Abstract
Enterovirus 71 (EV71) has emerged as a major cause of neurological disease following the near eradication of poliovirus. Accumulating evidence suggests that mammalian microRNAs (miRNAs), a class of noncoding RNAs of 18 to 23 nucleotides (nt) with important regulatory roles in many cellular processes, participate in host antiviral defenses. However, the roles of miRNAs in EV71 infection and pathogenesis are still unclear. Here, hsa-miR-296-5p expression was significantly increased in EV71-infected human cells. As determined by virus titration, quantitative real-time PCR (qRT-PCR), and Western blotting, overexpression of hsa-miR-296-5p inhibited, while inhibition of endogenous hsa-miR-296-5p facilitated, EV71 infection. Additionally, two potential hsa-miR-296-5p targets (nt 2115 to 2135 and nt 2896 to 2920) located in the EV71 genome (strain BrCr) were bioinformatically predicted and validated by luciferase reporter assays and Western blotting. Genomic alignment of various EV71 strains revealed synonymous mutations in hsa-miR-296-5p target sequences. Furthermore, the introduction of synonymous mutations into the EV71 BrCr genome by site-directed mutagenesis impaired the viral inhibitory effects of hsa-miR-296-5p and facilitated mutant virus infection. Meanwhile, compensatory mutations in corresponding hsa-miR-296-5p target sequences of the EV71 HeN strain (GenBank accession number JN256064) restored the inhibitory effects of the miRNA. These results indicate that hsa-miR-296-5p inhibits EV71 replication by targeting the viral genome. Our findings support the notion that cellular miRNAs can inhibit virus infection and that the virus mutates to escape suppression by cellular miRNAs.
INTRODUCTION
MicroRNAs (miRNAs) are a class of highly conserved 21- to 23-nucleotide (nt) RNA molecules that exist in almost all eukaryotes (1–3). They can regulate many cellular processes, including cell proliferation and differentiation, apoptosis, development, and host defense (4–6). miRNA precursors (preRNA) are cleaved by the RNase III Dicer, and one chain of the double-stranded RNA can direct the RNA-induced silencing complex (RISC) to target mRNA sequences for subsequent cleavage or translation inhibition (7). The result is determined by the degree of complementarity between the miRNA and target mRNA. In the case of a perfect sequence match, the recruitment of RISC mediated by miRNAs can lead to mRNA degradation. In the case of imperfect sequence complementarity, miRNA binding leads to inhibition of mRNA translation (8). In most common plants or lower animals, perfect or near-perfect complementarity results in the degradation of target mRNA (9). However, in most vertebrates, base pairing of miRNA and mRNA is imperfect, and although the mRNA is not cleaved, its translation efficiency is decreased. Under normal circumstances, perfect pairing between the 5′ end of the miRNA (nt 2 to 8, known as the seed sequence) and the 3′ untranslated region (UTR) of the mRNA is considered critical to the regulatory function of miRNA translation (10–12).
A growing body of evidence has demonstrated that miRNAs play a key role in the regulation of viral replication and gene expression (13, 14). Some viruses such as Epstein-Barr virus and herpes simplex virus (HSV) can regulate host and viral gene expression by virally encoded miRNAs (15, 16). Interestingly, recent studies have shown that a small RNA encoded by the West Nile virus has a role similar to that of cellular miRNA and that this small RNA can enhance viral replication in mosquito cells (17). In addition, as an antiviral defense mechanism, some intracellular miRNAs can target viral mRNA(s) to inhibit replication (18). For example, miR-32-1 targets the primate foamy virus (PFV) genome and can efficiently suppress viral replication (19). Two cellular miRNAs, miR-24 and miR-93, can inhibit vesicular stomatitis virus (VSV) replication through targeting the genes encoding the viral L protein and P protein, respectively (20). The interferon (IFN)-induced miRNAs (miR-196's of miR-296, miR-351, miR-431, and miR-448) have perfect complementarity to the hepatitis C virus (HCV) genome and can inhibit HCV replication in human hepatocellular carcinoma Huh7 cells (21). MiR-125-5p targets the S protein of hepatitis B virus (HBV) to inhibit its replication (22). Meanwhile, miR-122 can inhibit HBV replication by binding to a highly conserved region of the viral genome (23). By binding to the 3′ UTR region, miR-29 can inhibit the replication of human immunodeficiency virus type 1 (HIV-1) (24). However, some viruses suppressed by cellular miRNAs may in turn affect miRNA abundance by synthesis of viral RNAs or proteins, such as the Tas protein of PFV (19) and the HIV-1 Tat protein, which can abrogate the cellular RNA-silencing defense mechanism by subverting the ability of Dicer to process precursor double-stranded RNAs into small interfering RNAs (siRNAs) (25). Vaccinia virus protein E3L, influenza A virus protein NS1, and Ebola virus protein VP35 all act as broadly effective suppressors of RNA silencing, similar to the effect seen with the HIV-1 Tat protein (26). As the evidence for miRNA-mediated regulation of virus infection has only just begun to emerge, detailed investigations of the roles of miRNAs in virus replication and protein expression may contribute to a better understanding of host-pathogen interactions, the orientation of the virus, and development of new diagnostic biomarkers and treatments.
Enterovirus 71 (EV71) is a member of the Picornaviridae family and belongs to the genus Enterovirus (human enterovirus A). EV71 has become one of the most important neurotropic enteroviruses since the near eradication of poliovirus. It is known to cause hand, foot, and mouth disease (HFMD) in young children and lead to the development of severe neurological diseases, including aseptic meningitis, cerebellar encephalitis, and acute flaccid paralysis, culminating in fatality in some patients, especially children (27). EV71 infection can change the expression of miRNAs in host cells (28), which are likely to play a role in the viral pathogenesis. Indeed, EV71 has been shown to induce the expression of miRNA-141, which can target the eIF4E translation initiation factor to downregulate host protein synthesis (29). In addition, miR-342-5p suppresses the biosynthesis of coxsackievirus B3 (CVB3), another human enterovirus, by targeting the 2C-coding region on the viral genome (30). Since there is strong evidence that cellular miRNAs can be used by host cells to resist viral infection, it is possible that EV71 replication can be suppressed by host miRNAs. However, the involvement of miRNAs during EV71 infection or replication is still unclear.
In this study, we found elevated levels of miRNA-296-5p in EV71-infected cells and predicted by bioinformatic analysis that the miRNA could interact directly with the viral genomic RNA. Overexpression and inhibition of hsa-miR-296-5p and site-directed mutagenesis of the EV71 genome were carried out to investigate the effects and mechanism of this miRNA on viral replication. This study provides further insights into the role of miRNA in EV71 pathogenesis, which may aid in drug development for treating EV71 infections.
MATERIALS AND METHODS
Cells, synthetic oligonucleotides, and antibodies.
Vero cells (derived from African green monkey kidney cells) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% or 10% fetal bovine serum (FBS) and were used for RNA transfections and rescue of EV71 virus from an infectious clone. RD (rhabdomyosarcoma) and SK-N-SH (human neuroblastoma) cells were maintained in minimum essential medium (MEM) containing 2 or 10% FBS. Synthetic oligonucleotides representing the miR-296-5p mimics, the negative control for mimics, the miR-296-5p inhibitor (antisense oligonucleotides to the mature has-miR-296-5p according to sequences in the miRBase), and the negative control for the inhibitor all were obtained from RiBo Biotech (Guangzhou, China). Negative-control sequences are based on Caenorhabditis elegans miRNA cel-miR-67-3p. The sequences for these oligonucleotides are listed in Table 1. The EV71 VP1 antibody was obtained from Millipore (Billerica, MA, USA), and anti-actin antibodies were purchased from Proteintech (Wuhan, China). The mouse anti-IFNAR monoclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Table 1.
Primers used for plasmid construction and qRT-PCRa
| Primer name | Sequence (5′–3′) | Purpose |
|---|---|---|
| F2833 | ACC GAATTCACGCAGAGTTCACTTTTGTCG | Cloning miR-296 target sequence 2896–2920 |
| R3014 | GCA CTCGAG CTTGCAGGTGACATAAATGGC | |
| F2061 | ACC GAATTC GTCACTTTTATGTTCACCGGAT | Cloning miR-296 target sequence 2115–2135 |
| R2182 | GCA CTCGAG GATTGCAAGCCAAAGTCCC | |
| M2115F | CTTATAGCTTACACACCCCCAGGAGGTCCTTTGC | Generating nucleotide mutations in the miR-296 target sequence 2115–2135 |
| M2115R | GCAAAGGACCTCCTGGGGGTGTGTAAGCTATAAG | |
| M2896F | CTTCAATACATGTTTGTTCCACCCGGAGCCCCCAAAC | Generating nucleotide mutation in the miR-296 target sequence 2896–2920 |
| M2896R | GTTTGGGGGCTCCGGGTGGAACAAACATGTATTGAAG | |
| miR-296 mimics | UGUCCUAACUCCCCCCCGGGA | Overexpression of miR-296 |
| miR-296 inhibitor | UCCCGGGGGGGAGUUAGGACA | Inhibition of miR-296 expression |
| EVBC-1468F | CTCAGTTGACAGTGTGCCCTC | Generation of nucleotide mutations in the EV71 BrCr TR strain |
| EVBC-3320R | GATTGCCGTTCGGCTGGTAC | |
| BC2-2896mutF | TCAATACATGTTTGTTCCACCCGGAGCCCCCAAACCAGACTCCAGAG | |
| BC2-2896mutR | CTCTGGAGTCTGGTTTGGGGGCTCCGGGTGGAACAAACATGTATTGA | |
| BC2-2115mutF | TATAGCTTACACACCCCCAGGAGGTCCTTTGCCTAAAGATAGAGCCAC | |
| BC2-2115mutR | GTGGCTCTATCTTTAGGCAAAGGACCTCCTGGGGGTGTGTAAGCTATA | |
| VP3F | AACAAGCTTCAGGTTTCCCCACTGAATTGAAACCTGGC | Cloning VP3 ORF or generating mutation |
| VP3R | ATT CCCGGG TTA TTGAATAGTGGCCGTTTGCAAA | |
| VP1F | CCAGAATTCAGGGGACAGAGTGGCAGATGTGATTG | Cloning VP1 ORF or generating mutation |
| VP1R | AACGGATCCTTA GAGCGTAGTGATTGCCGTTCG | |
| HNMT-2896F | CCAATATATGTTTGTGCCACCCGGGGCCCCTAAGCCAGATTCTAGG | Generation of nucleotide mutations in the EV71-HeN luc infectious clone |
| EVHN4141F | GCGCGTGCTGAAGAACGAGC | |
| HNMT-2896R | CCTAGAATCTGGCTTAGGGGCCCCGGGTGGCACAAACATATATTGG | |
| EVHN6824R | GGTTGACCACTCTAAAGTTGCCC | |
| HNMT-2115F | TCATAGCCTATACACCGCCAGGGGGCCCTCTGCCCAAGGACCGGGCGA | |
| HNMT-2115R | CGCCCGGTCCTTGGGCAGAGGGCCCCCTGGCGGTGTATAGGCTATGA | |
| EvVP1F | GAGAGTTCTATA GGGGACAGT | qRT-PCR detection of EV71 genomic RNA |
| EvVP1R | AGCTGTGCTATG TGA ATTAGG AA | |
| GAPDH-F | GAAGGTGAAGGTCGGAGTC | qRT-PCR detection of GAPDH |
| GAPDH-R | GAAGATGGTGATGGGATTTC |
Bold, site mutant nucleotide; underlining, restriction endonuclease cutting sites.
Plasmid constructs.
To generate the parent construct pCMV-luc, the DNA fragment containing the Renilla luciferase open reading frame (ORF) was obtained from the pGL3-basic vector by digestion with HindIII and inserted into the corresponding site in pCDNA3.1. The cDNA sequences containing the hsa-miR-296-5p putative target sites, nt 2896 to 2920 and nt 2115 to 2135 in the EV71 genome, were obtained by PCR with primers introducing an upstream XhoI restriction site and a downstream EcoRI site and ligated into pCMV-luc, resulting in 2896-luc and 2115-luc, respectively. The m2896-luc and m2115-luc constructs carrying mutations in the miR-296 target were obtained by overlapping PCR using 2896-luc and 2115-luc as templates and the primers listed in Table 1.
To generate the Flag-VP1 and Flag-VP3 expression plasmids, VP1- and VP3-coding sequences were amplified by PCR and inserted into the pCMV-Flag vector. The corresponding VP1m and VP3m mutants containing point mutations in the hsa-miR-296-5p target were generated by overlapping PCR using the primers listed in Table 1.
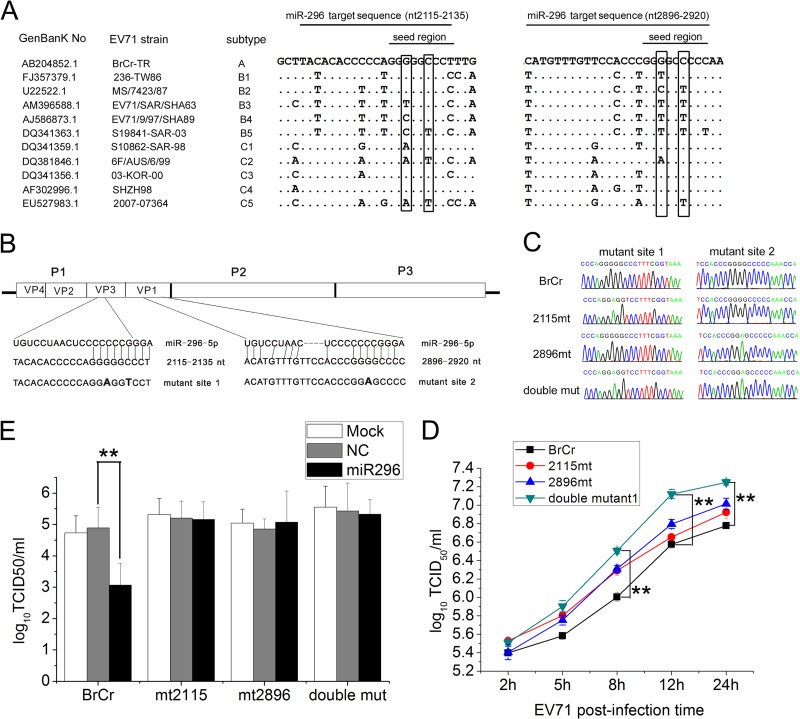
The miRNA-296 target mutant viruses (mt2115, mt2896, and the double mutant [double mut]) were generated using pEV71 (BrCr-TR) (a gift from Minetaro Arita, National Institute of Infectious Diseases, Tokyo, Japan) (31). The DNA fragment (from nt 1486 to 3320) amplified by overlapping PCR using primers listed in Table 1 was digested with EagI and MluI and used for replacement of the corresponding sequence in the full-length viral genome. The resulting cDNA clone contained mutations in the two hsa-miR-296-5p target seed sequences (see Fig. 5).
Fig 5.
Effects of synonymous mutations in hsa-miR-296-5p target sites on EV71 replication and inhibitory effects of hsa-miR-296-5p on EV71. (A) Genomic sequence alignment of EV71 strains of different subtypes. Synonymous nucleotide mutations in the seed region of the two hsa-miR-296-5p target sites are boxed. (B) Schematic representation of pEV71(BrCr-TR) constructs with the hsa-miR-296-5p target sites and their corresponding mutants. (C) Sequencing results of each mutant EV71 virus at mutation sites. (E) Effects of hsa-miR-296-5p on infectivity of WT (BrCr) and mutant (mt2115, mt2896, and double mutant) viruses. hsa-miR-296-5p-transfected RD cells were infected with each virus at an MOI of 5. Viral titers were determined at 12 h postinfection. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). **, P < 0.01. (D) Growth curves of WT and mutant viruses. SK-N-SH cells were infected with each virus at an MOI of 5, and EV71 titers of infected cells were determined. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). **, P < 0.01.
The EV71 luciferase reporter virus infectious clone pLR-EV71-HeN was a gift of Bo Zhang (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China). Point mutations in sequences corresponding to the target sequences in EV71 strains were generated by overlapping PCR using primers shown in Table 1.
Ago2-IP.
RD cells were transfected with miR-296 mimics or negative control and subsequently infected with EV71. At 12 h postinfection, the cells were subjected to Ago2 immunoprecipitation (Ago2-IP) using an miRNA isolation kit for human Ago2 (Wako, Osaka, Japan) according to the manufacturer's instructions. As a negative control, immunoprecipitation was performed using nonimmune IgG beads prepared with an antibody immobilization bead kit (Wako, Osaka, Japan).
Luciferase reporter assay.
Luciferase assays were performed using the dual-luciferase reporter assay system kit (Promega, Madison, WI, USA) according to the manufacturer's protocol. Briefly, RD cells were seeded in 24-well plates, and the luciferase reporter plasmid containing the hsa-miR-296-5p target was cotransfected with pLR-TK (Promega) and the hsa-miR-296-5p mimic or control mimic. Twenty-four hours later, the cells were collected and washed once with cold phosphate-buffered saline (PBS). Passive lysis buffer (Promega) was then added to the cells. After 15 min, supernatants were collected following centrifugation at 12,000 × g for 30 s, and relative luciferase expression levels were analyzed using the Modulus single-tube multimode reader (Promega).
For measuring luciferase activity of the recombinant EV71 virus, infected RD cells were harvested and lysed at 12 h postinfection. The Renilla luciferase activity expressed by the recombinant EV71 virus was measured by the Renilla Luciferase Assay System kit (Promega) according to the manufacturer's protocol.
Generation of EV71 infectious particles from an infectious clone.
RNA transcripts were obtained by using a RiboMAX large-scale RNA production system-T7 kit (Promega) with linearized DNA as the template according to the manufacturer's protocol. The in vitro-transcribed RNA was transfected into Vero cell monolayers in 6-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), which were then incubated at 37°C in 2 ml DMEM containing 2% FBS per well. Cytopathic effects (CPE) of Vero cells were observed 24 h posttransfection. The cell culture mixtures, including both cells and supernatants, were harvested when all of the cells exhibited CPE (4 to 7 days after transfection) and were stored at −70°C.
RNA extraction, reverse transcription, and qRT-PCR analysis.
Total RNA was obtained using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was reverse transcribed from 2 μg of total RNA with M-MLV Reverse Transcriptase (Promega). Random primers were used for reverse transcription of EV71 genomic RNA. GAPDH mRNA was measured as a control for the expression level of EV71 genomic RNA, and U6 rRNA served as an internal control for the expression level of hsa-miR-296-5p. hsa-miR-296-5p- and hsa-miR-196-specific bulge-loop miRNA quantitative real-time PCR (qRT-PCR) primers were purchased from RiboBio (Guangzhou, China), and other primers used for qRT-PCR are listed in Table 1.
qRT-PCR was carried out using the SYBR green I mix (Toyobo, Osaka, Japan) in a 25-μl reaction volume (12.5 μl SYBR green Real-time PCR Master Mix, 200 mM forward and reverse primers described in Table 1, 2.5 μl cDNA template) on an MJ Opticon I instrument (Bio-Rad, Hercules, CA, USA). The following cycling conditions were used to measure the EV71 genomic RNA: 95°C for 60 s and 40 cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 15 s. The conditions for measuring hsa-miR-296-5p were 95°C for 20 s and 40 cycles of 95°C for 10 s, 60°C for 20 s, and 70°C for 1 s. Data analysis was performed using the 2−ΔΔCt method.
Copy numbers of miR-296-5p per cell were determined using the method described by Sarasin-Filipowicz et al. (32). After counting cells under a microscope, RNA was extracted from various numbers of cells using TRIzol in triplicate. The average value of 15 pg of RNA per cell was determined and used for further calculations. This value resembled the 14.1 pg of RNA per cell used previously by Sarasin-Filipowicz et al. (32).
To determine miRNA copy numbers, standard curves were generated using high-performance liquid chromatography (HPLC) and polyacrylamide gel electrophoresis (PAGE)-purified oligoribonucleotides (RiBo Biotech, Guangzhou, China) corresponding to miR-196b and miR-296-5p sequences. The synthetic RNA input ranged from 2.5 aM (equivalent to 15 copies per reaction) to 25 pM (1.5 × 108 copies per reaction), and the reaction products were analyzed using a SYBR green I mix (Toyobo, Osaka, Japan) and the MJ Opticon I instrument (Bio-Rad, Hercules, CA, USA) as described above.
Western blot analyses.
Cells were collected and lysed using radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China), and 2 μg of extracted proteins was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane (0.45 μm; Millipore). The membranes were then blocked in 20 mM Tris-HCl buffer (pH 7.4) containing 37 mM NaCl (TBS) with 5% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, MO, USA) and incubated with primary antibody overnight at 4°C. After a standard washing cycle, the film was incubated with horseradish peroxidase (HRP)-labeled secondary antibody for 1 h at room temperature and washed again. The blots were stained using a Super Signal West Pico Chemiluminescent substrate (Pierce, Rockford, IL, USA) for signal detection and imaged with the Fluochem HD2 Imaging System (Alpha Innotech, San Leonardo, CA, USA). A multiclonal rabbit anti-EV71 VP1 antibody (Millipore) was used for VP1 detection (diluted 1:1,000). β-Actin, a loading control, was detected using a polyclonal antibody (sc-130301) (Santa Cruz Biotechnology).
Virus titration.
Virus titration was performed by seeding 104 RD cells per well in 96-well microtiter plates 1 day before infection. EV71 was added to the first set of wells in triplicate and serially diluted with DMEM containing 2% FBS (103- to 1010-fold dilutions). The plates were then incubated at 37°C in 5% CO2. CPE was observed under the microscope after 3 to 4 days. Determination of virus titer, expressed as the 50% tissue culture infectious dose (TCID50) was performed using the Reed-Münch endpoint calculation method (33).
Statistical analysis.
All experiments were reproducible and carried out in triplicate. Each set of experiments was repeated three times, and data from one representative experiment are shown. Results are presented as means ± standard deviations (SD). Student's t test for paired samples was used to determine statistical significance. Differences were considered statistically significant at a P value of <0.05.
RESULTS
EV71 infection upregulates expression of hsa-miR-296-5p.
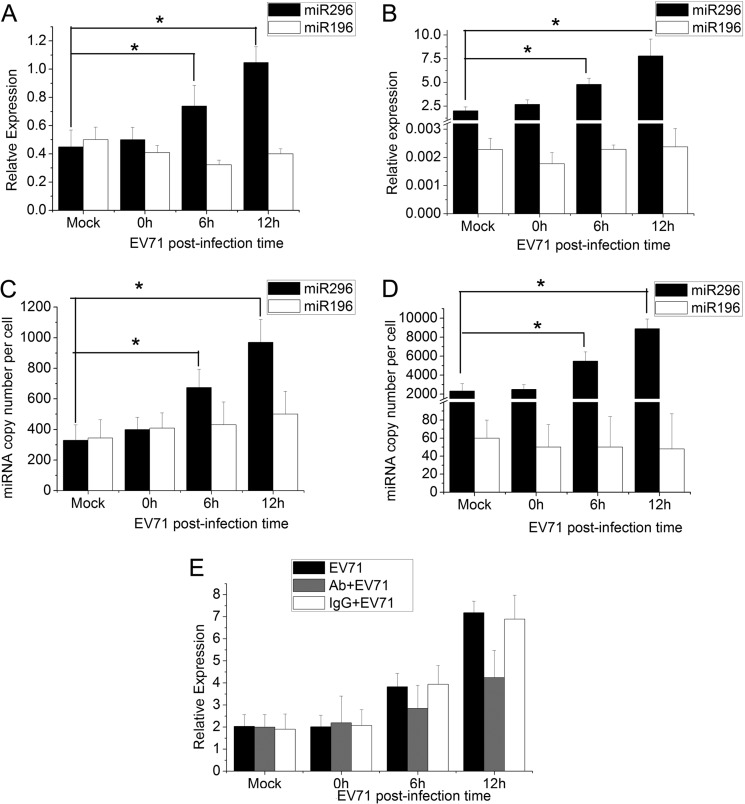
To determine whether cellular miRNAs are involved in the host response to EV71 infection, we performed a comprehensive miRNA profiling of EV71-infected cells by small RNA sequencing. And it was found that the expression of miR-296 increased significantly, 7-fold, and the miR-196 expression level increased about 0.2-fold. To confirm the response of hsa-miR-296-5p to EV71 infection, the expression patterns of this miRNA were analyzed by qRT-PCR analysis in RD and SK-N-SH cells. Both types of cells were infected with EV71 at a multiplicity of infection (MOI) of 1. At 6 and 12 h postinfection (hpi), the cells were harvested, and total RNA was extracted. Expression levels of hsa-miR-296-5p were determined by qRT-PCR using U6 rRNA as an internal control. Another miRNA, hsa-miR-196, was used as a control. The relative expression of hsa-miR-296-5p in RD cells increased as the infection progressed (Fig. 1A). Expression levels of hsa-miR-296-5p at 6 hpi and 12 hpi were 1.5- and 2-fold higher, respectively, than that of the mock-infected cells (0 hpi), while the level of miR-196 was not affected significantly by EV71 infection. Additionally, in another EV71-sensitive cell line, SK-N-SH, the expression levels of hsa-miR-296-5p were 2- and 3-fold higher than those of mock-infected cells at 6 and 12 hpi, while the expression of miR-196 did not change significantly during EV71 infection and its relative expression level was very low compared with that of miR-296-5p (Fig. 1B).
Fig 1.
Expression of hsa-miR-296-5p and hsa-miR-196 in EV71-infected human RD and SK-N-SH cells. (A, B) At different times after EV71 infection, levels of hsa-miR-296-5p and miR-196 were quantified by qRT-PCR, using U6 rRNA as an internal control. Values are means from triplicate experiments and represent relative levels of miRNA in RD (A) and SK-N-SH (B) cells. (C, D) To determine expression of miRNA on a per cell basis, 15 pg of total RNA extracted from EV71-infected cells at each time point was used to determine the copy numbers of miR-296-5p and miR-196 in RD (C) and SK-N-SH (D) cells. Chemically synthesized oligoribonucleotides corresponding to miR-196b and miR-296-5p were used to generate standard curves. (E) IFN receptors of EV71-infected SK-N-SH cells were blocked by mouse anti-IFNAR monoclonal antibody. Relative expression levels of hsa-miR-296-5p were determined by qRT-PCR at different times. The untreated group (EV71) and IgG-treated group (IgG+EV71) were used as a control. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). *, P < 0.05; **, P < 0.01.
It is known that a minimum of 100 miRNA copies per cell is required to provide biologically relevant qRT-PCR results. Therefore, we determined the copy number of miR-296 per cell. As shown in Fig. 1C, the copy number of miR-296 in an RD cell increased from 400 at 0 hpi to 970 at 12 hpi, while the copy numbers of miR-196 were not changed significantly. Furthermore, the copy number of miR-296 in SK-N-SH cell ranged from 2,500 to 9,000 during EV71 infection, while that of miR-196 was less than 100 (Fig. 1D).
IFN signaling has been reported to lead to miR-296 upregulation, and EV71 infection has been shown to induce IFN signaling. To investigate whether EV71 infection leads to miR-296 upregulation in the absence of IFN signaling, the IFN receptor was blocked by mouse anti-IFNAR1 monoclonal antibodies. Although miR-296 was still upregulated in EV71-infected cells with blockade of IFN receptors, its expression level was significantly lower than that in the untreated control group (Fig. 1E).
Overexpression of hsa-miR-296-5p inhibits EV71 replication.
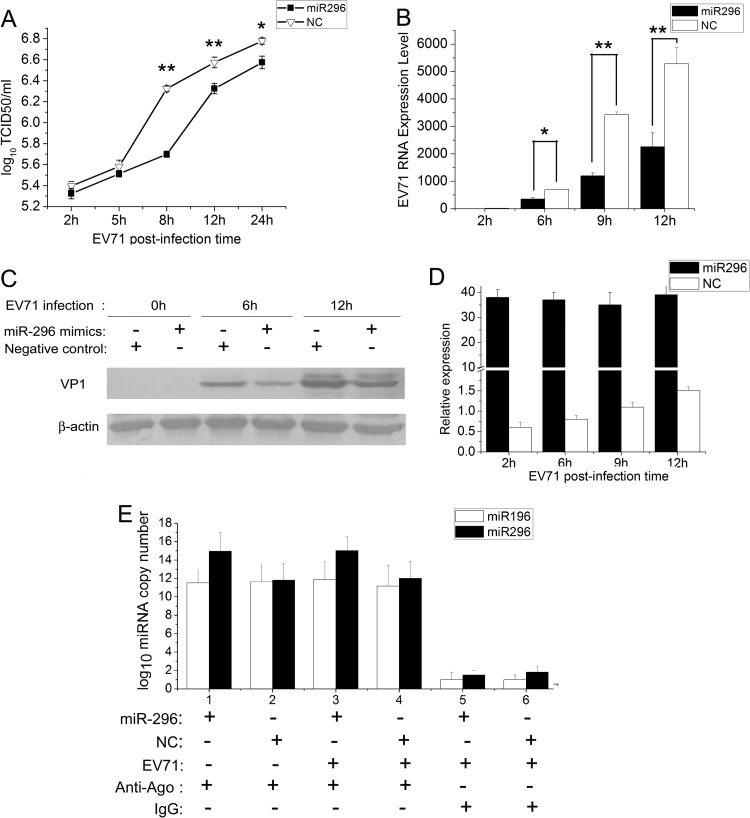
To investigate whether the activation of hsa-miR-296-5p affects EV71 replication, RD cells were transfected with chemically synthesized hsa-miR-296-5p oligonucleotides (hsa-miR-296-5p mimics), followed by infection with EV71 at an MOI of 5. The titers (TCID50) of EV71 harvested from the infected cells were determined at different times postinfection. The hsa-miR-296-5p mimic lowered EV71 titers at all time points postinfection compared to the negative control (Fig. 2A). Most significantly, the EV71 titer of the hsa-miR-296-5p-transfected sample was approximately 0.8 units lower than that of the negative-control sample at 8 hpi. This result indicated that the hsa-miR-296-5p mimic could suppress EV71 replication.
Fig 2.
Effects of hsa-miR-296-5p overexpression on EV71 infection. RD cells were transfected with synthetic hsa-miR-296-5p mimics or the negative-control mimics (NC). (A) Titration of EV71 in cells transfected with hsa-miR-296-5p or NC. Virus titers in infected cells were determined at the indicated times postinfection. (B, D) EV71 RNA expression in hsa-miR-296-5p-transfected cells during viral infection. After extraction of total RNA from infected cells in hsa-miR-296-5p- and NC-transfected groups, EV71 genomic RNA levels were determined by qRT-PCR and normalized against GAPDH transcript levels (B), and levels of hsa-miR-296-5p were quantified by qRT-PCR, using U6 rRNA as an internal control (D). (E) SK-N-SH cells transfected with miR-296-5p mimics and negative control were subjected to Ago2-IP, and copy numbers of miR-296-5p and miR-196 in the immunoprecipitates were determined by qRT-PCR. As a negative control, immunoprecipitation was performed using nonimmune IgG beads. The symbols “+” and “−” indicate treatment and no treatment, respectively, by the corresponding factor. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). *, P < 0.05; **, P < 0.01. (C) Viral protein expression in hsa-miR-296-5p-transfected cells during EV71 infection. Proteins of treated cells were extracted at the indicated times postinfection. Equal amounts of cell lysates were analyzed by immunoblotting with an anti-EV71-VP1 (upper panel) or anti-β-actin (lower panel) antibody. Results shown are representative of three independent experiments.
To further confirm this finding, the EV71 genomic RNA in infected cells was quantified by qRT-PCR. Over a series of time points, total-RNA samples were extracted, reverse transcribed into cDNA, and analyzed by qRT-PCR. The relative expression of EV71 genomic RNA was normalized against the human GAPDH gene. EV71 genomic RNA levels increased in both groups over the infection period. However, levels of EV71 RNA in the hsa-miR-296-5p-transfected group were reduced significantly at 9 and 12 hpi (70% and 60% reduction, respectively) compared to the negative control (Fig. 2B). Western blotting was performed to further examine whether this RNA inhibition corresponded to a reduction of viral proteins, and EV71 VP1 protein levels were found to decrease in hsa-miR-296-5p-transfected cells at 6 and 12 hpi (Fig. 2C). Collectively, these results suggested that the overexpression of hsa-miR-296-5p could suppress EV71 replication.
To determine the amount of miR-296-5p mimics to introduce into cells, cells transfected with miR-296-5p mimics and infected with EV71 were analyzed by qRT-PCR using U6 RNA as an internal control. As shown in Fig. 2D, the relative expression level of miR-296-5p in cells transfected with mimics was 60-fold higher than in the negative control at 0 hpi and showed a much higher level (more than 30-fold) than the negative control during EV71 infection. Furthermore, the functional levels of hsa-miR-296-5p assembled in RISC were quantified by Ago2-IP and qRT-PCR. In the uninfected group, the miR-296-5p copy number in Ago2 immunoprecipitates of miR-296-transfected cells was much higher (∼1,000-fold) than that of the negative control. In the EV71-infected group, the copy number of miR-296-5p in the transfected cells was about 800-fold higher than that of the negative control, although the miR-296-5p in Ago2 immunoprecipitates of the negative-control-transfected cells was slightly higher than in the uninfected control (Fig. 2E). This result indicated that transfection of miR-296-5p mimics significantly upregulated the amount of functional miR-296-5p.
Inhibition of hsa-miR-296-5p facilitates EV71 replication.
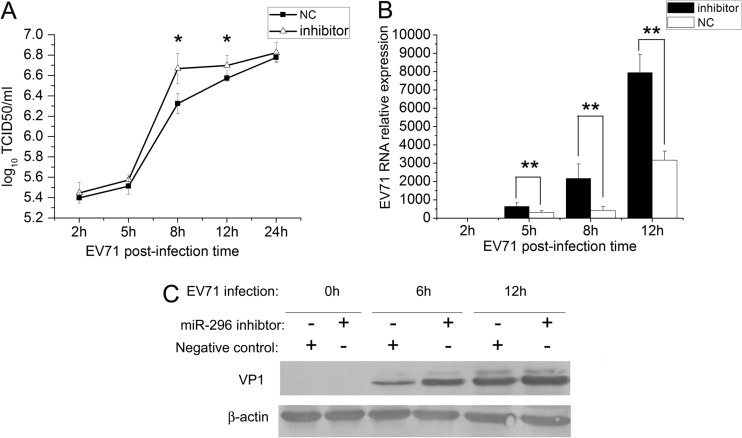
To determine the phenotypic consequence of the inhibition of EV71 gene expression and production by hsa-miR-296-5p, cells were transfected with the hsa-miR-296-5p inhibitor, a chemically synthetic oligonucleotide with complete sequence complementarity to endogenous hsa-miR-296-5p. Such oligonucleotide inhibitors have been shown to sequester intracellular miRNAs and to inhibit their activity in the RNA interference pathway (34). SK-N-SH cells transfected with the hsa-miR-296-5p inhibitor and negative control subsequently were infected with EV71. Differences in viral titers and expression levels of viral genomic RNA and proteins were determined by virus titration, qRT-PCR, and Western blotting, respectively. These analyses revealed that the hsa-miR-296-5p inhibitor enhanced the EV71 titers at each time point postinfection, compared to the negative control. The most significant enhancement of viral infection was observed at 8 hpi when the inhibitor-transfected sample titer was approximately 0.5 units higher than that of the negative-control sample (Fig. 3A). This result showed that the inhibition of hsa-miR-296-5p could enhance the viral replication of EV71. Furthermore, EV71 RNA levels in the various groups were examined by qRT-PCR and normalized against human U6 rRNA expression. The level of EV71 RNA in cells transfected with the hsa-miR-296-5p inhibitor was higher than that of the negative-control group at each time point evaluated (5, 8, and 12 hpi). EV71 RNA levels in the inhibitor-transfected group were 3- and 2.5-fold higher than those of the negative control at 8 and 12 hpi, respectively (Fig. 3B). Western blotting indicated that the amounts of EV71 VP1 protein increased in hsa-miR-296-5p inhibitor-transfected cells at 6 hpi and 12 hpi (Fig. 3C). Collectively, these results indicate that inhibition of hsa-miR-296-5p may facilitate EV71 infection.
Fig 3.
Effects of hsa-miR-296-5p-mediated inhibition of EV71 infection. SK-N-SH cells were transfected with a synthetic hsa-miR-296-5p inhibitor (inhibitor) or the NC inhibitor. (A) Titration of EV71 in infected cells transfected with hsa-miR-296-5p or NC. Virus titers in infected cells were determined at the indicated times postinfection. (B) EV71 RNA levels in hsa-miR-296-5p-transfected cells during viral infection. After extraction of total RNA from infected cells in inhibitor- and NC-transfected groups, EV71 genomic RNA levels were determined by qRT-PCR and normalized against GAPDH transcript levels. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). *, P < 0.05; **, P < 0.01. (C) Viral protein expression in hsa-miR-296-5p-transfected cells during EV71 infection. Proteins of treated cells were extracted at the indicated times postinfection. Equal amounts of cell lysates were analyzed by immunoblotting with an anti-EV71-VP1 (upper panel) or anti-β-actin (lower panel) antibody. Results shown are representative of three independent experiments.
hsa-miR-296-5p targets the EV71 genome.
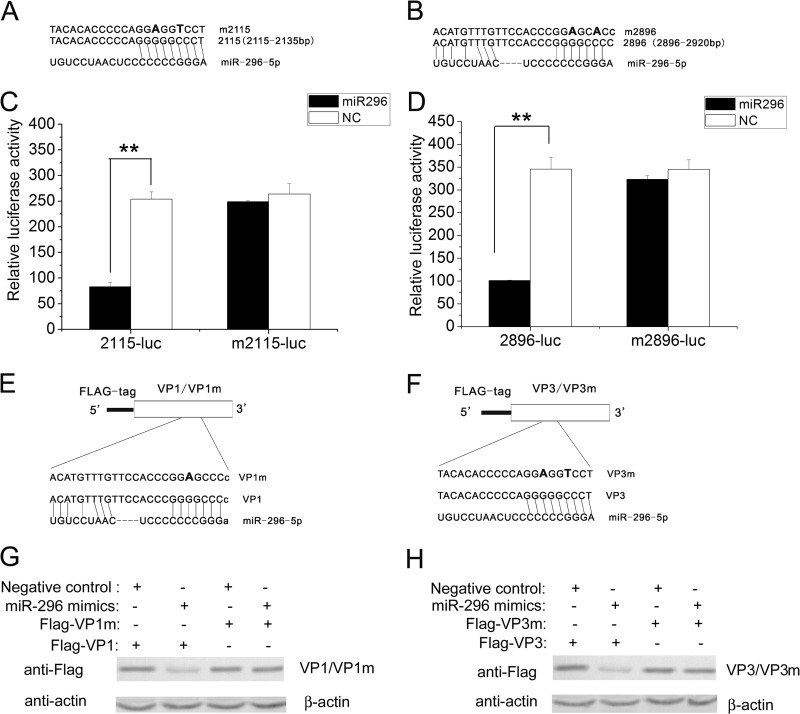
Identification of functional target sequences of hsa-miR-296-5p is necessary in order to define the regulatory mechanism of EV71 infection by this miRNA. By using ViTa algorithms (35) to predict viral targets of human miRNA, our analysis showed that the P1 region of the EV71 genome carries two putative hsa-miR-296-5p binding sites (nt 2115 to 2135 and nt 2896 to 2920) (Fig. 4A and B). The wild-type (WT) target sequences and mutated target sequences were cloned into the pCMV-luc vector. The resulting constructs (2115-luc, m2115-luc, 2896-luc, and m2896-luc) were used to confirm the binding sites of hsa-miR-296-5p by the luciferase reporter assay. These constructs were transfected into RD cells together with a control Renilla luciferase-expressing plasmid (pRL-TK) as a loading control in the presence of cotransfected synthetic hsa-miR-296-5p or negative control. Compared with cells treated with the negative-control mimic, luciferase activity was reduced in cells transfected with the 2115-luc and treated with the hsa-miR-296-5p mimic (Fig. 4C, left panel). In contrast, treatment with the hsa-miR-296-5p mimic had no effect on luciferase activity in cells transfected with m2115-luc, which contained a mutation in the target EV71 genome sequence of hsa-miR-296-5p (Fig. 4C, right panel). Similarly, luciferase activity was reduced in cells transfected with 2896-luc and treated with the hsa-miR-296-5p mimic, but treatment with the hsa-miR-296-5p mimic had no effect on luciferase activity in cells transfected with m2896-luc (Fig. 4D). These results suggest that hsa-miR-296-5p may suppress EV71 gene expression by interacting with nt 2115 to 2135 and nt 2896 to 2920 of the EV71 genome.
Fig 4.
Targeting of EV71 genomic RNA sequence by hsa-miR-296-5p. (A, B) The complementary sequences of two hsa-miR-296-5p candidate target “seed sequences” and mutated forms are indicated. Mutated nucleotides are bolded. The complementary sequences are from the EV71-BrCr-TR strain (GenBank number AB204852). (C, D) Luciferase reporter plasmids containing the hsa-miR-296-5p target sites (2115-luc and 2896-luc) and corresponding mutants (m2115-luc and m2896-luc) were cotransfected with the plasmid pRL-TK into hsa-miR-296-5p- or NC-treated RD cells. Reporter activities were determined 24 h posttransfection by dual-luciferase assays, and the resultant ratios are shown. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). *, P < 0.05; **, P < 0.01. (E, F) Schematic representation of constructs Flag-VP1 and Flag-VP3 and corresponding constructs Flag-VP1m and Flag-VP3m containing mutations in hsa-miR-296-5p target sites. (G, H) Western blot analysis of effects of hsa-miR-296-5p on expression of EV71 VP1 and its mutant VP1m (G) and VP3 and its mutant VP3m (H). RD cells were transfected with the indicated plasmids in the presence of hsa-miR-296-5p or randomized NC. Proteins were analyzed with an anti-FLAG antibody with β-actin as the internal control.
Because nt 2115 to 2135 and nt 2896 to 2920 are located in the coding regions for the VP3 and VP1 proteins, respectively, we cloned the VP1 and VP3 ORFs into the pCMV-FLAG vector (Sigma) and generated hsa-miR-296-5p-resistant mutations in corresponding sites of hsa-miR-296-5p target sites (Fig. 4E and F). The resulting constructs, Flag-VP1, Flag-VP1m, Flag-VP3, and Flag-VP3m, were cotransfected into RD cells with the hsa-miR-296-5p mimic or negative-control mimic. At 24 h posttransfection, the cells were harvested, lysed, and analyzed by Western blotting. Upon treatment with the hsa-miR-296-5p mimic, levels of both FLAG-tagged VP1 and VP3 decreased, whereas the mutant FLAG-tagged proteins (Flag-VP1m and Flag-VP3m) showed no change (Fig. 4G and H), suggesting that hsa-miR-296-5p inhibits EV71 replication by targeting the two viral sequences at nt 2115 to 2135 and nt 2896 to 2920.
Synonymous mutations impair viral inhibitory effects of hsa-miR-296-5p and facilitate infection of mutant EV71.
To determine whether the two putative hsa-miRNA-296-5p binding sites (nt 2115 to 2135 and nt 2896 to 2920) on the EV71 BrCr-TR genome are conserved in other strains of EV71, a sequence alignment was constructed with viral genomes of different subtypes. Among the mutations found in other EV71 subtypes, two synonymous mutations (Fig. 5A, boxed), which did not change the encoded amino acids, were found in the seed region of the hsa-miR-296-5p binding sites. To further determine the effects of the synonymous mutations on EV71 infection and inhibitory effects of hsa-miR-296-5p on the virus, the corresponding mutations were introduced into an EV71 infectious clone pEV71 (BrCr-TR) (Fig. 5B). RD cells infected with the rescued mutant virus were collected, and total RNA was extracted, reverse transcribed into cDNA, and inserted into a cloning vector. The expected mutated sites were confirmed by sequencing (ABI 3730 sequencer; Invitrogen) (Fig. 5C). One-step growth curves for each of the mutants as well as the WT virus were performed to determine the effects of the mutations on virus replication in SK-N-SH cells. Viruses containing single mutations (2115mt and 2896mt) showed enhanced growth at each time point compared to the WT virus (BrCr). The double mutant virus (double mut) grew better than any other viruses at each time point, and titers of the double mutant were significantly higher than those of the other three viruses at later time points (12 and 24 hpi) (Fig. 5D). These results indicated that mutations in both of the miRNA target sites may enhance EV71 replication in SK-N-SH cells. To confirm the effects of the mutations on inhibition of EV71 replication by miRNA-296, RD cells transfected with hsa-miR-296-5p mimics were infected with each of the mutant viruses (2115mt, 2896mt, and double mut) or WT virus (BrCr) at an MOI of 5. At 12 hpi, virus titers were determined and expressed as TCID50. The titer of the WT virus (BrCr) was significantly decreased by hsa-miR-296-5p, while all of the mutant viruses were insensitive to treatment by the hsa-miR-296-5p mimic (Fig. 5E). This result indicated that hsa-miR-296-5p inhibits EV71 replication by targeting the two putative binding sites on the viral genome.
Compensatory mutation in the corresponding regions of the hsa-miR-296-5p target sequence restores inhibitory effects of the miRNA on EV71 replication.
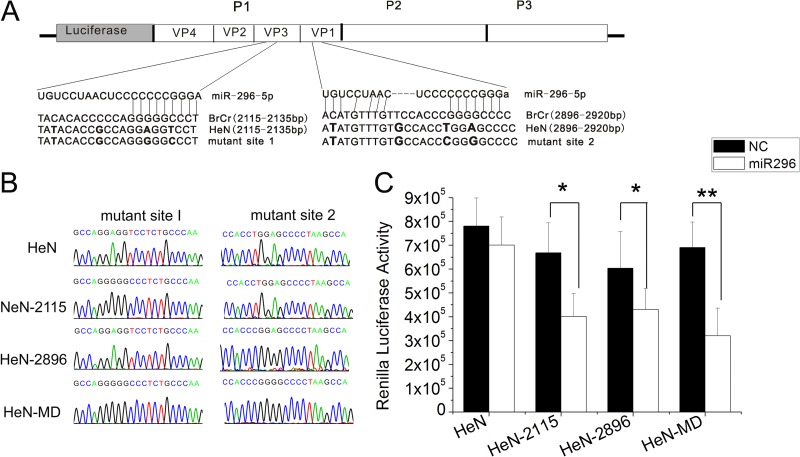
To further confirm that the putative binding sites were functionally responsible for the observed inhibitory effects of hsa-miR-296-5p on EV71, we introduced these hsa-miR-296-5p binding sites into an EV71 strain that did not contain these sites in the corresponding region of its genome. An infectious clone carrying the Renilla luciferase coding sequence and the full-length genome of the EV71 HeN strain (GenBank accession number JN256064) were used to confirm the effects of hsa-miR-296-5p. The corresponding sequences were mutated to match the potential binding sites of hsa-miR-296-5p (Fig. 6A). Genomic RNA was isolated from RD cells infected with the rescued virus, reverse transcribed to cDNA, cloned, and sequenced to confirm the mutation sites. The HeN-2115 and HeN-2896 mutant viruses carried the mutant sites 1 and 2, respectively, while the HeN-double mutant (HeN-MD) contained both sites (Fig. 6B). RD cells transfected with the hsa-miR-296-5p mimic were infected with the three mutant viruses (HeN-2115, HeN-2896, HeN-MD) or WT (HeN) virus at an MOI of 5. At 12 hpi, luciferase activities of the infected cells were measured. While the luciferase activity of the WT virus (HeN) was not affected, all three of the mutant viruses showed decreased luciferase activity upon treatment with the hsa-miR-296-5p mimic compared to the negative control. These results indicated that the introduction of hsa-miR-296-5p target sites into the genome of an EV71 strain can confer sensitivity of the virus to inhibitory effects of this miRNA.
Fig 6.
Effects of back mutations in hsa-miR-296-5p target sites on replication and inhibitory effects of hsa-miR-296-5p on EV71-HeN-luc. (A) Schematic representation of pEV71-HeN-luc constructs with hsa-miR-296-5p target sites and their corresponding back mutations. (B) Sequencing results of each mutant EV71 virus at the two mutation sites. (C) Effects of hsa-miR-296-5p on infectivity of WT (HeN) and mutant (HeN-2115, HeN-2896, and HeN-MD) viruses. hsa-miR-296-5p-transfected RD cells were infected with each virus at an MOI of 5. Viral titers were determined at 12 h postinfection. Data are representative of at least two independent experiments, with each determination performed in triplicate (mean ± SD of fold change). **, P < 0.01.
DISCUSSION
Recent studies have highlighted the role of miRNAs as critical effectors in intricate networks of host-pathogen interactions (36, 37). To address whether cellular miRNAs are involved in the host response to EV71 infection, miRNAs of EV71-infected cells were comprehensively profiled, and the miRNA hsa-miR-296-5p was found to be significantly increased in response to EV71 infection. In this study, we confirmed this finding by qRT-PCR in RD and SK-N-SH cells, which are both sensitive to EV71 infection. These results showed that EV71 infection indeed increased the expression of hsa-miR-296-5p. It is known that hsa-miR-296-5p is expressed in some human tissues, and its expression level varies in some tumors (34, 38–40). Pedersen et al. reported that type I IFN induced by HCV infection increases the expression of hsa-miR-296-5p and that the overexpression of hsa-miR-296-5p can suppress the expression of HCV genomic RNA (21). A minimum of 100 miRNA copies per cell has been reported to be required to produce biologically significant effects (32, 41). However, the copy number of miRNA per cell was not analyzed in that report. Subsequently, Sarasin-Filipowicz et al. demonstrated, by quantifying the miRNA copy number per cell, that miR-296-5p is stimulated by IFN in blood cells but not in perfused livers (32). We also confirmed that EV71 upregulated miR-296-5p by qRT-PCR with quantification of miRNA copy numbers per cell, and the results in this study were consistent with previous observations. EV71 infection has been reported to cause the activation of type I IFN (42). Antibody blockade of the IFN receptor led to a dramatic weakening of miR-296-5p upregulation caused by EV71 (Fig. 1E). This result indicated that upregulation of miR-296-5p may be related to IFN signaling. However, whether the IFN response or another underlying mechanism is involved in the expression of hsa-miR-296-5p requires further investigation. Because the genome of an RNA virus has a structure similar to that of the host mRNA, it is possible that cellular miRNAs are able to bind and degrade the RNA virus genome via the cell host machinery (43). We speculate that the elevated expression of miRNAs, including hsa-miR-296-5p, during virus infection may be part of the host antiviral response. The finding in this study that EV71 infection-induced expression of hsa-miR-296-5p in the host cell may negatively affect virus replication provides guidance for future studies of EV71 pathogenesis and host antiviral mechanisms.
To investigate the potential antiviral role of EV71-induced hsa-miR-296-5p, chemically synthesized oligonucleotides were used in our study to either overexpress or inhibit hsa-miR-296-5p in EV71-infected cells. Overexpression of hsa-miR-296-5p significantly inhibited replication of the EV71 virus, while inhibition of hsa-miR-296-5p facilitated viral replication. It has been reported that hsa-miR-296 inhibits apoptosis and promotes growth in HeLa cells (44) and can promote tumor angiogenesis by targeting hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) (40). The anti-EV71 activity of hsa-miR-296-5p may be a novel function described for cellular miRNA. Other cellular miRNAs have also been reported to inhibit replication of DNA viruses or retroviruses. For example, human miR-125-5p, miR-122, miR-210, and miR-190 were found to suppress HBV replication (22, 23, 45). miR-32 shuts down gene expression and viral replication of PFV (19), while miR-29 can inhibit HIV-1 replication (24). Meanwhile, replication of another RNA virus, VSV, can be inhibited by miR-93 and miR-24 (20). Replication of CVB3, which belongs to the same genus as EV71, has been shown to be inhibited by hsa-miR-342-5p (30). Our present work shows that cellular miRNAs can effectively exert antiviral activity during the host immune response to the infection. However, in other cases, cellular miRNA can facilitate virus replication. Some viruses, such as HIV-1 and HCV, take advantage of cellular miRNAs to facilitate their survival and propagation (36, 46, 47). Ultimately, an important role for hsa-miR-296-5p may be established in host-pathogen interactions as with other antiviral miRNAs (43).
Many miRNAs have been shown to interact directly with the genome of a virus to inhibit its replication (20, 48). As an important factor that regulates gene expression in host cells, hsa-miR-296-5p targets HRS, IκB kinaseε (IKKε), and Temd9 (49). However, whether hsa-miR-296-5p affects viral replication by interacting with these intracellular targets will require further investigation. Our present results illustrate that in addition to cellular mRNAs, hsa-miR-296-5p can also functionally interact with the EV71 genome. When we introduced mutations in its putative target sites in the EV71 genome, hsa-miR-296-5p could no longer inhibit viral replication (Fig. 5). Furthermore, these results showed that the effects of hsa-miR-296-5p on intracellular targets had little effect on viral replication.
By alignment of the target sequences of hsa-miR-296-5p in the genome of each EV71 subtype, we found synonymous mutations in the seed region of the target sites in some strains (Fig. 5A) and speculated that hsa-miR-296-5p would not be able to suppress the replication of these viruses. Subsequent experiments using viruses carrying mutations in these target sites (Fig. 5E) and the luciferase reporter assays (Fig. 6C) confirmed that the synonymous mutations in the target seed region were indeed responsible for the lack of sensitivity of those viruses to hsa-miR-296-5p. It has been reported that synonymous mutations may be a strategy by which RNA viruses can evade the suppression of host miRNAs as well as siRNAs (43, 50). Synonymous mutations in the viral genome can affect the replicative properties of many viruses. For example, the A426G mutation in a replication-competent HIV-1 clone dramatically reduces virion production without affecting viral protein synthesis in cells (51). Synonymous mutations artificially introduced into the poliovirus genome can attenuate the virus to generate a vaccine strain (52). Here, our results showed that synonymous mutations in the target sites of hsa-miR-296-5p in EV71 strain BrCr could facilitate virus infection and impair the inhibitory effects of hsa-miR-296-5p on the virus. This finding also suggests that virus escape from miRNA-mediated suppression can occur through mutations within the seed region of the miRNA target sequence, which is in line with data obtained for other viruses (53–56). Insertions in the miR-9, miR-124a, miR-128a, miR-218, or let-7c target sites of the neurovirulent chimeric tick-borne encephalitis/dengue virus (TBEV/DEN4) genome were found to be sufficient to prevent the development of otherwise lethal encephalitis in mice infected with a high dose of virus. Notably, all of those viruses isolated from the brains of mice contained single-nucleotide mutations located within the miRNA target sequences (57). Mutations including synonymous mutations of poliovirus, which also belongs to the human Enterovirus genus, were found to accumulate in sequentially isolated specimens of many individuals (58). Additionally, insertion of small miRNA homology sequences into the poliovirus genome can restrict its tissue tropism, prevent pathogenicity, and yield an attenuated viral strain (53). Therefore, we speculate that EV71 infection triggers the upregulation of miR-296-5p expression, which in turn suppresses viral replication; however, the virus can escape from the miRNA-mediated suppression via synonymous mutations to optimize cell tropism within the host or regulate viral replication and virulence.
To date, roles for miRNAs have been established in the antiviral defense of plants and invertebrates. Here, we provide evidence that human miRNAs can inhibit EV71 replication by targeting the EV71 genome. Our work reveals the existence of an intricate physiological interplay between cellular miRNAs and EV71 replication. Additional investigations into the role of miRNAs in EV71 infection may shed greater light on the viral-host interaction, ultimately providing a deeper understanding of the pathogenesis of EV71 infection and the host antiviral response.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Major State Basic Research Development Program of China (973 Program) (2012CB518904) and the National Natural Science Foundation of China (81071351).
We thank Minetaro Arita (National Institute of Infectious Diseases, Japan) and Bo Zhang (Wuhan Institute of Virology, Chinese Academy of Sciences, China) for providing the EV71 cDNA clone plasmid. We also thank Wenqian Hu (Whitehead Institute for Biomedical Research, Cambridge, MA, USA) for critical reading of the manuscript and helpful suggestions.
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cullen BR. 2009. Viral and cellular messenger RNA targets of viral microRNAs. Nature 457:421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shukla GC, Singh J, Barik S. 2011. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol. Cell. Pharmacol. 3:83–92 [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68 [DOI] [PubMed] [Google Scholar]
- 8. Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16:720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349 [DOI] [PubMed] [Google Scholar]
- 10. Brennecke J, Stark A, Russell RB, Cohen SM. 2005. Principles of microRNA-target recognition. PLoS Biol. 3:e85 doi:10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuersten S, Goodwin EB. 2003. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 4:626–637 [DOI] [PubMed] [Google Scholar]
- 12. Lai EC. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30:363–364 [DOI] [PubMed] [Google Scholar]
- 13. Berkhout B, Jeang KT. 2007. RISCy business: microRNAs, pathogenesis, and viruses. J. Biol. Chem. 282:26641–26645 [DOI] [PubMed] [Google Scholar]
- 14. Voinnet O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206–220 [DOI] [PubMed] [Google Scholar]
- 15. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736 [DOI] [PubMed] [Google Scholar]
- 16. Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, Khromykh AA, Asgari S. 2012. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 40:2210–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muller S. 2007. Dicing with viruses: microRNAs as antiviral factors. Immunity 27:1–3 [DOI] [PubMed] [Google Scholar]
- 19. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 20. Otsuka M, Jing Q, Georgel P, New L, Chen JM, Mols J, Kang YJ, Jiang ZF, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han JH. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- 21. Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. 2011. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 39:5157–5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen YN, Shen A, Rider PJ, Yu Y, Wu KL, Mu YX, Hao Q, Liu YL, Gong H, Zhu Y, Liu FY, Wu JG. 2011. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 25:4511–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 34:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennasser Y, Le SY, Benkirane M, Jeang KT. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607–619 [DOI] [PubMed] [Google Scholar]
- 26. Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang LY, Lin TY, Hsu KH, Huang YC, Lin KL, Hsueh C, Shih SR, Ning HC, Hwang MS, Wang HS, Lee CY. 1999. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 354:1682–1686 [DOI] [PubMed] [Google Scholar]
- 28. Cui L, Guo X, Qi Y, Qi X, Ge Y, Shi Z, Wu T, Shan J, Shan Y, Zhu Z, Wang H. 2010. Identification of microRNAs involved in the host response to enterovirus 71 infection by a deep sequencing approach. J. Biomed. Biotechnol. 2010:425939 doi:10.1155/2010/425939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, Li KC, Cheng CL, Cheng HW, Lee JY, Lee CN, Yang PC. 2011. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9:58–69 [DOI] [PubMed] [Google Scholar]
- 30. Wang L, Qin Y, Tong L, Wu S, Wang Q, Jiao Q, Guo Z, Lin L, Wang R, Zhao W, Zhong Z. 2012. MiR-342-5p suppresses coxsackievirus B3 biosynthesis by targeting the 2C-coding region. Antiviral Res. 93:270–279 [DOI] [PubMed] [Google Scholar]
- 31. Arita M, Shimizu H, Nagata N, Ami Y, Suzaki Y, Sata T, Iwasaki T, Miyamura T. 2005. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 86:1391–1401 [DOI] [PubMed] [Google Scholar]
- 32. Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. 2009. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat. Med. 15:31–33 [DOI] [PubMed] [Google Scholar]
- 33. Svensson L, Hjalmarsson A, Everitt E. 1999. TCID50 determination by an immuno dot blot assay as exemplified in a study of storage conditions of infectious pancreatic necrosis virus. J. Virol. Methods 80:17–24 [DOI] [PubMed] [Google Scholar]
- 34. Wei JJ, Wu X, Peng Y, Shi G, Basturk O, Yang X, Daniels G, Osman I, Ouyang J, Hernando E, Pellicer A, Rhim JS, Melamed J, Lee P. 2011. Regulation of HMGA1 expression by microRNA-296 affects prostate cancer growth and invasion. Clin. Cancer Res. 17:1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. 2007. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 35:D381–D385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ghosh Z, Mallick B, Chakrabarti J. 2009. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 37:1035–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsunetsugu-Yokota Y, Yamamoto T. 2010. Mammalian microRNAs: post-transcriptional gene regulation in RNA virus infection and therapeutic applications. Front. Microbiol. 1:108 doi:10.3389/fmicb.2010.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corbetta S, Vaira V, Guarnieri V, Scillitani A, Eller-Vainicher C, Ferrero S, Vicentini L, Chiodini I, Bisceglia M, Beck-Peccoz P, Bosari S, Spada A. 2010. Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr. Relat. Cancer 17:135–146 [DOI] [PubMed] [Google Scholar]
- 39. Hong L, Han Y, Zhang HW, Li MB, Gong TQ, Sun L, Wu KC, Zhao QC, Fan DM. 2010. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Ann. Surg. 251:1056–1063 [DOI] [PubMed] [Google Scholar]
- 40. Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. 2008. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14:382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 25:1457–1467 [DOI] [PubMed] [Google Scholar]
- 42. Lu J, Yi L, Zhao J, Yu J, Chen Y, Lin MC, Kung HF, He ML. 2012. Enterovirus 71 disrupts interferon signaling by reducing the level of interferon receptor 1. J. Virol. 86:3767–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gottwein E, Cullen BR. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng AM, Byrom MW, Shelton J, Ford LP. 2005. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. 2010. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 88:169–175 [DOI] [PubMed] [Google Scholar]
- 46. Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 47. Jopling CL, Yi MK, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 48. Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 49. Robson JE, Eaton SA, Underhill P, Williams D, Peters J. 2012. MicroRNAs 296 and 298 are imprinted and part of the GNAS/Gnas cluster and miR-296 targets IKBKE and Tmed9. RNA 18:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Novella IS, Zarate S, Metzgar D, Ebendick-Corpus BE. 2004. Positive selection of synonymous mutations in vesicular stomatitis virus. J. Mol. Biol. 342:1415–1421 [DOI] [PubMed] [Google Scholar]
- 51. Hamano T, Matsuo K, Hibi Y, Victoriano AF, Takahashi N, Mabuchi Y, Soji T, Irie S, Sawanpanyalert P, Yanai H, Hara T, Yamazaki S, Yamamoto N, Okamoto T. 2007. A single-nucleotide synonymous mutation in the gag gene controlling human immunodeficiency virus type 1 virion production. J. Virol. 81:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. 2008. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 4:239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gitlin L, Stone JK, Andino R. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heiss BL, Maximova OA, Thach DC, Speicher JM, Pletnev AG. 2012. MicroRNA targeting of neurotropic flavivirus: effective control of virus escape and reversion to neurovirulent phenotype. J. Virol. 86:5647–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin SS, Wu HW, Elena SF, Chen KC, Niu QW, Yeh SD, Chen CC, Chua NH. 2009. Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 5:e1000312 doi:10.1371/journal.ppat.1000312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heiss BL, Maximova OA, Pletnev AG. 2011. Insertion of microRNA targets into the flavivirus genome alters its highly neurovirulent phenotype. J. Virol. 85:1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kinnunen L, Huovilainen A, Poyry T, Hovi T. 1990. Rapid molecular evolution of wild type 3 poliovirus during infection in individual hosts. J. Gen. Virol. 71(Part 2):317–324 [DOI] [PubMed] [Google Scholar]