2032320009, 2029527007, and 2035918003. Sequences of all DGGE bands obtained in this study are available in GenBank under accession no. JF810915 to JF810926.
RESULTS
Microbial community analysis.
Principle component analysis of the DGGE data revealed no distinct clustering among beetle, gallery, or tree samples from either location or tree species (see Fig. S1 in the supplemental material). We sequenced bands corresponding to the 12 most commonly identified operational taxonomic units (OTU) in our DGGE analyses and found that 10 were most similar to Gammaproteobacteria, 1 to Betaproteobacteria, and 1 to Actinobacteria (see Table S1 in the supplemental material).
Community metagenome sequencing and phylogenetic binning.
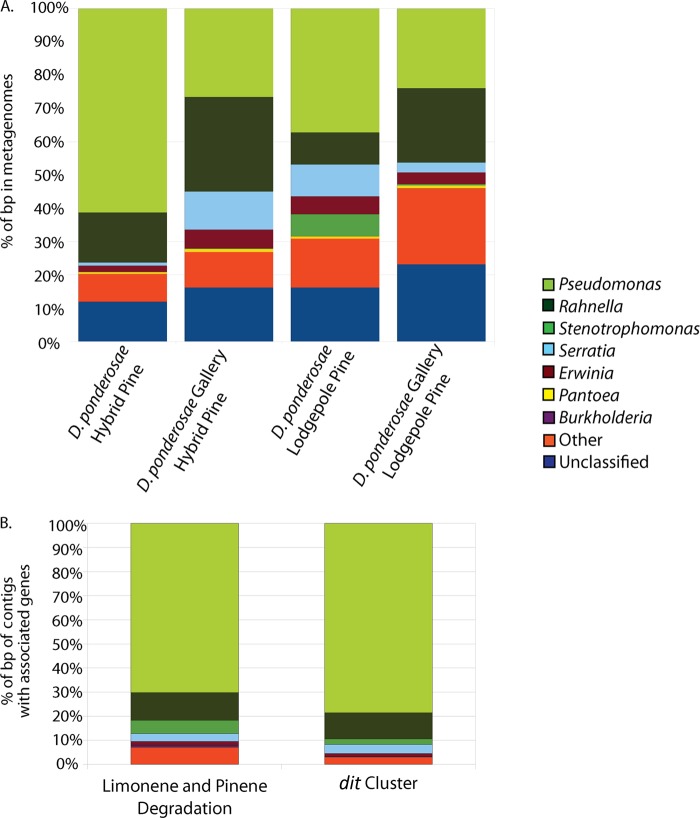
The four metagenomes generated using shotgun 454 pyrosequencing comprised 27.1 to 58.8 Mbp of sequence after assembly (Table 2). These were constructed from (i) whole D. ponderosae beetles from infested P. contorta, (ii) gallery material from infested P. contorta, (iii) whole D. ponderosae beetles from infested hybrid P. contorta-P. banksiana, and (iv) gallery material from infested hybrid P. contorta-P. banksiana. A phylogenetic binning analysis revealed similar taxonomic patterns across all four community metagenomes. Broadly, Gammaproteobacteria comprised the majority of all sequences in each sample, although Betaproteobacteria, Alphaproteobacteria, Firmicutes, and Actinobacteria were also represented (see Fig. S3 in the supplemental material). Analysis at the genus level identified abundant sequences matching those of Pseudomonas, Rahnella, Serratia, Erwinia, Stenotrophomonas, and Pantoea (Fig. 2A; for details, see Data Set S1 in the supplemental material).
Table 2.
Summary of sequencing statistics for the four D. ponderosae-associated metagenomes
| Parameter | Result for metagenome |
|||
|---|---|---|---|---|
| Hybrid pine |
Lodgepole pine |
|||
| D. ponderosae | D. ponderosae gallery | D. ponderosae | D. ponderosae gallery | |
| Size of assembly (Mbp) | 29 | 30.2 | 61.3 | 32.7 |
| No. of protein-coding genes | 42,426 | 53,181 | 86,776 | 66,202 |
| No. of contigs | 9,161 | 9,616 | 20,178 | 10,665 |
| No. of singleton reads | 24,220 | 47,597 | 56,588 | 60,200 |
| N50 contig size (bp) | 1,986 | 606 | 1,535 | 482 |
| Largest contig (bp) | 142,661 | 59,558 | 140,467 | 47,283 |
Fig 2.
Results of genus-level phylogenetic binning of the four beetle-associated community metagenome assemblies (A). Phylogenetic binning of only contigs containing genes annotated in the KEGG limonene and pinene degradation pathway or dit cluster of Pseudomonas abietaniphila BKME-9 (B). Only those genera with greater than 1% representation are shown.
Analysis of genes putatively involved in terpene degradation.
We performed a metabolic reconstruction analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (48) (Table 3). An analysis of proteins involved in the limonene and pinene degradation pathway yielded between 90 and 198 proteins in each community metagenome (Table 3; see Data Set S2 in the supplemental material). This comprises 0.17 to 0.27% of the predicted proteins in these data sets. Of the 20 enzymes comprising this pathway, 5 were found to be enriched in the pine beetle metagenomes compared to metagenomes of other plant biomass-associated microbial communities (Table 3; P < 0.05, Fisher's exact test). These included an aldehyde dehydrogenase (EC 1.2.1.3), an oxidoreductase (EC 1.14.13.-), an enoyl-coenzyme A (CoA) hydratase (EC 4.2.1.17), and two 3-hydroxyacyl-CoA epimerases (EC 1.1.1.35, 4.2.1.17, 5.1.2.3, and 5.3.3.8). All enzymes involved in the conversion of α-pinene to 3-isopropylbut-3-enoic acid, myrtenic acid, or pinocarvone, as described in KEGG, were detected. The beetle metagenome from the hybrid pine contained the highest proportion of potential terpene degradation enzymes (113 proteins, 0.27% of proteins), while the gallery metagenome from the hybrid pine contained the smallest (90 proteins, 0.17% of proteins).
Table 3.
Identification of genes involved in the KEGG limonene and pinene degradation pathway within the four mountain pine beetle metagenomes compared to a combined data set of publicly available metagenomesa
| Product annotation | EC no. | KEGG Orthology (KO) identifier | No. of genes |
||||
|---|---|---|---|---|---|---|---|
| Hybrid pine |
Lodgepole pine |
Plant biomass-degrading communities combined | |||||
| D. ponderosae | D. ponderosae gallery | D. ponderosae | D. ponderosae gallery | ||||
| Oxidoreductases | 1.1.-.- | K00120 | 0 | 0 | 0 | 0 | 62 |
| Aldehyde dehydrogenase | 1.2.1.3 | K00128 | 43 | 37 | 91 | 80 | 1,485 |
| Oxidoreductases | 1.2.1.- | K00155 | 0 | 0 | 1 | 1 | 23 |
| Oxidoreductases | 1.14.13.- | K00492 | 12 | 7 | 17 | 13 | 96 |
| Oxidoreductases | 1.14.-.- | K00517 | 4 | 2 | 6 | 5 | 378 |
| Acyltransferases with transferring groups other than amino-acyl groups | 2.3.1.- | K00680 | 1 | 0 | 3 | 0 | 271 |
| Hydrolases acting on ester bonds | 3.1.2.- | K01076 | 0 | 0 | 0 | 0 | 25 |
| Enoyl-CoA hydratase | 4.2.1.17 | K01692 | 32 | 19 | 51 | 34 | 768 |
| Carbon-oxygen lyases | 4.2.1.- | K01726 | 0 | 0 | 0 | 0 | 0 |
| 3-Hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase/3-hydroxybutyryl-CoA epimerase | 1.1.1.35; 4.2.1.17; 5.1.2.3 | K01782 | 10 | 16 | 15 | 15 | 182 |
| 3-Hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase/3-hydroxybutyryl-CoA epimerase/enoyl-CoA isomerase | 1.1.1.35; 4.2.1.17; 5.1.2.3; 5.3.3.8 | K01825 | 10 | 8 | 14 | 18 | 56 |
| Ligases forming carbon-sulfur bonds | 6.2.1.- | K01913 | 0 | 0 | 0 | 0 | 10 |
| (S)-Limonene 6-monooxygenase | 1.14.13.48 | K07381 | 0 | 0 | 0 | 0 | 0 |
| (S)-Limonene 7-monooxygenase | 1.14.13.49 | K07382 | 0 | 0 | 0 | 0 | 0 |
| Limonene 1,2-epoxide hydrolase | 3.3.2.8 | K10533 | 0 | 0 | 0 | 0 | 0 |
| trans-Carveol dehydrogenase | 1.1.1.275 | K12466 | 0 | 0 | 0 | 0 | 0 |
| trans-Carveol dehydrogenase | 1.1.1.243 | K14730 | 0 | 0 | 0 | 0 | 0 |
| Monoterpene epsilon-lactone hydrolase | 3.1.1.83 | K14731 | 0 | 0 | 0 | 0 | 0 |
| Limonene hydroxylase | 1.14.13.48; 1.14.13.49; 1.1.1.243; 1.1.1.144 | K14732 | 0 | 0 | 0 | 0 | 0 |
| Limonene 1,2-monooxygenase | 1.14.13.107 | K14733 | 1 | 1 | 0 | 0 | 49 |
| Total no. of proteins | 42,427 | 53,182 | 86,784 | 66,204 | 4,241,298 | ||
The publicly available metagenomes consist of plant biomass-degrading microbial communities associated with the termite hindgut, wallaby foregut, cow rumen, panda gut, leaf-cutter ant fungus garden, switchgrass compost community, and poplar biomass bioreactor (for details see Materials and Methods and Data Set S2 in the supplemental material). Results representing genes enriched relative to other plant biomass-degrading metagenomes are highlighted in boldface (P < 0.05, Fisher's exact test).
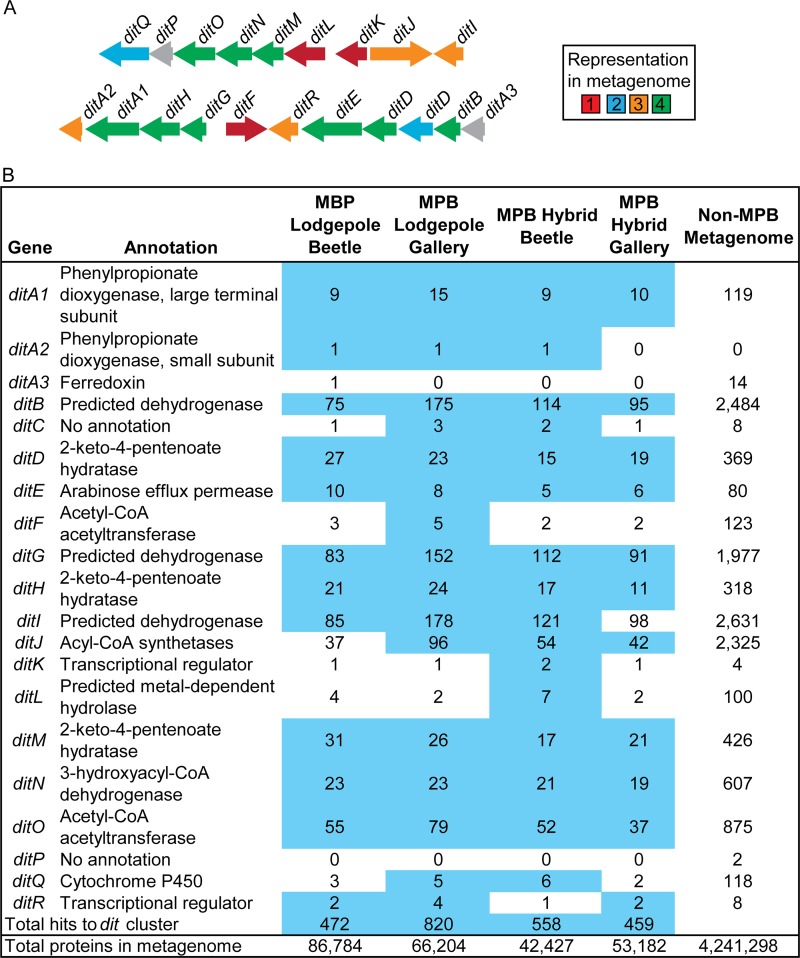
We also investigated genes involved in diterpene degradation using the well-characterized dit gene cluster found in the diterpene-degrading bacterium Pseudomonas abietaniphila BKME-9 (39, 49). Of the 20 proteins annotated as belonging to the dit gene cluster, 17 to 19 were present in each of the community metagenomes sequenced in our study (Fig. 3A). Of these, 12 to 16 were found to be enriched compared to metagenomes of other plant biomass-associated microbial communities (Fig. 3B; P < 0.05, Fisher's exact test). Between 459 and 920 proteins encoded by dit genes were identified in our metagenomes, which comprise 0.54 to 1.3% of the total proteins in the beetle-associated community metagenomes (see Data Set S2 in the supplemental material). Homologs of DitA1, an aromatic ring-hydroxylating dioxygenase, were identified in all metagenomes (9 to 15 proteins). Moreover, homologs to the predicted dehydrogenases DitB, DitG, and DitI were among the most abundant proteins identified by this approach.
Fig 3.
Representation of genes involved in diterpene degradation in mountain pine beetle-associated metagenomes. (A) In the diterpene degradation gene cluster dit, each gene is colored according to its representation within the four mountain pine beetle metagenomes. (B) Total copy numbers for each gene are shown, with those in blue enriched (Fisher's exact test, P < 0.05) relative to other plant biomass-degrading metagenomes.
Taxonomic classification of putative terpenoid degradation-related genes.
We characterized the putative taxonomic origins of those genes involved in pinene or limonene degradation or belonging to the dit gene cluster in our metagenomes. Consistent with the general phylogenetic composition of the metagenomes, we found that the majority of these genes were classified as belonging to bacteria in the genera Pseudomonas, Rahnella, Serratia, and Stenotrophomonas (Fig. 2B; see Fig. S4 in the supplemental material). In all cases, sequences from Pseudomonas and Rahnella comprised over 60% of the genes identified as belonging to these pathways (see Fig. S4).
DISCUSSION
This study provides the first community metagenomic analysis of a bark beetle. The results of both our community metagenomic and DGGE analyses indicate that Gammaproteobacteria are prevalent in both D. ponderosae beetles and their galleries from both lodgepole and hybrid lodgepole-jack pines (see Table S1 and Fig. S2 and S3 in the supplemental material). In particular, Gammaproteobacteria belonging to the genera Pseudomonas, Stenotrophomonas, Erwinia, and Serratia were particularly abundant in these metagenomes (Fig. 2A), indicating these groups are consistently associated with D. ponderosae or their host trees. Our DGGE-based analysis did not resolve differences between these environments (see Fig. S1 in the supplemental material), indicating that bacterial communities may be broadly similar among all of them. Moreover, no distinct differences were observed in the composition of metagenomes from the beetle and gallery samples from Alberta and British Columbia. Taken together, this suggests that a relatively consistent bacterial community is associated with D. ponderosae and its microenvironment and that the recent expansion of this insect's range will not be impeded by a lack of appropriate bacterial communities.
Conifers produce monoterpenes and diterpenes that are toxic to bark beetles and their fungal symbionts, both constitutively and in response to attack. We identified numerous bacterial genes associated with degradation of these compounds, including well-represented KEGG pathways for limonene and pinene degradation in each of the four metagenomes (Table 3). Moreover, we identified numerous genes homologous to the dit gene cluster of Pseudomonas abietophila BKME-9 (Fig. 3A), which is known to be involved in diterpene degradation. We found a significantly higher proportion of these genes in our beetle metagenomes than those from other plant biomass-associated microbial communities (Fig. 3B). The biomass-degrading communities used for comparison originate from a diversity of different environments where plant toxins would be likely encountered, suggesting that bacteria associated with pine beetles are particularly well adapted to metabolize the aromatic plant toxins in their environment.
The majority of genes involved in terpene degradation belong to bacteria in the genera Pseudomonas and Rahnella (Fig. 2B; see Fig. S4 in the supplemental material). The genus Pseudomonas contains numerous species that degrade a wide range of aromatic compounds, including plant toxins, xenobiotics, and pollutants (39, 50–52). The biodegradative capacities of Rahnella isolates have been less intensively studied, but numerous Enterobacteriaciae closely related to this genus are known to degrade a diversity of aromatic compounds (53, 54). Our finding that these genera are both associated with D. ponderosae and possess numerous genes putatively involved in terpenoid degradation suggests they may contribute to D. ponderosae's ability to attack live conifers. Future transcriptomic and culture-based work confirming the ability of these bacteria to degrade plant toxins is required.
These results suggest several testable models to explain associations of bacterial communities with D. ponderosae and their galleries. One hypothesis is that D. ponderosae vector terpene-degrading microbiota between trees. Bark beetles are known to consistently vector both fungi (16) and several genera of bacteria, including Streptomyces (19, 55), thereby explaining the similar composition of all metagenomes analyzed here. This would also be consistent with the high representation of Pseudomonas and Rahnella genes associated with terpene degradation in all four D. ponderosae-associated metagenomes we described. A second nonexclusive model is that these communities are associated primarily with host trees rather than the beetles themselves. This is likewise supported by our DGGE results showing similarities among bacteria in attacked and unattacked trees. Beetle colonization of a tree might induce proliferation of specific bacteria, such as Pseudomonas or Rahnella, that can exploit terpenoids and other carbon sources present in resin. Thus, even if not vectored by the beetles, the colonization attempts and subsequent tree responses may create an environment that promotes the growth of terpenoid-metabolizing bacteria. According to this model, resident microbial populations may influence bark beetles, with trees harboring fewer terpene-degrading bacteria posing more resistance to colonization.
This work provides insight into host colonization and range expansion of D. ponderosae by characterizing the microbiome associated with these beetles and host conifers. A combination of methods suggests that a relatively consistent bacterial community is associated with these beetles in lodgepole and hybrid lodgepole-jack pines. Our results identify bacteria of the genera Pseudomonas and Rahnella that may directly or indirectly contribute to the ability of beetles to overcome tree defenses. Future studies confirming and quantifying the ability of bacteria to degrade tree defenses are needed to understand the influences they may have on bark beetle biology.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Bleiker (CFS) for assistance with site selection, K. Aukema (University of Northern British Columbia) for assistance with and equipment for centrifugation and DNA extraction, J. Ariss (University of Alberta) as well as J. Koopmans and E. Teen (UNBC) for sample collection, J. Franz and J. Moeller (University of Wisconsin) for laboratory assistance, and S. Tringe, K. Barry, T. del Rio, and S. Malfatti (DOE Joint Genome Institute) for metagenome sequencing. We are also grateful to K. Jewell (University of Wisconsin—Madison) for assistance with figure layout and design. We thank three anonymous reviewers for helpful comments.
Funding for this study was obtained from the USDA NRI (2008-02438), the University of Wisconsin, CALS, and the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-Fc02-07ER64494). The work conducted by the U.S. DOE Joint Genome Institute was supported by the U.S. DOE Office of Science under contract no. DE-AC02-05CH1123.
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00068-13.
REFERENCES
- 1. Douglas AE. 2010. The symbiotic habit. Princeton University Press, Princeton, NJ [Google Scholar]
- 2. Moran NA. 2006. Symbiosis. Curr. Biol. 16: R866–R877 [DOI] [PubMed] [Google Scholar]
- 3. Currie CR. 2001. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu. Rev. Microbiol. 55: 357–380 [DOI] [PubMed] [Google Scholar]
- 4. Klepzig K, Adams AS, Handelsman J, Raffa KF. 2009. Symbioses, a key driver of insect physiological processes, ecological interactions, evolutionary diversification, and impacts on humans. Environ. Entomol. 36: 67–77 [DOI] [PubMed] [Google Scholar]
- 5. Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32: 723–735 [DOI] [PubMed] [Google Scholar]
- 6. Frago E, Dicke M, Godfray HC. 2012. Insect symbionts as hidden players in insect-plant interactions. Trends Ecol. Evol. 27: 705–711 [DOI] [PubMed] [Google Scholar]
- 7. Raffa KF, Aukema BH, Bentz BJ, Carroll AL, Hicke JA, Turner MG, Romme WH. 2008. Cross-scale drivers of natural disturbances prone to anthropogenic amplification: the dynamics of biome-wide bark beetle eruptions. Bioscience 58: 501–517 [Google Scholar]
- 8. Amman G, Cole W. 1983. Mountain pine beetle dynamics in lodgepole pine forests. II. Population dynamics. General Technical Report INT, vol 145 Intermountain Forest and Range Experiment Station, USDA, Forest Service, Ogden, UT [Google Scholar]
- 9. Hicke J, Logan J, Powell J, Ojima D. 2006. Changing temperatures influence suitability for modeled mountain pine beetle (Dendroctonus ponderosae) outbreaks in the western United States. J. Geophys. Res. 111: G02019 doi:02010.01029/02005JG000101 [Google Scholar]
- 10. Bentz B, Jaques R, Fettig C, Hansen E, Hayes J, Hicke J, Kelsey R, Negron J, Seybold S. 2010. Climate change and bark beetles of the western United States and Canada: direct and indirect effects. Bioscience 60: 602–613 [Google Scholar]
- 11. Sambaraju K, Carrol A, Zhu J, Stahl K, Moore R, Aukema B. 2012. Climate change could alter the distribution of mountain pine beetle outbreaks in western Canada. Ecography 35: 111–223 [Google Scholar]
- 12. de la Giroday H-MC, Carroll AL, Aukema BH. 2012. Breach of the northern Rocky Mountain geoclimatic barrier: initiation of range expansion by the mountain pine beetle. J. Biogeogr. 39: 1112–1123 [Google Scholar]
- 13. Cullingham CI, Cooke JE, Dang S, Davis CS, Cooke BJ, Coltman DW. 2011. Mountain pine beetle host-range expansion threatens the boreal forest. Mol. Ecol. 20: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lusebrink I, Evenden ML, Blanchet FG, Cooke JE, Erbilgin N. 2011. Effect of water stress and fungal inoculation on monoterpene emission from an historical and a new pine host of the mountain pine beetle. J. Chem. Ecol. 37: 1013–1026 [DOI] [PubMed] [Google Scholar]
- 15. Safranyik L, Carroll AL, Régnière J, Langor DW, Riel WG, Shore TL, Peter B, Cooke BJ, Nealis VG, Taylor SW. 2010. Potential for range expansion of mountain pine beetle into the boreal forest of North America. Can. Entomol. 142: 415–442 [Google Scholar]
- 16. Paine TD, Raffa KF, Harrington TC. 1997. Interactions among scolytid bark beetles, their associated fungi, and host conifers. Annu. Rev. Entomol. 42: 179–206 [DOI] [PubMed] [Google Scholar]
- 17. Six DL, Wingfield MJ. 2011. The role of phytopathogenicity in bark beetle-fungus symbioses: a challenge to the classic paradigm. Annu. Rev. Entomol. 56: 255–272 [DOI] [PubMed] [Google Scholar]
- 18. Adams AS, Currie CR, Cardoza YJ, Klepzig KD, Raffa KF. 2009. Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can. J. For. Res. 39: 1133–1147 [Google Scholar]
- 19. Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. 2008. Bacterial protection of beetle-fungus mutualism. Science 322: 63 doi:10.1126/science.1160423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cardoza YJ, Klepzig KD, Raffa KF. 2006. Bacteria in oral secretions of an endophytic insect inhibit antagonistic fungi. Ecol. Entomol. 31: 636–645 [Google Scholar]
- 21. Morales-Jimenez J, Zuniga G, Villa-Tanaca L, Hernandez-Rodriguez C. 2009. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb. Ecol. 58: 879–891 [DOI] [PubMed] [Google Scholar]
- 22. Adams AS, Adams SM, Currie CR, Gillette NE, Raffa KF. 2010. Geographic variation in bacterial communities associated with the red turpentine beetle (Coleoptera: Curculionidae). Environ. Entomol. 39: 406–414 [DOI] [PubMed] [Google Scholar]
- 23. Adams AS, Boone CK, Bohlmann J, Raffa KF. 2011. Responses of bark beetle-associated bacteria to host monoterpenes and their relationship to insect life histories. J. Chem. Ecol. 37: 808–817 [DOI] [PubMed] [Google Scholar]
- 24. Keeling C, Bohlmann J. 2006. Diterpene resin acids in conifers. Phytochemistry 67: 2415–2423 [DOI] [PubMed] [Google Scholar]
- 25. Raffa K, Smalley EB. 1995. Interaction of pre-attack and induced monoterpene concentrations in host conifer defense against bark beetle-fungal complexes. Oecologia 102: 285–295 [DOI] [PubMed] [Google Scholar]
- 26. Wallin K, Raffa KF. 2000. Influences of host chemicals and internal physiology on the multiple steps of postlanding host acceptance behavior of Ips pini (Coleoptera:Scolytidae). Environ. Entomol. 29: 442–453 [Google Scholar]
- 27. Kopper B, Illman B, Kersten P, Klepzig K, Raffa K. 2005. Effects of diterpene acids on components of a conifer bark beetle-fungal interaction: tolerance by Ips pini and sensitivity by its associate Ophiostoma ips. Environ. Entomol. 34: 486–493 [Google Scholar]
- 28. Boone C, Aukema B, Bohlmann J, Carroll A, Raffa K. 2011. Efficacy of tree defense physiology varies with bark beetle population density: a basis for positive feedback in eruptive species. Can. J. For. Res. 41: 1174–1188 [Google Scholar]
- 29. Raffa KR, Powell EN, Townsend PA. 2013. Temperature-driven range expansion of an irruptive insect heightened by weakly coevolved plant defenses. Proc. Natl. Acad. Sci. U. S. A. 110: 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clark EL, Carroll AL, Huber DPW. 2010. Differences in the constitutive terpene profile of lodgepole pine across a geographical range in British Columbia, and correlation with historical attack by mountain pine beetle. Can. Entomol. 142: 557–573 [Google Scholar]
- 31. Cudmore TJ, Björklund N, Carroll AL, Lindgren BS. 2010. Climate change and range expansion of an aggressive bark beetle: evidence of higher beetle reproduction in naïve host tree populations. J. Appl. Ecol. 47: 1036–1043 [Google Scholar]
- 32. Bleiker K, Carroll A. 2011. Rating introgression between lodgepole and jack pine at the individual tree level using morphological traits. North J. Appl. For. 28: 138–145 [Google Scholar]
- 33. Holben WE, Noto K, Sumino T, Suwa Y. 1998. Molecular analysis of bacterial communities in a three-compartment granular activated sludge system indicates community-level control by incompatible nitrification processes. Appl. Environ. Microbiol. 64: 2528–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- 36. Brady A, Salzberg SL. 2009. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat. Methods 6: 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25: 2271–2278 [DOI] [PubMed] [Google Scholar]
- 38. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35: W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin VJ, Yu Z, Mohn WW. 1999. Recent advances in understanding resin acid biodegradation: microbial diversity and metabolism. Arch. Microbiol. 172: 131–138 [DOI] [PubMed] [Google Scholar]
- 40. Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernandez M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–565 [DOI] [PubMed] [Google Scholar]
- 41. Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, McHardy AC, Cheng JF, Hugenholtz P, McSweeney CS, Morrison M. 2010. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc. Natl. Acad. Sci. U. S. A. 107: 14793–14798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331: 463–467 [DOI] [PubMed] [Google Scholar]
- 43. Zhu L, Wu Q, Dai J, Zhang S, Wei F. 2011. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 108: 17714–17719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aylward FO, Burnum KE, Scott JJ, Suen G, Tringe SG, Adams SM, Barry KW, Nicora CD, Piehowski PD, Purvine SO, Starrett GJ, Goodwin LA, Smith RD, Lipton MS, Currie CR. 2012. Metagenomic and metaproteomic insights into leaf-cutter ant fungus gardens. ISME J. 9: 1688–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allgaier M, Reddy A, Park JI, Ivanova N, D'Haeseleer P, Lowry S, Sapra R, Hazen TC, Simmons BA, VanderGheynst JS, Hugenholtz P. 2010. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One 5: e8812 doi:10.1371/journal.pone.0008812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van der Lelie D, Taghavi S, McCorkle SM, Li LL, Malfatti SA, Monteleone D, Donohoe BS, Ding SY, Adney WS, Himmel ME, Tringe SG. 2012. The metagenome of an anaerobic microbial community decomposing poplar wood chips. PLoS One 7: e36740 doi:10.1371/journal.pone.0036740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, Chen IM, Grechkin Y, Dubchak I, Anderson I, Lykidis A, Mavromatis K, Hugenholtz P, Kyrpides NC. 2008. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 36: D534–D538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mohn WW, Wilson AE, Bicho P, Moore ER. 1999. Physiological and phylogenetic diversity of bacteria growing on resin acids. Syst. Appl. Microbiol. 22: 68–78 [DOI] [PubMed] [Google Scholar]
- 50. Kanaly RA, Harayama S. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182: 2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whyte LG, Bourbonniere L, Greer CW. 1997. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl. Environ. Microbiol. 63: 3719–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Foght JM, Westlake DW. 1988. Degradation of polycyclic aromatic hydrocarbons and aromatic heterocycles by a Pseudomonas species. Can. J. Microbiol. 34: 1135–1141 [DOI] [PubMed] [Google Scholar]
- 53. Sarma PM, Bhattacharya D, Krishnan S, Lal B. 2004. Degradation of polycyclic aromatic hydrocarbons by a newly discovered enteric bacterium, Leclercia adecarboxylata. Appl. Environ. Microbiol. 70: 3163–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wongsa P, Tanaka M, Ueno A, Hasanuzzaman M, Yumoto I, Okuyama H. 2004. Isolation and characterization of novel strains of Pseudomonas aeruginosa and Serratia marcescens possessing high efficiency to degrade gasoline, kerosene, diesel oil, and lubricating oil. Curr. Microbiol. 49: 415–422 [DOI] [PubMed] [Google Scholar]
- 55. Hulcr J, Adams AS, Raffa K, Hofstetter RW, Klepzig KD, Currie CR. 2011. Presence and diversity of Streptomyces in Dendroctonus and sympatric bark beetle galleries across North America. Microb. Ecol. 61: 759–768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.