Abstract
Most human immunodeficiency virus (HIV) transmissions are initiated with CCR5 (R5)-using viruses across mucosal surfaces, with the majority in regions where HIV type 1 (HIV-1) clade C predominates. Mucosally transmissible, highly replication competent, pathogenic R5 simian-human immunodeficiency viruses (SHIVs) encoding biologically relevant clade C envelopes are therefore needed as challenge viruses in vaccine efficacy studies with nonhuman primates. Here we describe the generation of three lineage-related subtype C SHIVs through four successive rapid transfers in rhesus macaques of SHIVC109F.PB4, a molecular clone expressing the soluble-CD4 (sCD4)-sensitive CCR5-tropic clade C envelope of a recently infected subject in Zambia. The viruses differed in their monkey passage histories and neutralization sensitivities but remained R5 tropic. SHIVC109P3 and SHIVC109P3N were recovered from a passage-3 rapid-progressor animal during chronic infection (24 weeks postinfection [wpi]) and at end-stage disease (34 wpi), respectively, and are classified as tier 1B strains, whereas SHIVC109P4 was recovered from a passage-4 normal-progressor macaque at 22 wpi and is a tier 2 virus, more difficult to neutralize. All three viruses were transmitted efficiently via intrarectal inoculation, reaching peak viral loads of 107 to 109 RNA copies/ml plasma and establishing viremia at various set points. Notably, one of seven (GC98) and two of six (CL31, FI08) SHIVC109P3- and SHIVC109P3N-infected macaques, respectively, progressed to AIDS, with neuropathologies observed in GC98 and FI08, as well as coreceptor switching in the latter. These findings support the use of these new SHIVC109F.PB4-derived viruses to study the immunopathology of HIV-1 clade C infection and to evaluate envelope-based AIDS vaccines in nonhuman primates.
INTRODUCTION
Despite progress in treatment and understanding, the global human immunodeficiency virus type 1 (HIV-1) pandemic remains a public health problem of unprecedented proportions. A total of 33.3 million people are living with HIV, nearly half (48%) of whom are infected with clade C strains (www.unaids.org). The majority of new infections are also in regions where HIV-1 subtype C is prevalent and occur principally via heterosexual transmission (1). Although there is much interest in using antiretrovirals (ARVs) for prevention (2, 3), clinical trial results have been highly variable (4), and scaling up for use in the most afflicted regions to stop the spread of HIV requires substantial human and financial resources and political will (5, 6). The development of an effective vaccine therefore remains a major priority for the control of this pandemic (7, 8).
Studies with nonhuman primates (NHPs) are recognized as playing a critical part in basic vaccine discovery, potentially providing valuable information on the immunogenicity and efficacy of vaccine concepts and advancing candidate vaccines into human clinical trials (9). In particular, infection of rhesus macaques (RMs) with simian-human immunodeficiency viruses (SHIVs) containing envelopes (Envs) of HIV-1 could facilitate preclinical analysis of antibody-based candidate vaccines. This is because neutralizing antibody (NAb) responses in SHIV-infected macaques parallel those in HIV-1-infected humans: responses directed primarily against Env variable regions develop early, and cross-reactive antibodies develop late in a minority of infected animals (10–13). The specificity of the Env response in SHIV-infected macaques also mirrors that observed in HIV-1-infected individuals (12, 14, 15), with recent data showing the development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies and the isolation of monoclonal antibodies (MAbs) directed against quaternary neutralizing epitopes from rhesus monkeys infected with subtype B CCR5 (R5)-tropic SHIV (16, 17). Collectively, these findings support testing of the immunogenicity of candidate HIV-1 envelope immunogens and protection studies in NHPs.
Because most HIV-1 transmissions involve mucosal exposure and are initiated with R5-tropic viruses (18), pathogenic and mucosally transmissible R5 SHIVs would be the preferred tools for assessment of the effectiveness of envelope vaccine-induced immunity. However, most challenge stocks currently available express envelopes from culture-derived HIV-1 clade B strains, which represent ∼10% of all infections globally and are substantially different in sequence and envelope antigenic structure from the most predominant subtype C strains. Indeed, while coreceptor switching has been documented in 40 to 50% of individuals infected with subtype B and D viruses and is associated with faster disease progression, it is found less frequently in patients infected with subtypes A and C (19, 20), leading to the suggestion that intrinsic differences in V3 conformation and/or evolutionary pathways to efficient CXCR4 (X4) usage between subtypes may be playing a role (21–23). Several SHIVs containing subtype C envelopes (SHIVCs) have been described (24–28), but not all the SHIVCs utilize the CCR5 molecule exclusively as an entry coreceptor. Most replicated to low levels in vivo, with transmission across the mucosal barrier yet to be documented for some. The exceptions are the SHIV-1157i-derived viruses, including the late-stage, highly replication competent infectious molecular clone SHIV-1157ipd3N4. These SHIVC are mucosally transmissible and are pathogenic in RMs, but the animals progressed to AIDS slowly (29, 30). Moreover, although coreceptor switching and the development of central nervous system (CNS) disorders have been described for subtype B R5 SHIVSF162P3N-infected macaques (31, 32; S. Westmoreland, C. Harbison, K. Zhuang, A. Gettie, J. Blanchard, and C. Cheng-Mayer, presented at the 2012 Conference on HIV in the Nervous System), these biological and neurological sequelae have not been documented in R5 SHIVC-infected animals at terminal disease. Thus, there is a need for additional pathogenic SHIVs carrying the envelopes of dominant clade C strains as tools in NHPs to advance the discovery of effective and widely useful AIDS vaccines targeting the HIV-1 envelope glycoproteins. Studies of infection with pathogenic R5-using clade C SHIVs may also help in understanding why the development of CXCR4 usage is rare in HIV-1C-infected individuals.
With these objectives in mind, we constructed a SHIV molecular clone bearing ZM109F.PB4, a soluble-CD4 (sCD4)-sensitive CCR5-tropic clade C envelope from a recently infected subject in Zambia (33, 34). We used sCD4 sensitivity as a surrogate for exposure of the CD4 binding site (BS) and hypothesized that a SHIV molecular clone with an “open” HIV-1 envelope that exposes the receptor binding site for better CD4 binding will adapt more easily to differences in the levels and structure of viral receptors in the new host, allowing it to replicate to high levels and to generate variants with increased fitness and pathogenic properties. To adapt the newly constructed SHIV (designated SHIVC109F.PB4) to growth in vivo, four rapid serial passages in RMs were performed, with anti-CD8 antibody administered to the passage-3 (P3) and P4 animals during primary infection to dampen and/or delay the host immune response and to enhance viral replication. We report here the generation of a series of lineage-related R5 SHIVC109F.PB4-derived viruses that display biological, immunological, and pathological characteristics similar to those of HIV-infected patients, supporting the use of these viruses in studies of AIDS immunopathogenesis and vaccines in nonhuman primates.
MATERIALS AND METHODS
Cells.
293T cells and TZM-bl cells expressing CD4, CCR5, and CXCR4 and containing integrated reporter genes for firefly luciferase and β-galactosidase under the control of the HIV-1 long terminal repeat (LTR) (35) were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and l-glutamine. U87 cells stably expressing CD4 and one of the chemokine receptors (CCR5 or CXCR4) were maintained in DMEM supplemented with 10% FBS, antibiotics, 1 μg/ml puromycin (Sigma-Aldrich), and 300 μg/ml G418 (Geneticin; Invitrogen, Carlsbad, CA). Human peripheral blood mononuclear cells (PBMCs) and rhesus PBMCs (rhPBMCs) were prepared by Ficoll gradient centrifugation, stimulated with phytohemagglutinin (PHA; 3 μg/ml; Sigma, St. Louis, MO) and staphylococcal enterotoxin B (SEB; 3 μg/ml; Sigma), respectively, and cultured in RPMI medium containing 10% fetal calf serum (FCS), penicillin, streptomycin, l-glutamine, and 20 U/ml interleukin-2 (Novartis, Emeryville, CA).
Construction of a SHIVC molecular clone and preparation of inoculating stocks.
A plasmid expressing the soluble-CD4-sensitive Env (ZM109F.PB4) from an early, heterosexually acquired subtype C virus of an individual in Zambia was obtained from Cynthia Derdeyn. PCR amplification with forward primer WR30 (5′-TTATGGGGTACCTGTGTGGAAAGAAGC-3′ [KpnI site underlined]) and reverse primer WR32 (5′-AGCTAAGGATCCGTTCACTAATCGAATG-3′ [BamHI site underlined]) was performed to subclone most of gp120 and the entire extracellular domain and transmembrane region of gp41 of ZM109F.PB4 into the corresponding regions of the 3′ SHIVHXBc2vpu+ genome using the unique KpnI (nucleotide position 6347) and BamHI (nucleotide position 8475) sites (nucleotide position numbering based on that of HXBc2). The inserted sequence was confirmed, and the infectious SHIVC109F.PB4 molecular clone was recovered by cotransfection of 293T cells with a ligation product of the 3′ SHIVC109F.PB4 and 5′ simian immunodeficiency virus (SIV) hemigenomes, followed by cocultivation with PHA-stimulated human PBMCs. The SHIVC109F.PB4 molecular clone for virus inoculation was expanded, and titers were determined in SEB-stimulated rhPBMCs.
Serial passages, animal inoculation, and clinical assessments.
All inoculations were carried out in adult rhesus monkeys of Indian origin (Macaca mulatta) housed at the Tulane National Primate Research Center (TNRPC) in compliance with the Guide for the Care and Use of Laboratory Animals (36). Animals were confirmed to be serologically and virologically negative for simian type D retrovirus and serologically negative for SIV and simian T-cell lymphotropic virus prior to infection and were screened for the presence of the Mamu-A*01, Mamu-B*17, and Mamu-B*08 class I alleles, previously shown to be associated with control of pathogenic SIVmac239 replication, by using standard PCR with allele-specific primers (37). Serial passage of the molecular clone SHIV109F.PB4 was performed as follows. Two rhesus macaques (P1) were inoculated intravenously with 300 median tissue culture infectious doses (TCID50) of cell-free virus stock. Two weeks after inoculation, 10 ml each of peripheral blood and bone marrow aspirate was collected from each animal, combined, and used to transfuse two naïve recipients (P2). This inoculation schedule was performed two more times in naïve recipient groups P3 and P4, with the anti-CD8 antibody M-T807R1 administered at day −5 (10 mg/kg of body weight), day −1 (5 mg/kg), and day 14 posttransfusion (5 mg/kg) to delay/dampen host immune responses and promote virus replication. For cell-free infection with SHIVC109F.PB4-derived viruses, macaques received a single intrarectal (i.r.) inoculation with 5,000-TCID50 virus stocks. Whole blood from the inoculated animals was collected weekly for the first 8 weeks, biweekly for another 16 weeks, and monthly thereafter. Survival surgery was performed at peak viremia (2 weeks postinfection [wpi]) for collection of tissues from one external and one internal lymph node (LN) and from internal organs such as the gut, bone marrow, thymus, and spleen. Animals were euthanized at the end of the study period by intramuscular administration of telazol and buprenorphine, followed by an overdose of sodium pentobarbital. Euthanasia was considered to be AIDS related if the animal exhibited peripheral blood CD4+ T-cell depletion (<200/mm3), >25% loss of body weight, or combinations of the following conditions: diarrhea unresponsive to treatment, opportunistic infections, peripheral lymph node atrophy, and abnormal hematology. Plasma viremia was quantified by real-time PCR for SIVgag (Washington National Primate Research Center, Seattle, WA), and absolute CD4+ and CD8+ cell counts were monitored by TruCount absolute-count tubes (BD Biosciences, Palo Alto, CA). The lower limit of detection of the viral load (VL) assay is 30 RNA copies/ml plasma. The percentages of CD4+ T cells in the tissue cells were analyzed by flow cytometry (on a FACSCalibur system) using fluorescein isothiocyanate (FITC)-conjugated anti-CD3, phycoerythrin (PE)-conjugated anti-CD4, and peridinin chlorophyll protein (PerCP)-conjugated anti-CD8 antibodies. Except for FITC-conjugated anti-CD3 (BioSource, Camarillo, CA), all antibodies were obtained from BD Biosciences.
Neutralization assay.
Virus neutralization was assessed in TZM-bl assays using a panel of MAbs, soluble CD4, clade B sera, a panel of purified Igs from chronically infected patients from South Africa, and polyclonal anti-HIV immunoglobulin (Ig) preparations from clade B- and C-infected individuals. Briefly, equal volumes (50 μl) of SHIV (1 ng p27 Gag equivalent) and 4-fold serial dilutions of MAb/sera/Ig were incubated for 1 h at 37°C and were then added to cells, in duplicate wells, for an additional 2 h at 37°C. One hundred microliters of medium was then added to each well, and the virus-protein cultures were maintained for 48 to 72 h. Control cultures received virus in the absence of blocking agents. At the end of the culture period, the cells were lysed and were processed for β-galactosidase activity. A neutralization curve was generated by plotting the percentage of neutralization versus fusion protein dilution, and 50% inhibitory concentrations (IC50s) were determined using Prism 5 software (GraphPad Software Inc., San Diego, CA). Higher IC50s for the antibodies and purified Ig tested mean less neutralization sensitivity, whereas higher IC50 titers of the pooled clade B sera mean greater neutralization sensitivity.
Envelope sequence analysis, expression plasmid construction, and pseudotype virus production.
For sequence analysis, proviral DNA was extracted from 3 × 106 infected macaque PBMCs or tissue cells with a DNA extraction kit (Qiagen, Chatsworth, CA), while viral RNA was prepared from 300 to 500 μl of plasma or virus supernatant by using a commercially available RNA extraction kit (Qiagen), followed by reverse transcription (RT) with SuperScript III reverse transcriptase (Invitrogen) and random hexamer primers (Amersham Pharmacia, Piscataway, NJ). The V1-to-V5 region of gp120 was amplified from the viral DNA or RT products using Taq DNA polymerase with primers ED5 and ED12 as described previously (38). PCR products were cloned with the TOPO TA cloning kit (Invitrogen) according to the manufacturer's instructions, followed by direct automated sequencing (Genewiz, South Plainfield, NJ). The sequences were aligned with Clustal X (39), edited manually using BioEdit, version 7.0.9, and translated to amino acid sequences. For the generation of Env expression plasmids and luciferase reporter viruses, the full-length gp160 coding sequence was amplified with primers EnvC-1 (5′-AGAGACGGATCCACCATGAGAGTGAAGGAGAAATATC-3′) and EnvC-2 (5′-AGAGACGAATTCTCACAAGAGAGTGAGCTCGAGC-3′) (where underlining indicates BamHI and EcoRI sites, respectively) and was subcloned into the pCAGGS vector. An envelope trans-complementation assay was then employed to generate luciferase reporter viruses capable of only a single round of replication. The pseudovirions were quantified for p24gag content (Beckman Coulter, Fullerton, CA). For the construction of the V3 recombinant plasmid, PCR-based overlap extension methodology was employed to replace the V3 loop of ZM109F.PB4 with that of the variant bearing a 4-amino-acid (4-aa) deletion (ΔIGDI) in the V3 loop as described previously (40).
Determination of coreceptor usage.
For assessment of coreceptor usage, 7 × 103 U87.CD4.CCR5 or U87.CD4.CXCR4 cells were seeded in 96-well plates 1 day before use, infected in triplicate with 5 ng p24gag equivalent of the pseudovirions, and then incubated for 72 h at 37°C. At the end of the incubation period, the cells were harvested, lysed, and processed for activity according to the manufacturer's instructions (luciferase assay system; Promega, Madison, WI). Entry, as quantified by relative light units (RLU), was measured with an MLX microtiter plate luminometer (Dynex Technologies, Inc., Chantilly, VA). For assessment of coreceptor utilization in rhPBMCs, blocking with CCR5 (TAK779) and CXCR4 (AMD3100) inhibitors was performed. Briefly, 5 × 106 SEB-stimulated cells were infected with 200 TCID50 of the SHIV in the presence or absence of the chemokine receptor inhibitors at 1 μM. After incubation for 2 h at 37°C, cells were washed and cultured in 1.5 ml interleukin-2 and RPMI medium supplemented with the appropriate inhibitor in each well of a 24-well plate. Culture supernatants were collected over time, and the p27gag antigen content was quantified according to the manufacturer's instructions (ZeptoMetrix, Buffalo, NY). The percentage of blocking at 6 days postinfection was determined by calculating the amount of p27gag antigen production in the presence of the inhibitor relative to that in its absence.
Histopathological examination of the brain.
SHIV-infected cells were identified by double-label in situ hybridization (ISH) for SIVmac and immunohistochemistry (IHC) for the macrophage marker Iba-1. SIV ISH was performed on the Discovery Ultra platform (Ventana Medical Systems, Inc., Tucson, AZ). Briefly, tissue sections were pretreated with reagents from the RiboMap kit (Ventana, Tucson, AZ) and were digested with protease 3 (Ventana) for 4 min at 37°C. Sections were then prehybridized with RiboHyb (Ventana) and were hybridized with a digoxigenin (DIG)-labeled probe for 6 h at 55°C. Stringency washes were done using 1.0× saline-sodium citrate buffer (0.15 M NaCl plus 0.015 M sodium citrate) (Ventana) at 60°C. The bound probe was detected with rabbit anti-DIG (1:20,000; Sigma), an alkaline phosphatase-conjugated UMap anti-rabbit antibody (Ventana), and the chromogen NBT (nitroblue tetrazolium)–BCIP (5-bromo-4-chloro-3-indolylphosphate) (Ventana). Sections were then incubated with a rabbit polyclonal antibody against Iba-1 (catalog no. 019-19741; dilution, 1:1,000; Wako Chemicals, Richmond, VA) for macrophages for 30 min at room temperature, followed by a biotinylated secondary antibody (GAR-b; dilution, 1:200; Dako) for 30 min. Sections were detected using a standard avidin-biotin peroxidase complex technique (Vectastain ABC-Elite; Vector Laboratories, Burlingame, CA) and were counterstained with Nuclear Fast Red (Vector). Isotype-matched irrelevant controls were included for all runs.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism, version 5.0. Differences among groups in peak and cumulative viral load were examined using the Mann-Whitney t test. P values of <0.05 were considered statistically significant.
RESULTS
In vivo adaptation of SHIVC109F.PB4 in rhesus macaques.
With the goal of developing an R5-specific and pathogenic subtype C SHIV, we constructed SHIVC109F.PB4 (Fig. 1A), which contains most of the exterior envelope glycoprotein gp120, as well as the entire extracellular and transmembrane regions of gp41, from a subject in Zambia recently infected with subtype C (33, 34). We chose this envelope (ZM109F.PB4), which is sensitive to sCD4, to test the hypothesis that a glycoprotein that binds CD4 efficiently will adapt more readily to variations in the structure and levels of viral receptors in a new host. We subjected the SHIVC109F.PB4 molecular clone to rapid serial transfers in RMs, initially inoculating two rhesus macaques intravenously with 300 TCID50 of virus, followed by three sequential blood–bone marrow transfusions into naïve macaques, with transient CD8 depletion in the passage-3 and -4 monkeys to enhance virus replication (Fig. 1B).
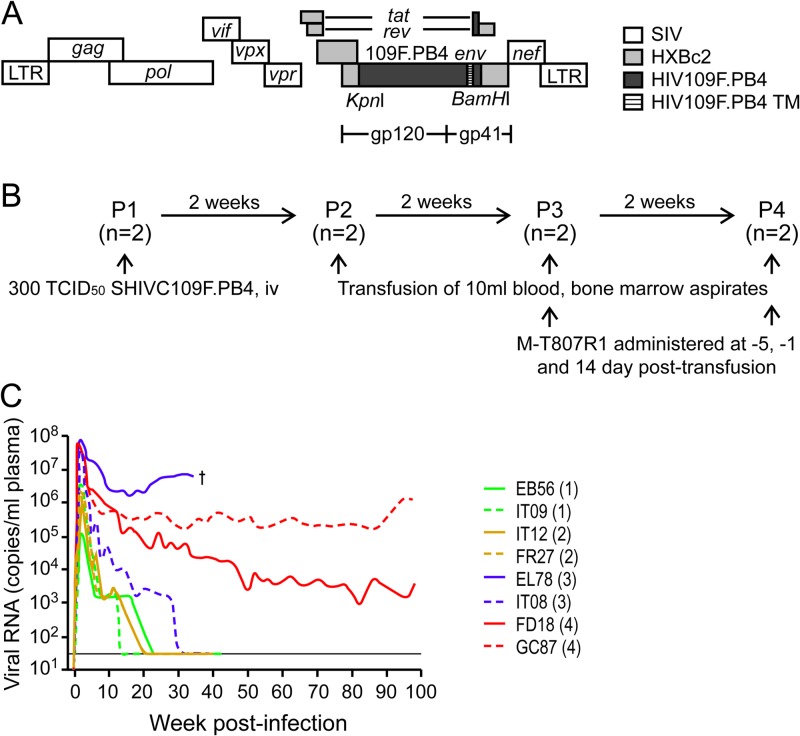
Fig 1.
Genomic organization (A), serial passaging (B), and replication (C) of the SHIVC109F.PB4 molecular clone in rhesus macaques. (A) SHIVC109F.PB4 was constructed by replacing a 2.07-kb KpnI/BamHI fragment of the 3′ SHIVHXBc2vpu+ plasmid with the corresponding env sequences of a recently transmitted CCR5-tropic, sCD4-sensitive HIV-1 subtype C isolate from Zambia. TM, transmembrane domain. (B) Two macaques were used for inoculation in each serial passage. The anti-CD8 antibody M-T807R1 was administered to passage-3 and -4 macaques at days −5 and −1 and at day 14 postinfection. (C) Virus replication in the passage-1 to passage-4 macaques. The passage number is given in parentheses after the designation of each animal. The dagger on the graph indicates euthanasia of P3 macaque EL78 at 34 wpi. The horizon line indicates the lower limit of sensitivity of the viral RNA detection assay (30 copies/ml plasma).
Significant amounts of viral replication were achieved during the acute phase of infection in the two passage-1 (P1) macaques, reaching peak viremia of 105 to 106 RNA copies/ml plasma at 2 wpi (Fig. 1C). Viremia declined thereafter and was undetectable after 12 and 20 weeks of infection. Similar kinetics of viral replication were observed in the P2 monkeys, but with slightly increased peak viral loads of 106 to 107 viral RNA copies/ml plasma. In comparison, substantial augmentation in peak viral replication was seen in the passage-3 and -4 animals treated with the anti-CD8 antibodies; viremia reached levels of >107 RNA copies/ml plasma in all four macaques at 1 to 2 wpi. Moreover, viremia was sustained in both P4 monkeys (GC87 and FD18) and in one of the two P3 animals (EL78), with a set point of 106 to 107 RNA copies/ml plasma in EL78 and 1- and 3-log-lower set points in GC87 and FD18. The exception was IT08, in which virus replication was undetectable at 30 wpi. Genotyping revealed that in contrast to the others, which tested negative for Mamu-A*01, -B*08, and -B*17, IT08 was positive for Mamu-B*17, a major histocompatibility complex (MHC) class I allele that has been shown to be associated with viral control in pathogenic SIVmac239 infection (41). All eight SHIVC109F.PB4-infected or -transfused macaques seroconverted by 3 wpi, with increases in SHIV binding antibody titers over time (unpublished data). The exception was EL78, whose titers peaked at 5 to 6 wpi and declined thereafter. This animal developed clinical symptoms consistent with AIDS, including chronic diarrhea, weight loss, and Pneumocystis carinii infection, and was euthanized at 34 wpi with a CD4+ T cell count of 59/μl blood. Histological examination revealed moderate to severe lymphoid depletion, thymic atrophy, and syncytial giant cells in the lung and lymph nodes (LNs).
Recovery and characterization of rhesus-adapted SHIVC109F.PB4-derived viruses.
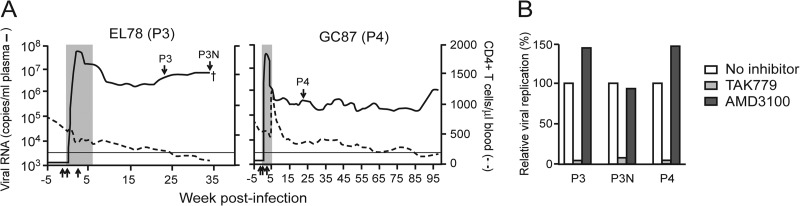
We recovered viruses from the plasma of viremic macaques EL78 (designated SHIVC109P3) and GC87 (designated SHIVC109P4) at 24 and 22 wpi, respectively (Fig. 2A), times that we and others have shown to be sufficient to allow the outgrowth of viral species with increased virulence (42, 43). Notably, neutralizing antibodies against the homologous SHIVC109F.PB4 molecular clone could be detected in GC87 (dilution of plasma inhibiting infection by 50% [ID50 titer], 1:1,000) but not in EL78 (ID50 titer, <1:20) at the time of virus isolation. Furthermore, because variants with increased pathogenicity and replication fitness have been identified late in infection in HIV-1-infected individuals as well as in SIV/SHIV-infected macaques that progressed to disease while maintaining an exclusively R5 tropic virus population (42, 44, 45), we also recovered viruses from the plasma of EL78 at the time of necropsy (designated SHIVC109P3N) for comparative studies with the early lineage-related virus SHIVC109P3. Infection of rhPBMCs with the three viruses was blocked by the CCR5 antagonist TAK779 but not by the CXCR4 inhibitor AMD3100, showing that these viruses maintained CCR5 specificity (Fig. 2B). The passage-3 SHIVC109P3 and SHIVC109P3N viruses were 3- to 10-fold more resistant to sCD4 than the inoculating molecular clone SHIVC109F.PB4, while the SHIVC109P4 virus was 53-fold less sensitive (Table 1). This contrasts with the findings for VRC01, a broadly neutralizing anti-CD4 binding site (BS) monoclonal antibody, for which the three passaged viruses displayed a rank order of sensitivity similar to that for sCD4 but the inoculating virus was more resistant. These differences in sCD4 and VRC01 sensitivity between SHIVC109F.PB4 and the passage-derived viruses are suggestive of structural alterations and/or adjustments in the receptor binding site with adaption in vivo. All three viruses, like SHIVC109F.PB4, were resistant to a number of neutralizing monoclonal antibodies, including those targeting the CD4BS (IgG1 b12), a mannan-dependent epitope in gp120 (2G12), and the gp41-specific membrane-proximal ectodomain region (MPER) epitope (2F5). There was no difference in sensitivity to neutralization by PG9, directed against a quaternary V1V2 epitope. However, using a panel of purified Igs from chronically infected patients from South Africa, a few anti-clade B sera, and polyclonal anti-HIV Ig preparations from clade B- and C-infected individuals, we observed that the passage-3-derived viruses were more neutralization sensitive than SHIV109F.PB4 (the late SHIVC109P3N virus was the most sensitive) while the SHIVC109P4 virus was more resistant. Overall, the passage-3 SHIVC109P3 and SHIVC109P3N viruses exhibit a tier 1B-like phenotype, whereas the SHIVC109P4 virus is a more neutralization resistant tier 2 strain (46). Sequence analysis of the gp120 V1-to-V5 region showed an overall envelope sequence identity of 95 to 99% among the three isolates, with amino acid changes primarily in the variable loop domains (see Fig. S1 in the supplemental material). Notable are changes in potential N-linked glycosylation sites (PNGS) that could explain their differences in immune recognition (Fig. 3). Compared to the parental SHIVC109F.PB4 clone, shifts in N-linked glycans in the V1V2, C3, and V4 regions of gp120, sites of early autologous NAb response in subtype C-infected individuals (47, 48), were observed in the neutralization-resistant SHIVC109P4 strain but not in the neutralization-sensitive SHIVC109P3 and SHIVC109P3N strains. Furthermore, both SHIVC109P3 and SHIVC109P3N lost PNGS just outside the C terminus of the V2 loop (N197D) as well as within the V4 loop, and this was not seen in SHIVC109P4.
Fig 2.
Recovery (A) and coreceptor usage (B) of viruses from P3 macaque EL78 and P4 animal GC87. (A) Arrows pointing down indicate the time points of virus recovery; arrows pointing up indicate the times of anti-CD8 antibody administration, at days −5 and −1 and day 14 postinfection. Shaded areas represent the periods when peripheral CD8+ T cell depletion was observed, and the dagger indicates the time of EL78 euthanasia. (B) The coreceptor usage of recovered viruses in rhesus PBMCs was determined by blocking with antagonists against CCR5 (TAK779) and CXCR4 (AMD3100). Results are representative of one of two independent experiments.
Table 1.
Neutralization profiles of SHIVC109F.PB4 and rhesus-adapted viruses SHIVC109P3, SHIVC109P3N, and SHIVC109P4
| Antibody | Neutralizing activitya against: |
|||
|---|---|---|---|---|
| SHIVC109F.PB4 | SHIVC109P3 | SHIVC109P3N | SHIVC109P4 | |
| sCD4 | 0.03 | 0.10 | 0.29 | 1.59 |
| VRC01 | 2.75 | 0.10 | 0.24 | 1.40 |
| PG9 | 0.65 | 0.50 | 0.37 | 0.425 |
| b12 | >50 | >50 | >50 | >50 |
| 2G12 | >50 | >50 | >50 | >50 |
| 2F5 | >50 | >50 | >50 | >50 |
| Polyclonal anti-HIV Ig | ||||
| Clade B | 666.80 | 153.06 | 44.38 | >2,500 |
| Clade C | 123.24 | 30.13 | 10.95 | 360.36 |
| Anti-clade C Igb | ||||
| SA-C2 | 316.15 | 13.46 | 4.31 | 1,329.72 |
| SA-C8 | 178.18 | 32.52 | 7.54 | 258.21 |
| SA-C44 | 167.51 | 6.46 | 2.33 | 258.21 |
| SA-C62 | 406.05 | 56.55 | 16.73 | 1,893.62 |
| SA-C72 | 219.15 | 30.82 | 7.48 | 826.72 |
| SA-C74 | 386.46 | 33.22 | 13.38 | 994.18 |
| Anti-clade B sera | ||||
| HIV-011 | 32 | 109 | 401 | 23 |
| HIV-014 | 127 | 225 | 372 | 117 |
Expressed as the IC50 (in micrograms per milliliter) for all antibodies except the anti-clade B sera HIV-011 and HIV-014, for which neutralizing activity is expressed as the ID50 titer (reciprocal of the dilution).
From chronically infected patients from South Africa.
Fig 3.
Amino acid sequence comparison of the Env gp120 V1V2, C3, and V4 regions of SHIVC109P3, SHIVC109P3N, and SHIVC109P4. Dots denote identical residues in the sequence, and PNGS in the reference ZM109F.PB4 sequence are highlighted by red boxes. Deletions of PNGS are indicated by dashed red boxes, while those that are shifted are bracketed in blue. Arrows mark the borders of the loop regions, and the numbers of clones matching the indicated sequences per total number of clones sequenced are given in parentheses. The numbering of amino acids is based on that in the HXBc2 subtype B reference strain.
Mucosal transmissibility and induction of disease by SHIVC109P3, SHIVC109P4, and SHIVC109P3N in Indian rhesus macaques.
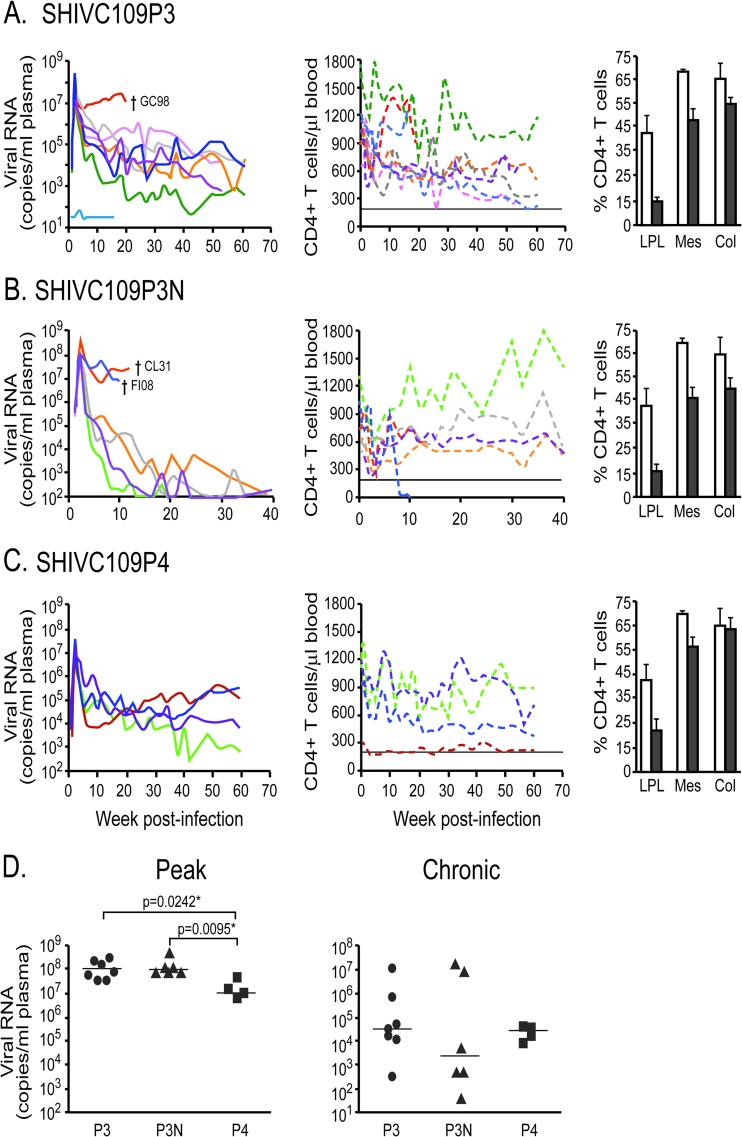
We next assessed the mucosal transmissibility and pathogenicity of the rhesus-adapted SHIVC viruses by intrarectal inoculation of RMs with SHIVC109P3 (n = 8), SHIVC109P3N (n = 6), and SHIVC109P4 (n = 4). Virus exposure resulted in systemic infection in all animals except one (IT11) in the SHIVC109P3 challenge group (Fig. 4). Peak viremia in the SHIVC109P3- and SHIVC109P3N-infected macaques reached 107 to 109 RNA copies/ml plasma at 2 wpi (Fig. 4A and B), with low, intermittent viral blips detected for macaque IT11, which resisted systemic infection. In comparison, while the kinetics of early virus spread in SHIVC109P4-infected macaques were similar, with peak viremia detected at 2 wpi (Fig. 4C), the levels reached were 1 log lower than those seen in the SHIVC109P3- and SHIVC109P3N-infected rhesus macaques. Virus replication declined in all infected macaques postpeak, establishing various set points. Sustained viremia of 104 to 106 RNA copies/ml plasma for close to 40 weeks was seen in five of the seven SHIVC109P3-infected animals; of the remaining two, one had control levels of 102 to 103 RNA copies/ml plasma and the other (GC98) had high virus replication (>107 RNA copies) (Fig. 4A). GC98 developed clinical symptoms of AIDS (weight loss, diarrhea, lymphadenopathy) and was euthanized at 20 wpi. Levels of virus replication were also persistently high in two (CL31 and FI08) of the six SHIVC109P3N-infected monkeys but dropped precipitously in the other four to low and intermittent (103 RNA copies/ml plasma) or undetectable levels at 20 wpi (Fig. 4B). CL31 and FI08 progressed to disease at 13.5 and 10 wpi, respectively, and were euthanized. In contrast, all four SHIVC109P4-infected macaques showed sustained viremia of 104 to 106 RNA copies/ml plasma for >30 weeks of infection, with a gradual decline thereafter in one monkey. None of the chronically SHIVC109P4 infected macaques, however, progressed to disease over a 1-year infection period.
Fig 4.
(A to C) Plasma viral loads, peripheral CD4+ T cell counts, and percentages of CD4+ T cells in the tissues of macaques infected with SHIVC109P3 (A), SHIVC109P3N (B), or SHIVC109P4 (C). (Left) Daggers indicate times of necropsy. (Center) Horizontal lines indicate CD4+ T cell counts of 200 cells per microliter of blood. (Right) Open bars, mean baseline percentages of tissue CD4+ T cells generated from three uninfected macaques; filled bars, mean percentages of tissue CD4+ T cells in infected animals during peak viremia. Error bars, standard deviations. LPL, lamina propria lymphocytes; Mes, mesenteric lymph nodes; Col, colonic lymph nodes. (D) Comparison of peak and steady-state viremias in macaques infected with different SHIVC viruses. Statistical analysis was performed using a two-tailed Mann-Whitney test. Asterisks indicate significant differences (P < 0.05). P3, SHIVC109P3; P3N, SHIVC109P3N; P4, SHIVC109P4.
In agreement with observations of HIV-1-infected individuals and macaques infected with subtype B R5-tropic SHIV, a transient drop in peripheral CD4+ T cell counts accompanied acute infection in all the viremic SHIVC-infected animals, with drastic (50 to 80%) depletion of CD4+ T cells in the gut lamina propria but minimal loss in the gut-associated lymph nodes (Fig. 4A to C). Levels of CD4+ T cells in the blood rebounded close to preinfection values in most of the infected animals by 10 wpi and fluctuated during the chronic phase of infection. The exception was FI08, where a precipitous drop in peripheral CD4+ T cell numbers was seen at 8 wpi, and only 2 cells/μl blood were detected at the time of euthanasia 2 weeks later. Seroconversion was observed at 3 to 5 wpi in all animals except for the three with AIDS (GC98, CL31, FI08). No anti-SHIV antibodies could be detected in GC98 and FI08 during the entire course of infection, and such antibodies were weak and transient in CL31 (data not shown). High levels of virus replication and undetectable or weak antiviral antibody responses, with disease development within 20 weeks of infection, classified GC98, FI08, and CL31 as rapid progressors (RPs).
Comparison of peak viremia among the three groups of SHIVC-infected macaques showed significantly lower levels in the SHIVC109P4-infected animals than in those infected with SHIVC109P3 (P = 0.0242) or SHIVC109P3N (P = 0.0095), with no differences in set point viremia (Fig. 4D). However, when the rapid progressors in the SHIVC109P3 (n = 1) and SHIVC109P3N (n = 2) infection groups were excluded, steady-state viremia in macaques chronically infected with SHIVC109P3N was significantly lower than that in SHIVC109P4-infected animals (P = 0.0286), and the difference from animals infected with the earlier SHIVC109P3 virus approached significance (P = 0.0667). Thus, the late-stage isolate SHIVC109P3N appeared to be less capable of establishing persistence following the development of an antiviral antibody response.
Coreceptor switching in one of two macaques with AIDS that had been infected intrarectally with SHIVC109P3N.
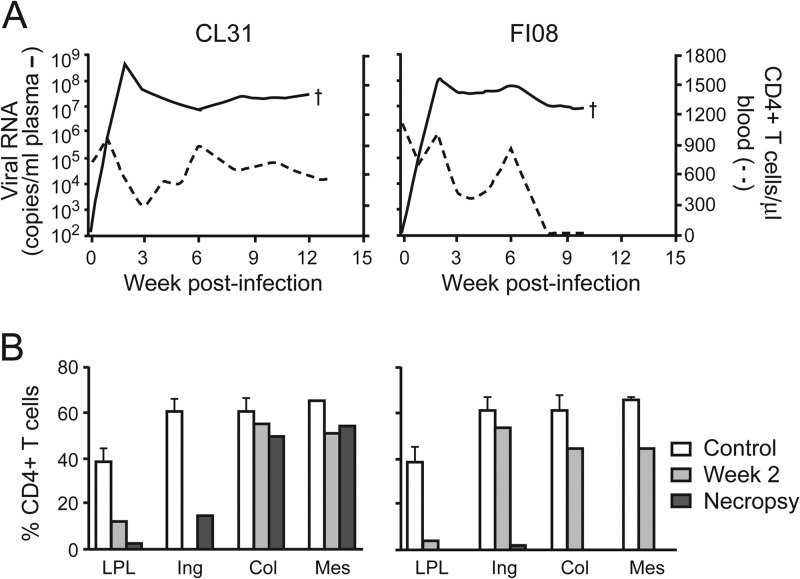
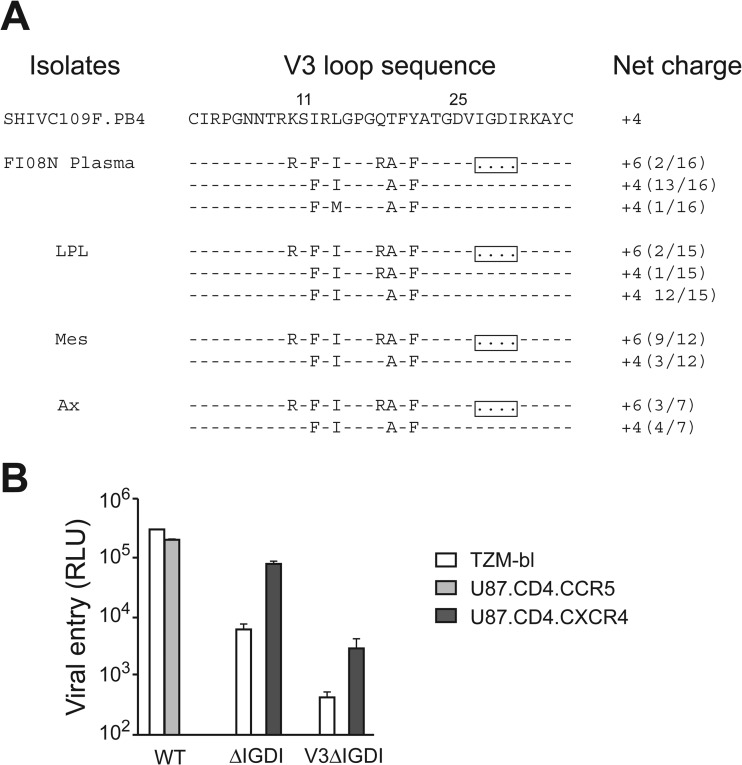
The precipitous drop in the peripheral CD4+ T cell counts of SHIVC109P3N-infected macaque FI08 as it approached end-stage disease prompted us to examine tissue CD4+ T cell loss in this animal (Fig. 5). The results showed severe depletion of CD4+ T cells in the gut as well as the lymph nodes of FI08 at the time of necropsy. In contrast, whereas CD4+ T cells were depleted in the gut lamina propria of CL31, the other SHIVC109P3N-infected AIDS macaque, >80% of these target cells were preserved in secondary lymphoid tissues, such as the colonic and mesenteric LNs, with greater loss (>70%) in the inguinal LN at the time of euthanasia. Envelope sequence analysis of variants in the plasma, lamina propria lymphocytes (LPL), and LNs of FI08 at the time of euthanasia revealed the presence of viruses with a 4-amino-acid deletion in the V3 loop that increased the net positive charge of this region (Fig. 6A). Two of 16 and 2 of 15 clones sequenced from the plasma and LPL, respectively, harbored this V3 deletion, with greater representation in the mesenteric (9 of 12 clones) and axillary (3 of 7 clones) LNs. Functional analysis showed that viruses expressing FI08 envelope gp160 with a 4-aa deletion in the V3 loop (ΔIGDI) used CXCR4 but not CCR5 for entry, while those with wild-type (WT) V3 Env sequences functioned with CCR5 and not CXCR4 (Fig. 6B). A V3 recombinant with a 4-aa deletion in the backbone Env ZM109F.PB4 (V3ΔIGDI) used CXCR4 exclusively, indicating that, in agreement with findings for HIV-1 subtypes, V3 is a principal determinant for coreceptor usage for SHIVC as well as SHIVB strains.
Fig 5.
Severe lymphoid CD4+ T cell loss in SHIVC109P3N-infected macaque FI08. (A) Plasma viral loads and peripheral CD4+ T cell counts in SHIVC109P3N-infected RMs that developed clinical symptoms of AIDS. (B) Percentages of CD4+ T cells in the LPL from the jejunum and in the inguinal (Ing), colonic (Col), and mesenteric (Mes) lymph nodes during acute viremia (week 2) and at the time of necropsy. Means and standard deviations of baseline data generated from three uninfected macaques (controls) are shown for reference.
Fig 6.
V3 sequences and coreceptor usage of Env variants in FI08 at the time of necropsy. (A) Comparison of the V3 loop sequence of the parental SHIVC109F.PB4 molecular clone with those of representative viruses in the plasma, jejunal LPL, and mesenteric (Mes) and axillary (Ax) LNs of macaque FI08. Dashes stand for identity in sequences, and the net positive charge of this region is shown on the right. Positions 11 and 25 within the V3 loop are indicated, and the 4-aa deletion in the V3 loop is boxed. The number of clones matching the indicated sequence per total number of clones sequenced is given in parentheses to the right of the net charge. (B) Entry into TZM-bl, U87.CD4.CCR5, and U87.CD4.CXCR4 indicator cells of pseudovirions from FI08 bearing WT Env gp160, a variant with a 4-aa deletion in the V3 loop (ΔIGDI), and a recombinant in which the ZM109F.PB4 V3 sequence was replaced with that of the ΔIGDI variant (V3ΔIGDI). RLU, relative light units. Data are means and standard deviations from triplicate wells and are representative of two independent experiments.
S/HIVE in SHIVC109P3- and SHIVC109P3N infected-macaques.
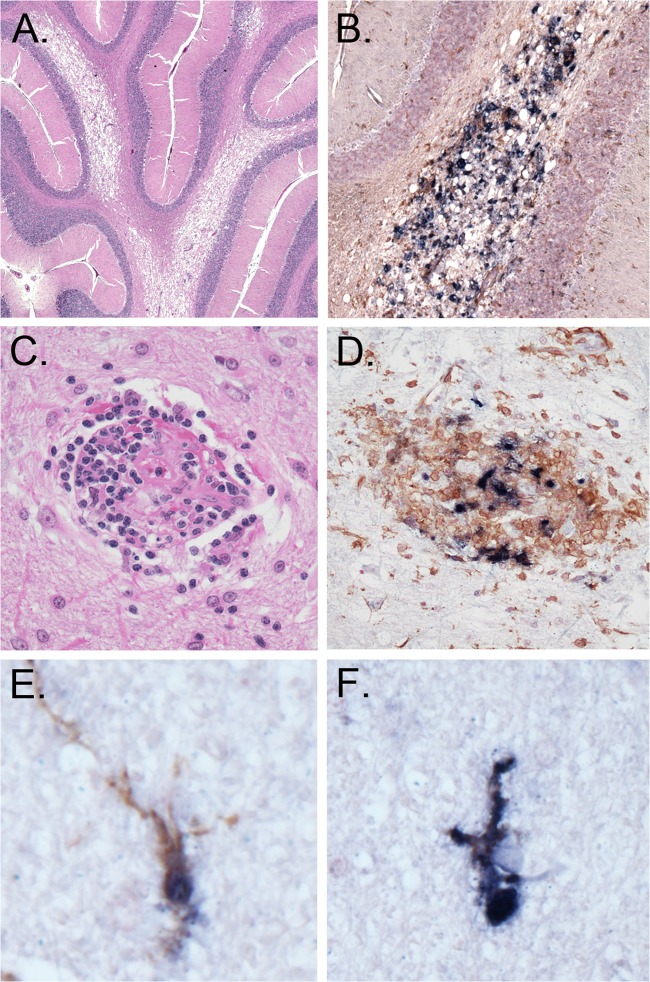
The development of central nervous system (CNS) disorders during AIDS has been documented for HIV-1-infected individuals and for SIV- and X4 and R5X4 SHIV-infected macaques (49–52). We recently observed severe, multifocal SHIV-induced giant cell encephalitis (S/HIVE) in macaques infected with the pathogenic subtype B R5 SHIVSF162P3N, with several unique and classical HIVE features, including expansive lesions with spongiosis and necrosis, deep gray matter involvement, microglial infection with high levels of HLA-DR expression, and burnt-out lesions (Westmoreland et al., presented at the 2012 Conference on HIV in the Nervous System). Histopathologic examination of the brains from SHIVC109P3- and SHIVC109P3N-infected macaques with AIDS (GC98 and FI08, respectively) revealed vacuolar leukoencephalopathy in white-matter tracts, but few multinucleated giant cells (MNGCs) were observed (Fig. 7A to D). Furthermore, infected microglia in the SHIVC-infected animals remained ramified, indicative of a quiescent or semiactivated state (Fig. 7E and F). These observations demonstrate that subtype C R5 SHIVs can also induce CNS disease in rhesus monkeys, but the lesions appeared to differ between the subtype B- and C-infected animals.
Fig 7.
Neuropathological lesions in the brain of macaque GC98. Histological staining, immunohistochemistry, and double-label in situ hybridization staining of brain sections were carried out. (A) Vacuolar leukoencephalopathy (hematoxylin and eosin staining; cerebellum); (B) vacuolar change with associated infected macrophages (SIV ISH and IHC for the macrophage marker Iba-1; cerebellum); (C) representative perivascular lesion of infected macrophages devoid of MNGCs (hematoxylin and eosin staining; frontal cortex); (D) perivascular lesion of infected macrophages (SIV ISH and IHC for Iba-1; frontal cortex); (E and F) ramified microglia (SIV ISH and IHC for Iba-1; thalamus).
DISCUSSION
CCR5-tropic, mucosally transmissible, pathogenic SHIVs carrying the envelopes of strains belonging to clade C, the most prevalent HIV-1 subtype worldwide, are needed as tools in NHPs to advance the discovery of effective and widely useful antibody-based AIDS vaccines. We report in this study the construction of the SHIVC109F.PB4 molecular clone, expressing the envelope of a recently transmitted, CCR5-tropic, sCD4-sensitive HIV-1C isolate, for serial passaging in rhesus monkeys to generate pathogenic strains. We showed that while the early (SHIVC109P3) and late-stage (SHVC109P3N) viruses recovered from a passage-3 macaque with a weak antiviral humoral response remained sensitive to sCD4 and antibody neutralization, an early SHIV (SHIVC109P4) isolated from a chronically infected passage-4 animal that mounted a strong neutralizing antibody response is now resistant. The three rhesus-adapted, lineage-related viruses remained exclusively R5 tropic, were transmitted efficiently via intrarectal exposure, replicated to high peak viral RNA levels, and induced acute depletion of CD4+ lamina propria T lymphocytes, with persistent infection in a majority of the SHIVC109P3- and SHIVC109P4-infected animals. Moreover, progression to AIDS was seen in macaques infected with SHIVC109P3 and SHIVC109P3N, with coreceptor switching and the development of multifocal SIV-induced giant cell encephalitis at end-stage disease. Collectively, the data support the use of these three newly developed lineage-related SHIVC viruses to assess vaccine efficacy, with prevention of virus acquisition, lowering of peak viremia, or depletion of CD4+ T cells in the gut as virologic and immunologic endpoints. The evolutionary pathway needed for efficient CXCR4 usage and induction of neuroAIDS by viruses with a subtype C R5 envelope can also be investigated.
Progression to AIDS was seen in two of six macaques infected with the late-stage isolate SHIVC109P3N, suggesting that protection from disease progression can be an additional criterion of vaccine efficacy when this virus is used for challenge. However, in contrast to macaques infected with SHIVC109P3 and SHIVC109P4, the other four SHIVC109P3N-infected monkeys controlled their infection to low or undetectable levels at 15 to 20 wpi. Additional animals will be required in order to determine if this is a common outcome of R5 SHIVC109P3N chronic infection, which could limit the utility of this virus for evaluating vaccines that rely on reductions in set points for viral load. Although there is still controversy regarding the role of autologous NAb responses in viral control, significant increases in postacute viremia have been reported for SIV-infected macaques depleted of B cells by treatment with an anti-CD20 antibody (53, 54). Clinical treatment for lymphocytic B cell lymphoma with B cell depletion also resulted in increased viremia in an HIV-1-infected individual (55). Thus, a possible reason for spontaneous viral control in the SHIVC109P3N-infected macaques could be the neutralization sensitivity of this isolate. Compared to SHIVC109P3 and SHIVC109P4, SHIVC109P3N is 3- to 300-fold more sensitive to neutralization by purified Igs and sera from patients chronically infected with clade B or C (Table 1), suggesting that the late isolate gained virulence at the expense of immune defense. In this scenario, SHIVC109P3N can induce disease in monkeys that mount a weak or undetectable antiviral antibody response but will be readily suppressed when humoral immune selection pressure develops. The absolute CD4+ T cell count has remained below 200/μl of blood for more than 3 months in the passage-4 macaque GC87, with a sustained viral load of >105 RNA copies/ml plasma that has increased by 1 log unit recently. SHIVC109P4, the early isolate from GC87, is highly resistant to neutralization by antibodies. It will be of interest to determine if viruses recovered from this animal at terminal disease become more pathogenic without a tradeoff in immune evasion. Regardless, the relatively high steady-state viremia (104 to 106 RNA copies/ml plasma) in a significant portion of the SHIVC109P3- and SHIVC109P4-infected macaques indicates that these viruses may be useful for testing antiviral inhibitors or vaccines aimed at reducing chronic viral replication.
Genetic analysis suggests that differences in carbohydrate moieties on the envelope glycoproteins of the three rhesus-adapted SHIVC109F.PB4-derived viruses contributed to the differences in their neutralization sensitivities (Fig. 3). Both SHIVC109P3 and SHIVC109P3N lost an N-linked glycan (N197D) just outside the carboxy terminus of the V2 loop, which has been shown to render HIV-1B viruses sensitive to antibodies directed at the CD4BS, V3, CD4-induced, and MPER epitopes by altering the position of the V1V2 loop (56–58). An additional loss of a PNGS within the V1V2 loop of SHIVC109P3N was seen in comparison to the early isolate SHIVC109P3; this loss could have played a role in conferring on SHIVC109P3N a more neutralization sensitive phenotype overall. In contrast, the neutralization-resistant virus SHIVC109P4 did not lose or gain any PNGS. Rather, the pattern of glycosylation was altered as a result of shifts in the sugar moieties in the V1V2, C3, and V4 regions, highlighting the importance of the positioning of the glycan shield in immune evasion. Notable is the shift in glycan from position 334 to 332 in the C3 region of SHIVC109P4. The N332 glycan is important for the formation of the epitopes recognized by the glycan-specific monoclonal antibody 2G12 (59) and the potent broadly cross neutralizing (BCN) PGT NAbs (60), the latter often found in HIV-1-infected individuals who have developed neutralization breadth (60–62). SHIVC109P4 is resistant to neutralization by 2G12 (Table 1), consistent with the dependency of this epitope on glycans at other amino acids that are uncommon among subtype C viruses (63). It will be of interest to determine whether, like that occurring naturally in HIV-1C human infection (64), the 334-to-332 glycan shift in SHIVC109P4 created the BCN PGT epitope as a means of immune escape and whether this epitope elicits BCN responses in SHIVC109P4-infected macaques.
Despite vigorous in vivo replication and adaptation, the passaged viruses maintained CCR5 specificity. Moreover, as in our findings with the late R5 subtype B SHIVSF162P3N, induction of S/HIVE and tropism switching were observed in one of the two SHIVC109P3N-infected RPs, indicating that these pathological sequelae of RM infection are shared by passaged CCR5-tropic SHIVs irrespective of the clade of origin of the HIV-1 envelope glycoprotein. As seen in HIV-1-infected individuals and in subtype B R5 SHIVSF162P3N-infected rhesus macaques, amino acid changes in the V3 loop of SHIVC109F.PB4 determine CXCR4 use (Fig. 6A). Furthermore, in agreement with findings for CXCR4 usage in HIV-1C infection, there was no evidence of positively charged amino acids at positions 11 and 25 (65), but the Q at position 18 in the V3 loop of FI08 was replaced with R, and there were deletions near the C terminus of the V3 loop (positions 27 to 30) that could influence coreceptor choice (22, 66–68). However, in contrast to findings in which deletions within the V3 loop render the virus dualtropic in SHIVSF162P3N-infected rhesus macaques and typically result in loss of R5 HIV-1 B Env function (69–72), the V3 deletion variant (ΔIGDI) in FI08 uses CXCR4 exclusively but is less efficient at entry than WT viruses (Fig. 6B). The V3 loop of HIV-1C has been suggested to possess a rigid or non-β-strand structure that differs from subtype B V3 conformations and could impose functional constraints (21, 73). Thus, it is tempting to speculate that the rigid V3 configuration of subtype C R5 Envs limits the flexibility to evolve so as to use CXCR4 and that deletions in this domain release such constraints. Studies in a larger cohort of SHIVC-infected animals, in comparison to subtype B R5 SHIVSF162P3N-infected monkeys (45, 74), may help us to understand if the reported lower rate of coreceptor switching in subtype C-infected humans is indeed related to differences in envelope antigenic structure and evolutionary pathways to efficient CXCR4 usage.
The development of S/HIVE was observed in two of the three macaques with SHIVC AIDS, one each infected with SHIVC109P3 (GC98) and SHIVC109P3N (FI08). The incidence and severity of neurological impairment have been reported to be lower among subtype C-infected patients than among subtype B-infected individuals (75–78), with one study showing that MNGCs and multifocal microglial nodules, frequently found in clade B-infected patients, are absent in the brains of clade C-infected individuals (79). Initial analysis of a limited number of macaques with AIDS that had been infected i.r. with subtype B (n = 10) (Westmoreland et al., presented at the 2012 Conference on HIV in the Nervous System) or subtype C (n = 3) (this study) failed to support a lower incidence of S/HIVE in the latter (40 and 75%, respectively). However, CNS lesions appeared to differ, with more vacuolar leukoencephalopathy in white-matter tracts but fewer MNGCs observed in SHIVC-infected rhesus macaques (Fig. 7) (Westmoreland et al., presented at the 2012 Conference on HIV in the Nervous System). Infected microglia were found in both subtype B- and subtype C-infected monkeys, but the pattern differed in that subtype C-infected microglia remained ramified, whereas subtype B-infected microglia had rounded and amoeboid morphology, consistent with activation with high levels of HLA-DR expression. These preliminary observations of subtype differences in the neuropathology of SHIV-infected macaques are intriguing and merit further analysis.
We chose an envelope that is sCD4 sensitive for SHIV construction and adaption in RMs, hypothesizing that it would replicate efficiently and thrive in a new host. We reasoned that a clonal virus with an “open” envelope structure that exposes the receptor binding site for efficient CD4 binding would be able to adapt more easily to variations in expression levels of the viral receptors in the new host, enabling it to replicate to high peak levels in an immune-naïve environment. An “open” Env would also provide greater flexibility in accommodating and accumulating the mutations necessary to generate the degree of genetic diversity needed for virus adaptation and to escape early selective pressure through recombination. Adaptation of subtype A HIV-1 Envs to pig-tailed macaque cells in vitro has been reported to render them sCD4 sensitive (80), and this correlated with the ability of the envelope to mediate entry using macaque CD4 (81), leading to the suggestion that sensitivity to sCD4 may provide a means to identify representative transmitted Env variants that would be the best candidates for a successful SHIV. Our success in generating a series of highly replication competent SHIVC through serial passaging in RMs of the sCD4-sensitive molecular clone SHIVC109F.PB4 lends further support to this notion. Moreover, the significantly higher peak viral loads (week 2; P < 0.05) (Fig. 4D) and progressive disease observed in RMs infected intrarectally with the sCD4-sensitive SHIVC109F.PB4-derived strains (SHIVC109P3 and SHIVC109P3N) than in RMs infected with a lineage-related but more resistant strain (SHIVC109P4) suggest that an “open” Env structure may indeed predispose to high levels of virus replication. Construction and testing of additional SHIVs will be required to determine whether the use of sCD4-sensitive Envs from diverse HIV-1 subtypes provides a greater chance of success in generating highly infectious and pathogenic SHIVs.
In conclusion, we show that Asian macaques infected with the newly generated R5 SHIVC109F.PB4-derived viruses exhibited many similarities to HIV-infected individuals, including CCR5 coreceptor usage, mucosal transmission, high acute viremia and persistent infection, depletion of mucosal CD4+ T cells, and progression to AIDS with neurological disorders and coreceptor switching. Our study has significance for HIV-1/AIDS prevention research, providing useful tools in NHPs to assess the efficacy of candidate vaccines and therapeutics targeting the envelope of the most prevalent HIV-1 subtype.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Derdeyn the Vaccine Center at Yerkes National Primate Research Center for the gift of ZM109F.PB4. The TZM-bl cells (catalog no. 8129; from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc.), U87.CD4 indicator cell lines (catalog no. 4035 and 4036; from HongKui Deng and Dan Littman), AMD100 (catalog no. 8128), and TAK779 (catalog no. 4983) were obtained through the NIH AIDS Research and Reagent Program, Division of AIDS, NIAID, NIH, and the rhesus anti-CD4 antibody M-T807R1 was obtained through the NIH Nonhuman Primate Regent Resource.
This work was supported by National Institutes of Health grant R01AI46980. Additional support was provided by the Tulane National Primate Research Center Base grant RR00164.
Footnotes
Published ahead of print 20 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00178-13.
REFERENCES
- 1. Hemelaar J. 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 18:182–192 [DOI] [PubMed] [Google Scholar]
- 2. Kim SC, Becker S, Dieffenbach C, Hanewall BS, Hankins C, Lo YR, Mellors JW, O'Reilly K, Paxton L, Roffenbender JS, Warren M, Piot P, Dybul MR. 2010. Planning for pre-exposure prophylaxis to prevent HIV transmission: challenges and opportunities. J. Int. AIDS Soc. 13:24 doi:10.1186/1758-2652-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Underhill K, Operario D, Mimiaga MJ, Skeer MR, Mayer KH. 2010. Implementation science of pre-exposure prophylaxis: preparing for public use. Curr. HIV/AIDS Rep. 7:210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen MS, Baden LR. 2012. Preexposure prophylaxis for HIV—where do we go from here? N. Engl. J. Med. 367:459–461 [DOI] [PubMed] [Google Scholar]
- 5. De Cock KM, Crowley SP, Lo YR, Granich RM, Williams BG. 2009. Preventing HIV transmission with antiretrovirals. Bull. World Health Organ. 87:488–488A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gostin LO, Kim SC. 2011. Ethical allocation of preexposure HIV prophylaxis. JAMA 305:191–192 [DOI] [PubMed] [Google Scholar]
- 7. Global HIV/AIDS Vaccine Enterprise 2005. The Global HIV/AIDS Vaccine Enterprise: scientific strategic plan. PLoS Med. 2:e25 doi:10.1371/journal.pmed.0020025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klausner RD, Fauci AS, Corey L, Nabel GJ, Gayle H, Berkley S, Haynes BF, Baltimore D, Collins C, Douglas RG, Esparza J, Francis DP, Ganguly NK, Gerberding JL, Johnston MI, Kazatchkine MD, McMichael AJ, Makgoba MW, Pantaleo G, Piot P, Shao Y, Tramont E, Varmus H, Wasserheit JN. 2003. Medicine. The need for a global HIV vaccine enterprise. Science 300:2036–2039 [DOI] [PubMed] [Google Scholar]
- 9. Morgan C, Marthas M, Miller C, Duerr A, Cheng-Mayer C, Desrosiers R, Flores J, Haigwood N, Hu SL, Johnson RP, Lifson J, Montefiori D, Moore J, Robert-Guroff M, Robinson H, Self S, Corey L. 2008. The use of nonhuman primate models in HIV vaccine development. PLoS Med. 5:e173 doi:10.1371/journal.pmed.0050173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Etemad-Moghadam B, Karlsson GB, Halloran M, Sun Y, Schenten D, Fernandes M, Letvin NL, Sodroski J. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J. Virol. 72:8437–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kraft Z, Derby NR, McCaffrey RA, Niec R, Blay WM, Haigwood NL, Moysi E, Saunders CJ, Wrin T, Petropoulos CJ, McElrath MJ, Stamatatos L. 2007. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J. Virol. 81:6402–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyazaki Y, Kuwata T, Takehisa J, Hayami M. 2002. Analysis of a primary isolate-like virus from simian and human immunodeficiency virus-infected macaque having broad neutralizing activity. AIDS Res. Hum. Retroviruses 18:469–475 [DOI] [PubMed] [Google Scholar]
- 13. Montefiori DC, Reimann KA, Wyand MS, Manson K, Lewis MG, Collman RG, Sodroski JG, Bolognesi DP, Letvin NL. 1998. Neutralizing antibodies in sera from macaques infected with chimeric simian-human immunodeficiency virus containing the envelope glycoproteins of either a laboratory-adapted variant or a primary isolate of human immunodeficiency virus type 1. J. Virol. 72:3427–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malenbaum SE, Yang D, Cheng-Mayer C. 2001. Evidence for similar recognition of the conserved neutralization epitopes of human immunodeficiency virus type 1 envelope gp120 in humans and macaques. J. Virol. 75:9287–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derby NR, Kraft Z, Kan E, Crooks ET, Barnett SW, Srivastava IK, Binley JM, Stamatatos L. 2006. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J. Virol. 80:8745–8762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robinson JE, Franco K, Elliott DH, Maher MJ, Reyna A, Montefiori DC, Zolla-Pazner S, Gorny MK, Kraft Z, Stamatatos L. 2010. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. J. Virol. 84:3443–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker LM, Sok D, Nishimura Y, Donau O, Sadjadpour R, Gautam R, Shingai M, Pejchal R, Ramos A, Simek MD, Geng Y, Wilson IA, Poignard P, Martin MA, Burton DR. 2011. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc. Natl. Acad. Sci. U. S. A. 108:20125–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaleebu P, Nankya IL, Yirrell DL, Shafer LA, Kyosiimire-Lugemwa J, Lule DB, Morgan D, Beddows S, Weber J, Whitworth JA. 2007. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D: results from a rural Ugandan cohort. J. Acquir. Immune Defic. Syndr. 45:28–33 [DOI] [PubMed] [Google Scholar]
- 20. Schuitemaker H, van 't Wout AB, Lusso P. 2011. Clinical significance of HIV-1 coreceptor usage. J. Transl. Med. 9(Suppl. 1):S5 doi:10.1186/1479-5876-9-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel MB, Hoffman NG, Swanstrom R. 2008. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J. Virol. 82:903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coetzer M, Cilliers T, Ping LH, Swanstrom R, Morris L. 2006. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology 356:95–105 [DOI] [PubMed] [Google Scholar]
- 23. Raymond S, Delobel P, Mavigner M, Ferradini L, Cazabat M, Souyris C, Sandres-Saune K, Pasquier C, Marchou B, Massip P, Izopet J. 2010. Prediction of HIV type 1 subtype C tropism by genotypic algorithms built from subtype B viruses. J. Acquir. Immune Defic. Syndr. 53:167–175 [DOI] [PubMed] [Google Scholar]
- 24. Cayabyab M, Rohne D, Pollakis G, Mische C, Messele T, Abebe A, Etemad-Moghadam B, Yang P, Henson S, Axthelm M, Goudsmit J, Letvin NL, Sodroski J. 2004. Rapid CD4+ T-lymphocyte depletion in rhesus monkeys infected with a simian-human immunodeficiency virus expressing the envelope glycoproteins of a primary dual-tropic Ethiopian clade C HIV type 1 isolate. AIDS Res. Hum. Retroviruses 20:27–40 [DOI] [PubMed] [Google Scholar]
- 25. Chen Z, Huang Y, Zhao X, Skulsky E, Lin D, Ip J, Gettie A, Ho DD. 2000. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina). J. Virol. 74:6501–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ndung'u T, Lu Y, Renjifo B, Touzjian N, Kushner N, Pena-Cruz V, Novitsky VA, Lee TH, Essex M. 2001. Infectious simian/human immunodeficiency virus with human immunodeficiency virus type 1 subtype C from an African isolate: rhesus macaque model. J. Virol. 75:11417–11425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y, Hong K, Chenine AL, Whitney JB, Xu W, Chen Q, Geng Y, Ruprecht RM, Shao Y. 2005. Molecular cloning and in vitro evaluation of an infectious simian-human immunodeficiency virus containing env of a primary Chinese HIV-1 subtype C isolate. J. Med. Primatol. 34:101–107 [DOI] [PubMed] [Google Scholar]
- 28. Siddappa NB, Song R, Kramer VG, Chenine AL, Velu V, Ong H, Rasmussen RA, Grisson RD, Wood C, Zhang H, Kankasa C, Amara RR, Else JG, Novembre FJ, Montefiori DC, Ruprecht RM. 2009. Neutralization-sensitive R5-tropic simian-human immunodeficiency virus SHIV-2873Nip, which carries env isolated from an infant with a recent HIV clade C infection. J. Virol. 83:1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J. Virol. 80:8729–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Humbert M, Rasmussen RA, Song R, Ong H, Sharma P, Chenine AL, Kramer VG, Siddappa NB, Xu W, Else JG, Novembre FJ, Strobert E, O'Neil SP, Ruprecht RM. 2008. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology 5:94 doi:10.1186/1742-4690-5-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho SH, Tasca S, Shek L, Li A, Gettie A, Blanchard J, Boden D, Cheng-Mayer C. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 81:8621–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ren W, Tasca S, Zhuang K, Gettie A, Blanchard J, Cheng-Mayer C. 2010. Different tempo and anatomic location of dual-tropic and X4 virus emergence in a model of R5 simian-human immunodeficiency virus infection. J. Virol. 84:340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 34. Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC [Google Scholar]
- 37. Goulder PJ, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delwart EL, Gordon CJ. 1997. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 12:348–354 [DOI] [PubMed] [Google Scholar]
- 39. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, Engl.) 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 40. Ho SH, Shek L, Gettie A, Blanchard J, Cheng-Mayer C. 2005. V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques. J. Virol. 79:12296–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kimata JT, Kuller L, Anderson DB, Dailey P, Overbaugh J. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535–541 [DOI] [PubMed] [Google Scholar]
- 43. Harouse JM, Gettie A, Eshetu T, Tan RC, Bohm R, Blanchard J, Baskin G, Cheng-Mayer C. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIVSF162P3. J. Virol. 75:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blaak H, Brouwer M, Ran LJ, de Wolf F, Schuitemaker H. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600–610 [DOI] [PubMed] [Google Scholar]
- 45. Shakirzyanova M, Tsai L, Ren W, Gettie A, Blanchard J, Cheng-Mayer C. 2012. Pathogenic consequences of vaginal infection with CCR5-tropic simian-human immunodeficiency virus SHIVSF162P3N. J. Virol. 86:9432–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, Abdool Karim SS, Williamson C, Morris L, CAPRISA 002 Study Team 2008. The C3-V4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, Robinson JE, Gnanakaran S, Derdeyn CA. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594 doi:10.1371/journal.ppat.1000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Buch S, Pinson D, Hou Y, Adany I, Li Z, Mukherjee S, Jia F, Mackay G, Silverstein P, Kumar A, Narayan O. 2000. Neuropathogenesis of chimeric simian human immunodeficiency virus infection in rhesus macaques. J. Med. Primatol. 29:96–106 [DOI] [PubMed] [Google Scholar]
- 50. Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. 1988. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science 239:586–592 [DOI] [PubMed] [Google Scholar]
- 51. Lackner AA, Dandekar S, Gardner MB. 1991. Neurobiology of simian and feline immunodeficiency virus infections. Brain Pathol. 1:201–212 [DOI] [PubMed] [Google Scholar]
- 52. Raghavan R, Stephens EB, Joag SV, Adany I, Pinson DM, Li Z, Jia F, Sahni M, Wang C, Leung K, Foresman L, Narayan O. 1997. Neuropathogenesis of chimeric simian/human immunodeficiency virus infection in pig-tailed and rhesus macaques. Brain Pathol. 7:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miller CJ, Genesca M, Abel K, Montefiori D, Forthal D, Bost K, Li J, Favre D, McCune JM. 2007. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J. Virol. 81:5024–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmitz JE, Kuroda MJ, Santra S, Simon MA, Lifton MA, Lin W, Khunkhun R, Piatak M, Lifson JD, Grosschupff G, Gelman RS, Racz P, Tenner-Racz K, Mansfield KA, Letvin NL, Montefiori DC, Reimann KA. 2003. Effect of humoral immune responses on controlling viremia during primary infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 77:2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang KH, Bonsall D, Katzourakis A, Thomson EC, Fidler SJ, Main J, Muir D, Weber JN, Frater AJ, Phillips RE, Pybus OG, Goulder PJ, McClure MO, Cooke GS, Klenerman P. 2010. B-cell depletion reveals a role for antibodies in the control of chronic HIV-1 infection. Nat. Commun. 1:102 doi:10.1038/ncomms1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:e1001028 doi:10.1371/journal.ppat.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, CAPRISA 002 Study Team 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gray ES, Moore PL, Pantophlet RA, Morris L. 2007. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J. Virol. 81:10769–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 18:1688–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jensen MA, Coetzer M, van 't Wout AB, Morris L, Mullins JI. 2006. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J. Virol. 80:4698–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cormier EG, Dragic T. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953 [DOI] [PubMed] [Google Scholar]
- 68. Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang ZY, Chakrabarti BK, Xu L, Welcher B, Kong WP, Leung K, Panet A, Mascola JR, Nabel GJ. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 78:4029–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laakso MM, Lee FH, Haggarty B, Agrawal C, Nolan KM, Biscone M, Romano J, Jordan AP, Leslie GJ, Meissner EG, Su L, Hoxie JA, Doms RW. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3:e117 doi:10.1371/journal.ppat.0030117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shakirzyanova M, Ren W, Zhuang K, Tasca S, Cheng-Mayer C. 2010. Fitness disadvantage of transitional intermediates contributes to dynamic change in the infecting-virus population during coreceptor switch in R5 simian/human immunodeficiency virus-infected macaques. J. Virol. 84:12862–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Almond D, Krachmarov C, Swetnam J, Zolla-Pazner S, Cardozo T. 2012. Resistance of subtype C HIV-1 strains to anti-V3 loop antibodies. Adv. Virol. 2012:803535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhuang K, Finzi A, Tasca S, Shakirzyanova M, Knight H, Westmoreland S, Sodroski J, Cheng-Mayer C. 2011. Adoption of an “open” envelope conformation facilitating CD4 binding and structural remodeling precedes coreceptor switch in R5 SHIV-infected macaques. PLoS One 6:e21350 doi:10.1371/journal.pone.0021350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kolson DL, Gonzalez-Scarano F. 2000. HIV and HIV dementia. J. Clin. Invest. 106:11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Clifford DB, Mitike MT, Mekonnen Y, Zhang J, Zenebe G, Melaku Z, Zewde A, Gessesse N, Wolday D, Messele T, Teshome M, Evans S. 2007. Neurological evaluation of untreated human immunodeficiency virus infected adults in Ethiopia. J. Neurovirol. 13:67–72 [DOI] [PubMed] [Google Scholar]
- 77. Riedel D, Ghate M, Nene M, Paranjape R, Mehendale S, Bollinger R, Sacktor N, McArthur J, Nath A. 2006. Screening for human immunodeficiency virus (HIV) dementia in an HIV clade C-infected population in India. J. Neurovirol. 12:34–38 [DOI] [PubMed] [Google Scholar]
- 78. Robertson K, Kopnisky K, Hakim J, Merry C, Nakasujja N, Hall C, Traore M, Sacktor N, Clifford D, Newton C, Van Rie A, Holding P, Clements J, Zink C, Mielk J, Hosseinipour M, Lalloo U, Amod F, Marra C, Evans S, Liner J, Second Assessment of NeuroAIDS in Africa Conference Participants 2008. Second assessment of neuroAIDS in Africa. J. Neurovirol. 14:87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mahadevan A, Shankar SK, Satishchandra P, Ranga U, Chickabasaviah YT, Santosh V, Vasanthapuram R, Pardo CA, Nath A, Zink MC. 2007. Characterization of human immunodeficiency virus (HIV)-infected cells in infiltrates associated with CNS opportunistic infections in patients with HIV clade C infection. J. Neuropathol. Exp. Neurol. 66:799–808 [DOI] [PubMed] [Google Scholar]
- 80. Humes D, Overbaugh J. 2011. Adaptation of subtype A human immunodeficiency virus type 1 envelope to pig-tailed macaque cells. J. Virol. 85:4409–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Humes D, Emery S, Laws E, Overbaugh J. 2012. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J. Virol. 86:12472–12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.