Abstract
The sooty mangabey-derived simian immunodeficiency virus (SIV) strain E660 (SIVsmE660) is a genetically heterogeneous, pathogenic isolate that is commonly used as a vaccine challenge strain in the nonhuman primate (NHP) model of human immunodeficiency virus type 1 (HIV-1) infection. Though it is often employed to assess antibody-based vaccine strategies, its sensitivity to antibody-mediated neutralization has not been well characterized. Here, we utilize single-genome sequencing and infectivity assays to analyze the neutralization sensitivity of the uncloned SIVsmE660 isolate, individual viruses comprising the isolate, and transmitted/founder (T/F) viruses arising from low-dose mucosal inoculation of macaques with the isolate. We found that the SIVsmE660 isolate overall was highly sensitive to neutralization by SIV-infected macaque plasma samples (50% inhibitory concentration [IC50] < 10−5) and monoclonal antibodies targeting V3 (IC50 < 0.01 μg/ml), CD4-induced (IC50 < 0.1 μg/ml), CD4 binding site (IC50 ∼ 1 μg/ml), and V4 (IC50, ∼5 μg/ml) epitopes. In comparison, SIVmac251 and SIVmac239 were highly resistant to neutralization by these same antibodies. Differences in neutralization sensitivity between SIVsmE660 and SIVmac251/239 were not dependent on the cell type in which virus was produced or tested. These findings indicate that in comparison to SIVmac251/239 and primary HIV-1 viruses, SIVsmE660 generally exhibits substantially less masking of antigenically conserved Env epitopes. Interestingly, we identified a minor population of viruses (∼10%) in both the SIVsmE660 isolate and T/F viruses arising from it that were substantially more resistant (>1,000-fold) to antibody neutralization and another fraction (∼20%) that was intermediate in neutralization resistance. These findings may explain the variable natural history and variable protection afforded by heterologous Env-based vaccines in rhesus macaques challenged by high-dose versus low-dose SIVsmE660 inoculation regimens.
INTRODUCTION
Similarities between simian immunodeficiency virus (SIV) infection of rhesus macaques and human immunodeficiency virus type 1 (HIV-1) infection of humans were first recognized in the 1980s, when captive Asian-origin macaques were found to be infected with an immunodeficiency-causing retrovirus originating with African-origin sooty mangabeys (1–6). Whereas SIVsmm viruses cause a nonpathogenic infection in their natural hosts (7), they produce a pathogenic infection in macaques, with virologic and clinical outcomes that parallel those of HIV-1 infection in humans. Like HIV-1, SIVsmm and SIVmac infect CD4+ T cells, utilize CCR5 as a coreceptor, establish high peak and setpoint viremia, and cause generalized immune activation and a profound acute and sustained loss of intestinal CD4+ T cells (3, 8, 9). As in HIV-1 infection, these events lead to progressive immune deficiency, opportunistic infections, AIDS-defining neoplasms, and death in the majority of infected animals (10).
Given these parallels with HIV infection, the SIV-nonhuman primate (NHP) model has been utilized as an important component of HIV vaccine development efforts. There are many iterations of this NHP model, with options in animal species, challenge viruses, inoculation routes, dosing strategies, and intrinsic host genetic restriction factors. Though there is no single standardized SIV-NHP infection model, much of the recent work in antibody- and cell-based vaccine design and assessment has been conducted with Indian-origin rhesus macaques challenged with SIVsmm and SIVmac viruses. Two of the most commonly used challenge viruses in the macaque model are the isolates SIVsmE660 and SIVmac251, along with the latter's derivative clone SIVmac239. SIVsmE660 was originally isolated from a rhesus macaque (Rh660) with a terminal AIDS-defining illness after it had been infected with virus previously passaged through three rhesus macaques. The Rh660 spleen homogenate was then cocultured with human CEMx174 cells and passaged through pigtail macaque peripheral blood mononuclear cells (PBMCs) to obtain the virus challenge stock (9, 11). SIVmac251 was also isolated terminally from SIV-infected macaque spleen cells after the macaque had been infected with virus cocultured with human PBMCs and serially passaged through rhesus PBMC cultures (12). Based on uncorrected mean character differences, the SIVmac251 and SIVsmE660 virus swarms are each approximately 77% identical in amino acid sequence to HIV-2, which originated from a cross-species transmission of SIVsmm from sooty mangabeys to humans, but only 52 to 57% identical to HIV-1 (9, 13–16). Despite these differences in primary amino acid sequence, SIVsmE660, SIVmac251, HIV-2, and HIV-1 are all highly related from an envelope (Env) structure-function perspective (15, 16). SIVsmE660 and SIVmac251 each exhibit modest within-isolate genetic heterogeneity, with 1.8% and 2.6% maximum env diversity for SIVsmE660 and SIVmac251, respectively. The genetic distance between the SIVsmE660 and SIVmac251 lineages in env is substantially greater, 14.6% in nucleotide sequence and a 13.5% difference in amino acid sequence, based on uncorrected mean character differences. This interlineage diversity has made these virus strains attractive candidates for heterologous challenge studies of candidate HIV vaccines.
Uncloned challenge stocks of SIVsmE660 and SIVmac251 have been used to test various vaccine strategies, and the neutralization profiles of these viruses were examined using methods commensurate with neutralizing antibody assay strategies available at the time. SIVmac251 and its derivative clone SIVmac239 consistently demonstrated high-level resistance to heterologous antibody neutralization; lab adaptation or site-directed mutagenesis was required to confer enhanced neutralization sensitivity (17–19). In addition, compared to humans acutely infected by HIV-1, SIVmac251-infected rhesus macaques showed delayed and generally low-titer autologous neutralizing antibody (NAb) responses (20). Due to this neutralization resistance, it has been suggested that SIVmac251 and SIVmac239 infection models may be too stringent to detect antibody-based vaccine activities that could translate to vaccine-elicited HIV-1 prevention in humans (20, 21).
Compared with SIVmac251, SIVsmE660 is generally perceived to be more neutralization sensitive, but published assessments of this key virological property have been inconsistent. In 2000, Ourmanov and colleagues ascribed intermediate neutralization sensitivity to SIVsmE660 compared to that of the genetically related, nonpathogenic, and highly neutralization-sensitive SIVsmH-4 strain (22). Using a cell-killing assay, these investigators detected NAb titers at 12 weeks postinfection that peaked at 20 to 28 weeks in the SIVsmE660-infected animals. More recently, Ourmanov and colleagues reexamined the neutralization sensitivity of the uncloned SIVsmE660 isolate by the same plasma specimens from SIVsmE660-infected, vaccinated, or control animals using the TZM-bl single cycle entry assay and observed much higher NAb titers (23). Two other studies have examined the neutralization sensitivity of molecular virus clones derived from the SIVsmE660 isolate with conflicting results. Letvin and colleagues performed an assessment of two transmitted/founder (T/F) Env clones derived from animals infected by repeated low-dose mucosal inoculation with the SIVsmE660 isolate (24). In TZM-bl assays, one of the T/F Envs demonstrated exquisite sensitivity to four different SIVsmE660-infected macaque plasma samples, while the second Env displayed greater resistance. The SIVsmE660 isolate itself was sensitive to neutralization by the four SIVsmE660-infected macaque plasma samples in the TZM-bl assay but was purportedly more resistant when tested in a rhesus PBMC target cell assay. The authors concluded that the SIVsmE660 isolate exhibited intermediate neutralization sensitivity (24). Wu and colleagues recently reported on the role of NAb in rhesus macaques infected with infectious molecular clones (IMCs) derived from the SIVsmE660 isolate (21). The two SIVsmE660 IMCs were shown to be extremely sensitive to neutralization by homologous and heterologous sera. Following intravenous inoculation of the IMCs into four TRIM5 permissive rhesus macaques, NAb titers arose by 4 weeks postinfection and peaked at high titers of 1:2.7 × 105 by 16 weeks with concurrent evidence of virus escape from these NAbs. Based on these studies, the neutralization sensitivity of the SIVsmE660 isolate, and the individual viruses comprising it, remains unclear.
In addition to exhibiting variability in neutralization sensitivity, SIVsmE660 has also been associated with variable clinical pathogenicity, especially following low-dose mucosal inoculation (25, 26). Quantitative estimates of the relative transmission barrier posed by rectal mucosa compared to intravenous inoculation of SIV in rhesus macaques are about 2,000-fold (27), with low-dose mucosal inoculation generally resulting in productive clinical infection by one or few T/F viruses (27). While this feature of the SIV-NHP infection model recapitulates the numbers of viruses that typically lead to productive infection in humans following sexual transmission of HIV-1 (28–32), it leads to the potential for variability in viral pathogenesis due to transmission of viruses with distinct genetic features. This problem may be more of an issue with SIVsmE660, which has shown substantial variability in clinical outcomes following low-dose mucosal challenge, than with SIVmac251 or SIVmac239. This variability in natural history and pathogenicity has been largely attributed to the SIVsmE660 strain's sensitivity to host restriction factors, such as TRIM5α (25, 26, 33), but heterogeneity in replication fitness and neutralization sensitivity of viruses comprising the SIVsmE660 isolate could contribute. Consistent with this hypothesis is the recent finding by Wu and colleagues that two infectious molecular virus clones derived from the SIVsmE660 stock that were initially very sensitive to autologous antibody neutralization evolved over 24 weeks of infection in rhesus macaques to acquire neutralization resistance (21).
Despite continued interest in SIVsmE660 as a challenge stock in vaccine protection studies and natural history studies (21, 24, 34), no studies have directly assessed and compared the neutralization sensitivity of the uncloned SIVsmE660 virus stock, individual virus Envs comprising this stock, and T/F Envs and T/F infectious molecular virus clones (IMCs) derived from rhesus macaques infected by low-dose rectal mucosal challenge with the uncloned virus stock. The objective of the present study was to perform this analysis so as to better inform the interpretation of SIVsmE660-based vaccine trials (24, 35) and as a first step toward obtaining data relevant to the construction of molecular SIVsmm challenge stocks that could more closely reflect the neutralization sensitivity of primary tier 2 strains of HIV-1 (36). Single-genome amplification (SGA) and direct amplicon sequencing, otherwise known as single-genome sequencing (SGS) (27, 30, 37, 38), were chosen as the experimental strategy for identifying the relevant env genes and T/F virus genomes because this method proportionally represents viruses within a genetically complex mixture and precludes the introduction of nucleotide misincorporations or recombination artifacts between genetically distinct target templates (27, 30, 37, 38), all of which could cloud the phenotypic characterization of the virus isolate and viral clones derived from it. Our studies were enabled by the use of recently described neutralizing monoclonal antibodies (MAb) that target discrete SIVsmm/HIV-2 V3, V4, CD4 binding site (CD4bs), and CD4-induced (CD4i) Env epitopes (39, 40).
MATERIALS AND METHODS
SIVsmE660 isolate.
The cell-free SIVsmE660 isolate used was initially generated by Vanessa Hirsch (11) and then expanded on rhesus macaque PBMCs and used as a virus challenge stock in studies conducted to test the transmission bottleneck and adaptive immune responses in mucosally infected rhesus macaques in naive and vaccinated rhesus macaques (27, 41). For this study, the SIVsmE660 isolate was expanded on rhesus macaque CD4+ T cells.
Viral RNA extraction and cDNA synthesis.
From plasma specimens and inoculum virus stock (culture supernatant) specimens, 20,000 viral RNA copies were extracted using the QIAamp viral RNA minikit (Qiagen). RNA was eluted and immediately subjected to cDNA synthesis. Reverse transcription of RNA to single-stranded cDNA was performed using SuperScript III reverse transcriptase (RT) according to the manufacturer's recommendations (Invitrogen). In brief, each cDNA reaction included 1× RT buffer, 0.5 mM (each) deoxynucleoside triphosphate, 5 mM dithiothreitol, 2 U/ml RNaseOUT (RNase inhibitor), 10 U/ml of SuperScript III reverse transcriptase, and 0.25 mM antisense primer SIVsm/macEnvR1 5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′ (nt 9454 to 9479 in SIVmac239). The mixture was incubated at 50°C for 60 min, followed by an increase in temperature to 55°C for an additional 60 min. The reaction was then heat inactivated at 70°C for 15 min and treated with 2 U of RNase H at 37°C for 20 min. The newly synthesized cDNA was used immediately or frozen at −80°C.
Single-genome amplification.
cDNA was serially diluted and distributed among wells of replicate 96-well plates so as to identify a dilution where PCR-positive wells constituted <30% of the total number of reactions, as previously described (30, 42). At this dilution, most of the PCR-positive wells contain amplicons derived from a single cDNA molecule. This was confirmed in every positive well by direct sequencing of the amplicon and inspection of the sequence chromatograms for mixed bases (double peaks), which would be indicative of priming from more than one original template or the introduction of PCR error in early cycles. Any sequence with mixed bases was excluded from further analysis. PCR amplification was performed in the presence of 1× High Fidelity Platinum PCR buffer, 2 mM MgSO4, 0.2 mM (each) deoxynucleoside triphosphate, 0.2 μM (each) primer, and 0.025 U/μl Platinum Taq High Fidelity polymerase in a 20-μl reaction mixture (Invitrogen). First-round PCR primers included the sense primer SIVsm/macEnvF1 (5′-CCTCCCCCTCCAGGACTAGC-3′; nucleotides [nt] 6127 to 6146 in SIVmac239) and antisense primer SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′; nt 9454 to 9479 in SIVmac239), which generated a 3.3-kb amplicon. PCR was carried out in MicroAmp 96-well reaction plates (Applied Biosystems) with the following PCR parameters: 1 cycle of 94°C for 2 min, 35 cycles of a denaturing step of 94°C for 15 s, an annealing step of 55°C for 30 s, and an extension step of 68°C for 4 min, followed by a final extension of 68°C for 10 min. Next, 2 μl from the first-round PCR product was added to a second-round PCR that included the sense primer SIVsmEnvF2 (5′-TATGATAGACATGGAGACACCCTTGAAGGAGC-3′; nt 6292 to 6323 in SIVmac239) or SIVmacEnvF2 (5′-TATAATAGACATGGAGACACCCTTGAGGGAGC-3′; nt 6292 to 6323 in Mac239) and antisense primer SIVsmEnvR2 (5′-ATGAGACATRTCTATTGCCAATTTGTA-3′; nt 9413 to 9439 in SIVmac239). The second-round PCR was carried out under the same conditions used for first-round PCR, but with a total of 45 cycles. Amplicons were inspected on precast 1% agarose E-gels 96 (Invitrogen). All PCR procedures were performed under PCR clean-room conditions using procedural safeguards against sample contamination, including prealiquoting of all reagents, use of dedicated equipment, and physical separation of sample processing from pre- and post-PCR amplification steps.
DNA sequencing.
Amplicons were directly sequenced by cycle sequencing using BigDye Terminator chemistry (Applied Biosystems). Sequencing reaction products were analyzed using an ABI 3730xl genetic analyzer (Applied Biosystems). Both DNA strands were sequenced using overlapping fragments. Individual sequence fragments for each amplicon were assembled and edited using the Sequencher 4.8 software program (Gene Codes; Ann Arbor, MI). Chromatograms containing mixed bases (double peaks) indicative of multiple templates were excluded.
Sequence alignments and diversity estimates.
All sequences were aligned manually or using the software tool CLUSTALW (43) and aligned in the program MacClade 4.08 (44) to optimize alignments. The T/F env sequences and some of the isolate env sequences were previously published (27). To illustrate the array of uncorrected pairwise distances among the SIVsmE660 viruses, a neighbor-joining tree (45) was constructed using mean pairwise distances for nucleotide sequences using the software program PAUP* version 4.0b10 (46). To illustrate the relationships of the SIVsmE660 and SIVmac251 strains, along with other SIVsmm lineages, in comparison to HIV-1 group M subtype reference sequences (47), phylogenetic trees based on Env amino acid sequences were constructed using the program PhyML version 3 (48) with HIVb+I+G (49), with the best-fit model of protein amino acid replacement chosen using the ProtTest version 2.4 software program (50) based on the Akaike information criterion (51). Regions that could not be unambiguously aligned were removed from the analysis. Sequence data sets include published SIVsmE660 and SIVmac251 sequences (32, 52, 53), published SIVsmm sequences (5, 45–47, 54–64), and HIV-1 group M sequences from the Los Alamos National Laboratories Subtype Reference set (47, 65, 66).
Antibodies and sera.
The antibodies 3.11H, 6.10F, 1.10A, 1.9c, 1.7A, 1.4B, 6.10B, and 1.4H were provided by J. Robinson (Tulane University Medical Center, New Orleans, LA), and their neutralization properties and target epitopes have been reported (39, 40). The SIV sera were provided by N. Letvin (Harvard University, Boston, MA) and by D. Watkins (University of Wisconsin, Madison, WI) from chronically infected rhesus macaques that were intrarectally challenged and productively infected with SIVsmE660 or SIVmac251.
Env gene cloning.
SGA-derived isolate env sequences and T/F env sequences identified previously (30) were molecularly cloned for the production of pseudovirus and phenotypic analyses. The primers used to amplify these genes had been designed such that the DNA amplicon would contain a complete rev/env cassette. To reduce the probability of generating molecular env clones with Taq polymerase errors, we reamplified from the first-round PCR product under the same nested PCR conditions but used only 35 cycles. Correctly sized amplicons identified by gel electrophoresis were gel purified by using the QIAquick gel purification kit according to the recommendations of the manufacturer (Qiagen), ligated into the pcDNA3.1 Directional Topo vector (Invitrogen Life Technologies), and transformed into TOP10 competent bacteria. Bacteria were plated on LB agar plates supplemented with 100 μg/ml ampicillin and cultured overnight at 30°C. Single colonies were selected and grown overnight in liquid LB broth at 30°C with 225-rpm shaking followed by plasmid isolation. Finally, each molecular clone was sequence confirmed to be identical to the previously determined env sequence of the T/F env or isolate env amplicon.
Pseudovirus preparation and titration.
Pseudovirus was prepared by transfecting 3 × 106overnight-cultured 293T cells in 10-cm2 tissue culture dishes with 4 μg of rev/env expression plasmid and 4 μg of the HIV-2 backbone construct pJK7312AΔEnv using Fugene 6 (Roche Applied Science, Indianapolis, IN). Pseudovirus-containing culture supernatants were harvested 2 days after transfection, cleared of cellular debris by low-speed centrifugation, and stored in aliquots at −80°C. Viruses were titrated on TZM-bl reporter cells (8129; NIH AIDS Research and Reference Reagent Program), which contain a Tat-inducible luciferase and a β-galactosidase gene expression cassette. Infectious titers were measured on 24-well plates based on β-galactosidase production, representing the number of infection events per μl of virus stock (IU/μl), as described previously (67, 68). Viruses determined to have low IU/μl were subsequently produced in the presence of 10 mM sodium butyrate (TCI America, Portland, OR) at the time of transfection. Viruses with infectious titers below 25 IU/μl were not used.
IMC generation.
The full-length T/F IMCs, pE660.CG7G.ir1 and pE660.CG7V.ir1, were constructed from five overlapping pieces amplified from PBMC DNA isolated during peak infection following low-dose rectal transmission (27) using the primers SIVsmE660-1F (5′-TGGAAGGGATTTATTACAATGAGAAAAGAC-3′), SIVsmE660-1R (5′-GTATTGGATCAGGGTCACATC-3′), SIVsmE660-2F (5′-GGAAGCCCCTCCAACCAATCC-3′), SIVsmE660-2R (5′-CCCAGGACCTTCGCCAAACC-3′), SIVsmE660-3F (5′-CATGTCAGATCCCAGGGAGAGG-3′), SIVsmE660-3R (5′-CCTTCTTACACTTGTGGGGGCC-3′), SIVsmE660-4F (5′-GAGACCAATATCACCATGAGTGC-3′), SIVsmE660-4R (5′-GCTGAATAGCCAAGTCAAGAGGCG-3′), SIVsmE660-5F (5′-CCCCTCCCGCTTATGTTCAGC-3′), and SIVsmE660-5R (5′-TGCTAGGGATTTTCCTGCYTCGG-3′). Each segment was designed around a unique restriction site, amplified separately using the Phusion high-fidelity DNA polymerase (Finnzymes, Vantaa, Finland) and subcloned into pCR-XL-TOPO. Individual segments were sequenced to ensure an identical match to the T/F virus sequence, as previously identified (M. Lopker and G. M. Shaw, unpublished). Full-length IMCs were assembled in pBR-322-MCS (69) and sequenced again to confirm a match to the T/F sequence.
IMC virus stocks.
293T-derived virus was produced by transfecting 3 × 106 293T cells cultured overnight in 10-cm2 tissue culture dishes with 6 μg of pE660.CG7G.ir1 and pE660.CG7V.ir1 DNA. Culture supernatants were harvested 2 days after transfection, cleared of cellular debris by low-speed centrifugation, and stored in aliquots at −80°C. Viruses were titrated on TZM-bl reporter cells. CD4+ T cell stocks were derived by isolating rhesus PBMCs from whole blood by gradient centrifugation over Ficoll-Hypaque Plus medium (GE Healthcare, Piscataway, NJ). CD4+ T cells were positively selected using magnetic nonhuman primate CD4+ microbeads (Miltenyi Biotec, Auburn, CA) and an autoMACS cell separator (Miltenyi Biotec, Auburn, CA). CD4+ T lymphocytes were activated in RPMI 1640 medium supplemented with 15% fetal bovine serum (FBS) and 3 μg/ml of staphylococcal enterotoxin B (SEB) (Sigma-Aldrich) and 30 U/ml interleukin-2 (IL-2) (Roche Diagnostics) for 48 h at 37°C. Activated CD4+ cells were infected in a small volume (<2 ml) at a multiplicity of infection (MOI) of 0.1, washed 3 times, and then suspended at a concentration of 1 × 106/ml in RPMI 1640 medium, 15% FBS, and 30 U/ml of IL-2. On day four postinfection and every 2 days thereafter, cells were collected by low-speed centrifugation, and a complete medium exchange was performed and saved. Viral replication was assessed retrospectively by SIV p27 antigen enzyme-linked immunosorbent assay (ELISA) (Zeptometrix Corporation, Buffalo, NY), and a final stock was created by combining the three time points with the highest viral load: days 6, 8, and 10.
Neutralization assays.
Virus neutralization by sera and MAbs was assessed on TZM-bl cells as described previously (68, 70). TZM-bl cells were seeded and cultured in 96-well plates for 24 h. Virus stocks dilutions were made to final concentrations of Dulbecco's modified Eagle medium (DMEM) containing 6% FBS and 40 μg/ml DEAE-dextran (Sigma-Aldrich, St. Louis, MO) to achieve 2,000 IU/well. Viruses with low numbers of IU/μl were added at no less than 1,500 IU/well. Validation experiments showed no difference in neutralization results for viruses at 1,500 IU/well or 2,000 IU/well. Equal-volume virus dilutions and 5-fold serially diluted sera or MAbs were mixed and incubated at 37°C for 1 h. Supernatants were then removed, and 100 μl of these mixtures were added. Medium-only and virus-only control wells were included as background and 100% infectivity, respectively. Luciferase activity was measured after 48 h of incubation in 37°C. Neutralization of SIVsmE660 virus grown in rhesus CD4+ T cells and tested on rhesus CD4+ T cells in a multi-replication-cycle assay format was performed as described by Montefiore and colleagues for HIV-1 in human CD4+ T cells (71, 72). Rhesus macaque PBMCs were isolated from blood of 5 animals by gradient centrifugation over Ficoll-Hypaque Plus medium, and CD4+ T cells were positively selected, combined, and activated using staphylococcal enterotoxin B (50 ng/ml) for 3 days. Cells were then cultured for 3 days in 15% FBS-RPMI and 30 U/ml IL-2 to allow for expansion. Five hundred 50% tissue culture infective doses (TCID50) of CD4+ T cell-derived virus and test or control plasma (1 × 10−4 dilution) or MAbs (0.1 μg/ml) were combined and incubated for 1 h before the addition of 3 × 105 PBMCs per well in a total volume of 154 μl. All plasma dilutions were tested in triplicate in a 96-well plate format, and on the next day, three complete medium exchanges were performed. Thirty microliters of medium was removed daily from day 2 through day 7 and replaced with fresh medium each day. Virus replication was measured by RT assay (reverse transcriptase assay, colorimetric; Roche Applied Science, Indianapolis, IN). Virus neutralization was assessed during exponential virus growth, generally at 4 days postinoculation.
Nucleotide sequence accession numbers.
Sequences determined in this work have been deposited in GenBank (accession numbers KC595632 to KC595664).
RESULTS
Genetic diversity of the SIVsmE660 isolate.
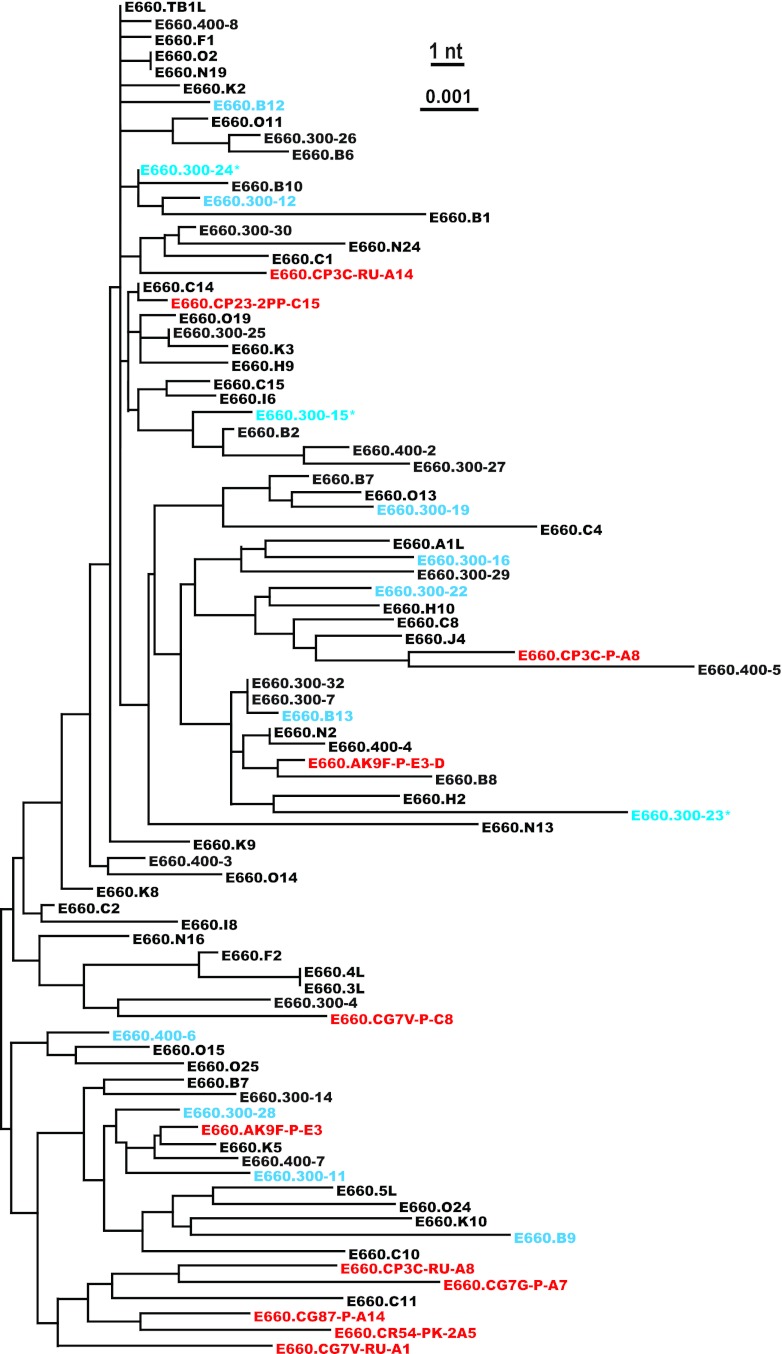
We performed SGS of vRNA extracted from the SIVsmE660 isolate to obtain 75 gp160 env sequences. These sequences exhibited 1.61% maximum within-env diversity, which is similar to previous determinations of SIVsmE660 diversity and similar to HIV-1 diversity in humans infected for 1 to 2 years (27, 30, 73, 74). Figure 1 illustrates the genetic relationship of the SIVsmE660 Env amino acid sequences compared with SIVmac251 and other SIVsmm lineages (32, 52–54). In addition to SIVsmE660 isolate env sequences, we also analyzed env sequences of SIVsmE660 viruses that were transmitted by low-dose intrarectal inoculation of the SIVsmE660 isolate in Indian rhesus macaques (27). Eleven of these T/F env genes are depicted in a phylogenetic tree containing the SIVsmE660 isolate env sequences (Fig. 2). As shown here and elsewhere (27), the T/F sequences are relatively evenly distributed throughout the tree, indicating that no particular sublineage of SIVsmE660 was preferentially transmitted mucosally from the isolate to naive monkeys. In addition, the isolate clones that we generated for phenotypic analysis are randomly dispersed throughout the tree, highlighting the genetic heterogeneity and lack of phylogenetic clustering of the envelope glycoprotein sequences.
Fig 1.
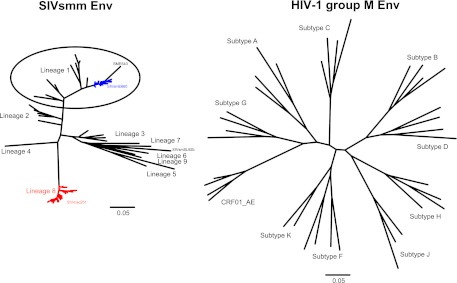
Maximum-likelihood (ML) phylogenetic trees inferred from representative SIVsmm and HIV-1 group M Env amino acid sequences. The computed ML genetic distances include correction for multiple substitutions and thus show greater genetic distance between divergent clades than does the uncorrected pairwise sequence difference. There is less overall diversity among naturally occurring SIVsmm lineages than among HIV group M subtypes at an amino acid level but not at a nucleotide level, a finding that may reflect the more ancient origin of SIVsmm (54). Sequence data sets were derived from published SIVsmm and HIV-1 sequences, and SIVsmm lineages were designated according to the method of Apetrei et al. (103), as described in Materials and Methods. The scale bars represent 5% diversity.
Fig 2.
Neighbor-joining phylogeny of SIVsmE660 uncloned isolate-derived env genes, cloned isolate env genes, and T/F env genes. Uncloned isolate env sequences are represented in black, cloned isolate env sequences in blue, and T/F env sequences in red. The isolate clones with asterisks were nonfunctional in cell entry assays. The SIVsmE660 isolate env sequences displayed 1.61% maximum within-env diversity. The scale bar represents 0.1% diversity.
The SIVsmE660 isolate is neutralization sensitive.
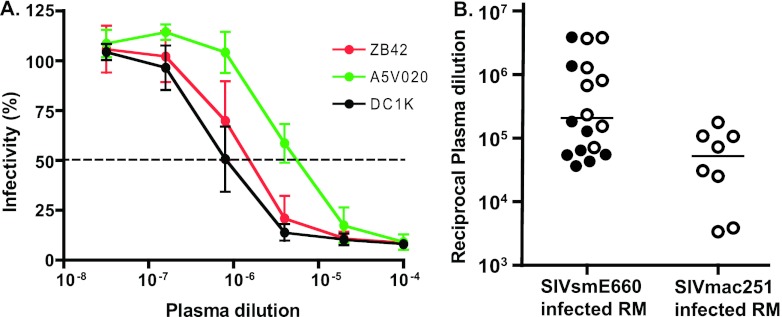
The SIVsmE660 isolate was expanded on rhesus CD4+ T cells and tested in the TZM-bl infectivity assay (68, 75, 76) for neutralization sensitivity to SIV-infected rhesus macaque plasma samples. As shown in Fig. 3A, the SIVsmE660 isolate was highly neutralization sensitive to SIVsmE660-infected macaque plasmas, with typical sigmoidal neutralization curves asymptotically approaching >90% neutralization. The neutralization sensitivities of the SIVsmE660 isolate to plasma specimens from SIVsmE660-infected macaques from two different primate centers were comparable (P > 0.8, Wilcoxon test), with a median reciprocal 50% inhibitory concentration (IC50) of 1.8 × 105 (range, 3.7 ×104 to 3.9 × 106) (Fig. 3B). The SIVsmE660 isolate was also sensitive to heterologous neutralization by SIVmac251-infected macaque plasmas (median reciprocal IC50 of 5.2 × 104), although SIVmac251 plasma titers were lower than those for plasma of SIVsmE660-infected animals (P = 0.016, Mann-Whitney).
Fig 3.
Neutralization sensitivity of the uncloned SIVsmE660 isolate. (A) Neutralization sensitivity of the uncloned SIVsmE660 isolate, expanded on rhesus CD4+ T cells and tested with three SIV-infected macaque plasmas (ZB42, A5V020, and DC1K) on TZM-bl cells. Values represent means ± SD for three separately performed experiments. (B) Mean IC50s of isolate tested with SIVsmE660- and SIVmac251-infected macaque plasma samples on TZM-bl cells. Horizontal lines indicate median values. Titers from SIVsmE660-infected rhesus macaques from New England (shown in open circles) and Wisconsin (shown in filled circles) primate centers were statistically similar; P > 0.8 by Wilcoxon test. Titers from SIVsmE660-infected macaques were statistically greater than those from SIVmac251-infected macaques; P = 0.016 (Mann-Whitney).
SIVsmE660 isolate cloned Envs and T/F Envs are variable in their neutralization sensitivity to macaque plasma samples.
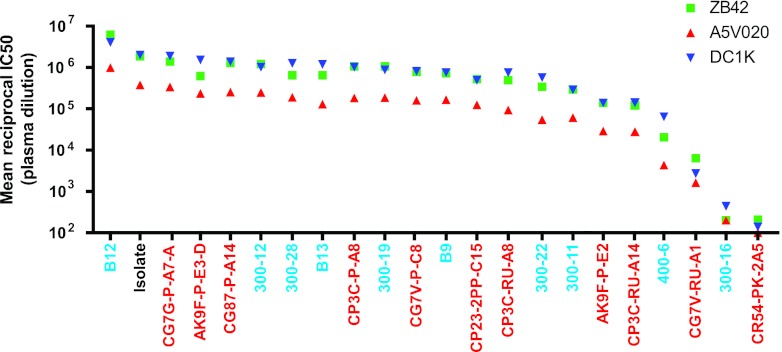
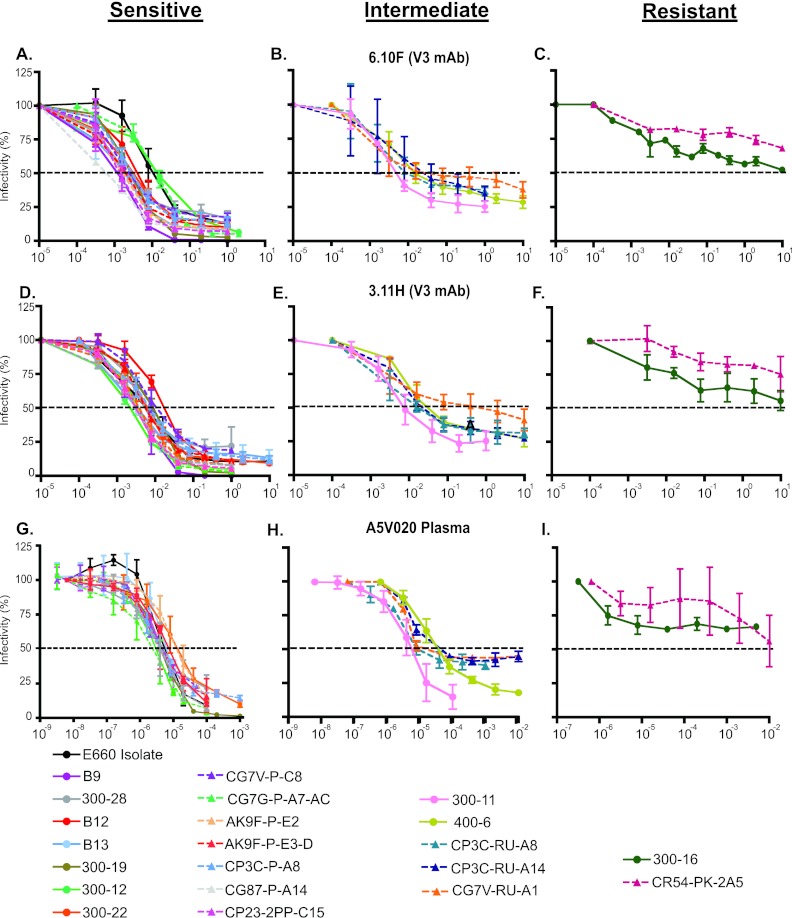
We next considered the neutralization sensitivities of the individual viruses comprising the isolate. Thirteen env sequences distributed throughout the phylogenetic tree (Fig. 2) were molecularly cloned, expressed in 293T cells, and pseudotyped with an Env-minus HIV-2 backbone (pJK7312AΔEnv). As expected, not all cloned Envs were biologically functional: only 10 of 13 could confer efficient entry into the TZM-bl target cells. This finding is consistent with those of previous studies (77) and can be explained by lethal mutations that occur in the most recent replication cycle but which still allow for viral gene expression and virion production and release in vivo. This is not the case for env genes and full-length genomes of T/F viruses, which by definition are biologically functional and capable of establishing productive clinical infection (27, 30, 42, 78, 79). We studied 11 T/F Envs identified and molecularly cloned from 7 rhesus macaques that were productively infected with between one and five T/F viruses through low-dose mucosal inoculation (27, 41). All 11 T/F Envs were functional in the TZM-bl assay. When tested for neutralization sensitivity to SIV-infected macaque plasma samples, most of the isolate-derived Env clones and T/F Env clones were highly neutralization sensitive, displaying mean IC50s similar to that of the uncloned isolate (Fig. 4). A minority of isolate-derived Env clones and T/F Envs exhibited significantly greater resistance to neutralization; one isolate-derived Env (300-16) and one T/F Env (CR54-PK-2A5) were highly resistant to the panel of macaque plasma samples, and additional Envs (CG7V-RU-A1 and 400-6) exhibited intermediate IC50s to the macaque plasma samples. The differences in IC50s of rhesus polyclonal anti-SIV antibodies between sensitive and resistant Envs spanned more than 4 orders of magnitude, from 3.8 × 106 for the isolate-derived Env clone B12 to 1.5 × 102 for T/F Env clone CR54-PK-2A5 (Fig. 4).
Fig 4.
Mean reciprocal IC50 titers for three SIV-infected macaque plasmas (ZB42, A5V020, and DC1K) for the uncloned SIVsmE660 isolate, Env clones derived from the isolate, and T/F Env clones tested on TZM-bl cells. Clone designation (x axis) and color coding correspond to Fig. 2.
SIVsmE660 isolate cloned Envs and T/F Envs vary in their neutralization sensitivities to V3, CD4i, CD4bs, and V4-targeting MAbs.
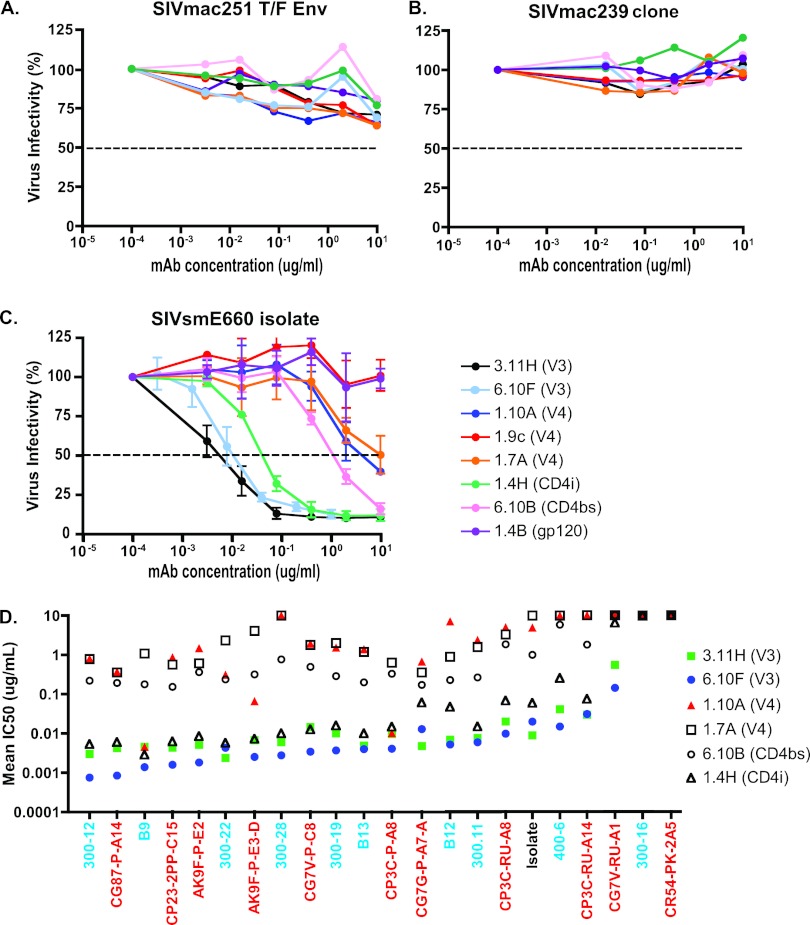
To further assess the neutralization properties of the uncloned SIVsmE660 isolate, isolate-derived Env clones, and T/F virus-derived Env clones, we tested each for sensitivity to neutralization by a panel of well-characterized neutralizing MAbs. The MAbs were obtained from Epstein-Barr virus (EBV)-transformed B lymphocytes from HIV-2-infected humans and SIV-infected macaques. The MAb neutralization specificities were defined by peptide binding, site-directed mutagenesis, and antibody competition. They targeted a range of gp120 epitopes, including V3 (3.11H, 6.10F), V4 (1.10A, 1.9c, 1.7A), the CD4 binding site (CD4bs) (6.10B), a CD4-induced epitope (CD4i) (1.4H), or other gp120 epitopes (1.4B) (18, 40, 80, 81). Each of the MAbs, with the exception of 1.4H, demonstrated binding to SIVmac239 and other SIV Env monomers by ELISA; 1.4H bound some Envs alone, while other virus strains required gp120 to be complexed with CD4 for efficient binding, as has been described for CD4-induced epitopes (18, 80, 81). As a control, we tested the MAb panel's capacity to neutralize a T/F Env clone derived from low-dose mucosal inoculation with SIVmac251 (27) and the SIVmac239 clone. Consistent with previous reports describing these viruses as resistant to neutralization by polyclonal plasma IgG and neutralizing MAbs, we found the SIVmac251 T/F Env and SIVmac239 isolate to be resistant to neutralization by the entire MAb panel, even at 10-μg/ml concentrations (Fig. 5A and B). The SIVsmE660 isolate, in contrast, demonstrated sensitivity to neutralization by most of the MAbs (Fig. 5C). The MAbs targeting V3 (3.11H and 6.10F) and CD4i (1.4H) neutralized the SIVsmE660 isolate potently with IC50s below 0.1 μg/ml. In addition, the isolate was moderately sensitive to MAbs targeting the CD4 binding site (6.10B) and V4 (1.10A and 1.7A) but was resistant to two other MAbs targeting gp120 (1.9C and 1.4B). As with the uncloned isolate, the majority of the individual isolate Envs and T/F cloned Envs displayed exquisite sensitivity to neutralization by the V3- and CD4i-targeting MAbs, with IC50s of <0.1 μg/ml for one or more of these MAbs (Fig. 5B). As was seen with plasma neutralization, two of the Env clones, a single T/F Env (CR54-PK-2A5) and a single isolate Env (300-16), were outright resistant to neutralization by the panel of MAbs. Additionally, Envs that displayed intermediate IC50s to plasma (CG7V-RU-A1 and 400-6) also displayed intermediate IC50s to the V3 and CD4i MAbs (Fig. 5D). Similar to the uncloned isolate, the isolate-derived Env clones and T/F Env clones were neutralized by CD4bs and V4 MAbs at higher concentrations (Fig. 5D).
Fig 5.
Neutralization sensitivity to monoclonal antibodies. Neutralization sensitivity of the SIVmac251 T/F Env clone PBE (27, 41) (A), the SIVmac239 clone (B), or the uncloned SIVsmE660 isolate (C). IC50 titers for the uncloned SIVsmE660 isolate, isolate Env clones, and T/F Env clones of selected MAbs (D). Clone designation (x axis) and color coding correspond to Fig. 2. Results represent the means (A, B, and D) or mean ± SD (C) for at least three independent neutralization experiments in the TZM-bl assay.
Neutralization sensitivity to V3 MAbs correlates with sensitivity to plasma polyclonal antibodies.
Figure 6 depicts the virus neutralization curves for plasma and MAbs, grouped based on the percent infectivities remaining at high concentrations of MAb. The top two rows show responses to the two V3 MAbs (6.10F and 3.11H), and the bottom row shows neutralization by a representative SIVsmE660-infected macaque plasma sample. The majority (66%) of the Envs demonstrated the highly sensitive phenotype shown in the left column, with typical sigmoidal curves with less than 25% retained infectivity with both MAbs and plasma antibody (Fig. 6A, D, and G). In the right column, the two resistant viruses (CR54-PK-2A5 and 300-16) retained >50% baseline infectivity at high concentrations of both MAb and plasma antibody (Fig. 6C, F, and I). In between these two phenotypes, there were five Envs with flattened neutralization curves and plateaus of retained infectivity between 25% and 50% for the V3 MAbs (Fig. 6B and E). Four of these five Envs also displayed similar plateaus of retained infectivity with plasma (Fig. 6H). Thus, for the uncloned isolate and each of the Env clones, the sensitivity to neutralization by V3-targeting MAbs closely paralleled their neutralization sensitivity to polyclonal macaque plasma. In addition, the frequencies of sensitive, intermediate, and resistant Envs between the isolate Envs and T/F Envs were similar (P > 0.8 by the Wilcoxon test), with 70% versus 64% sensitive, 20% versus 27% intermediate, and 10% versus 9% resistant in the isolate versus T/F Envs, respectively. By inspecting the primary sequences of neutralization-resistant, -intermediate, and -sensitive viruses, we could identify no salient differences in number or position of potential N-linked glycosylation sites, variable loop length, or sequence motifs that could distinguish them.
Fig 6.
Neutralization curve of the isolate and Env clones for V3-targeting MAbs and immune plasma. Three distinct profiles are depicted: sensitive (infectivity < 25% at a high MAb concentration), intermediate (infectivity 25 to 50% at a high MAb concentration), and resistant (infectivity > 50% at a high MAb concentration). Antibody concentrations are expressed as concentrations (μg/ml) or plasma dilutions (x axis). Experimental values depicted represent means ± SD for at least three independently performed experiments with TZM-bl cells.
Neutralization sensitivity is only modestly influenced by the source of virus production.
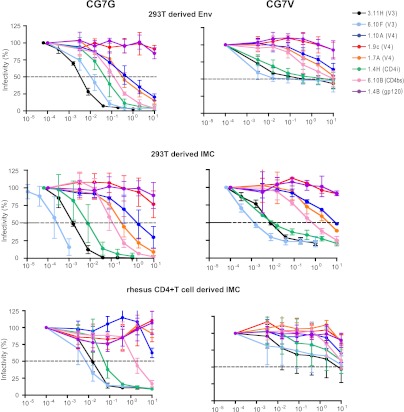
Previous studies have demonstrated differences in neutralization sensitivity based on the cell type of virus derivation and the use of pseudotyped virus compared with full-length IMCs (39, 82–84). To assess differences in SIVsmE660 neutralization sensitivity based on the cell type from which the Envs were derived, we utilized the T/F Env pseudoviruses CG7G-P-A7-A and CG7V-RU-A1 and their isogenic full-length IMCs pE660.CG7G.ir1 and pE660.CG7V.ir1. CG7G-P-A7-A is a neutralization-sensitive virus, representative of the majority of the SIVsmE660 clones tested, whereas CG7V-RU-A1 has a more intermediate sensitivity to neutralization, with higher IC50s and a 40 to 50% plateau of retained infectivity with V3 MAbs and plasma (Fig. 6). Importantly, the IMCs and Env clones contained identical gp160 nucleotide sequences. The IMC viruses were generated in two ways, transfection of IMC DNA into 293T cells or short-term expansion in rhesus macaque CD4+ T cells. The Env pseudoviruses were generated by transfection of 293T cells. Compared with the 293T-derived Env neutralization pattern, the 293T-derived IMC viruses displayed similarly shaped neutralization curves and a modest increase in neutralization sensitivity as determined by IC50. With the exception of neutralization of CG7G by the MAb 6.10F, the relative order of MAb sensitivities was consistent, with V3 MAbs the most potent, followed by CD4i-, CD4bs-, and V4-specific MAbs. For CG7G, generating the IMCs in rhesus macaque CD4+ T cells shifted the neutralization curves slightly to the right, increasing the IC50s of the V3 and CD4i MAbs by approximately 10-fold. CD4+ T cell-generated virus also rendered additional MAbs targeting V4 incapable of neutralization at any concentration; however, the order of MAb specificities remained intact. For CG7V, the order of specificity was also unchanged, while the neutralization plateau was raised such that some MAbs no longer reached 50% neutralization at high antibody concentrations (Fig. 7). Thus, the phenotypes of neutralization-sensitive (CG7G) and -resistant (CG7V) viruses were retained regardless of the Env expression system (Env-pseudotype versus IMC) or cell of derivation (293T versus rhesus CD4 T cell).
Fig 7.
Effect of virus cell source on neutralization profile of SIVsmE660. Neutralization sensitivity of SIVsmE660 T/F Env pseudotyped virus versus full-length infectious molecular clone-derived virus by cell of derivation. Isogenic Env clones and IMCs were generated by transfection of 293T cells (top and middle rows) or infection of primary rhesus CD4+ T cells (bottom row). The left column is T/F virus CG7G, and the right column is T/F virus CG7V, both tested with a panel of MAbs in TZM-bl cells. Results represent the means ± SD for at least three independent experiments.
SIVsmE660 is neutralization sensitive when grown and tested in primary rhesus CD4+ T cells.
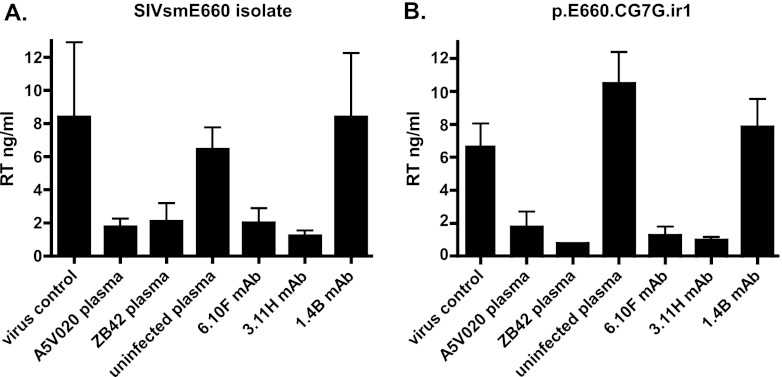
HIV-1 and SIV isolates, including SIVsmE660, when grown and tested in primary PBMCs or isolated CD4+ T cells, have been reported to be modestly less sensitive to neutralization than when tested in immortalized human cells, including 293T and TZM-bl cells. Such differences, however, have been variable (24, 39). To determine whether the exquisite neutralization sensitivity of the SIVsmE660 isolate (and T/F viruses derived from it) that we observed in the TZM-bl system would also be seen with virus grown and tested in primary rhesus CD4+ T cells, we employed a conventional multi-replication-cycle virus neutralization assay described by Montefiore and colleagues (71, 72). The SIVsmE660 isolate and pE660.CG7V.ir1 virus stocks grown on primary rhesus macaque CD4+ T cells were preincubated with low concentrations of polyclonal immune or control nonimmune rhesus plasmas (1 × 10−4 dilution) or low concentrations (0.1 μg/ml) of neutralizing (6.10F and 3.11H) or nonneutralizing control (1.4B) MAbs. These concentrations of plasma and MAbs effectively neutralized the uncloned isolate, isolate- and T/F-derived Envs, and T/F IMCs in the TZM-bl assay (Fig. 2, 6, and 7 and data not shown). As shown in Fig. 8A, the polyclonal immune plasmas A5V020 and ZB42, but not the nonimmune control plasma, effectively mediated virus neutralization (P < 0.05 for both; Student's t test) for the SIVsmE660 isolate. Similarly, low concentrations (0.1 μg/ml) of the V3-targeting neutralizing MAbs 6.10F and 3.11H but not the nonneutralizing control MAb 1.4B effectively neutralized the SIVsmE660 isolate (P < 0.005 for both). The findings were similar for the pE660.CG7V.ir1 IMC (Fig. 8B); the virus strain was potently neutralized by immune plasma samples and neutralizing MAbs but not by control antibodies (P < 0.005 for all comparisons).
Fig 8.
Neutralization sensitivity of the SIVsmE660 isolate and pE660.CG7G.ir1 grown in primary rhesus CD4+ T lymphocytes and tested with the same target cells. Neutralization was assessed at low concentrations (1 × 10−4) of the chronic immune plasmas A5V020 and ZB42 and uninfected control rhesus plasma and at low concentrations (0.1 μg/ml) of the V3-specific MAbs 6.10F and 3.11H and a control nonneutralizing gp120 binding MAb, 1.4B. Both the SIVsmE660 isolate (A) and pE660.CG7G.ir1 (B) were sensitive to neutralization by polyclonal immune plasma (P < 0.05 for each virus) and V3 MAbs (P < 0.005 for each virus) compared with results for mock-treated virus or virus incubated with control plasma or MAb. Values depicted represent means ± SD.
DISCUSSION
Optimizing a rational design and development strategy for HIV vaccines requires critical evaluation of all components of the NHP model of HIV infection, including challenge viruses. In this study, we explored the neutralization profiles of viruses comprising the SIVsmE660 isolate and vaccine challenge stock. We show that the uncloned SIVsmE660 isolate, a major proportion of viruses comprising the isolate, and the majority of T/F viruses arising from low-dose mucosal inoculation of the SIVsmE660 isolate are all remarkably sensitive to antibody-mediated neutralization by antibodies that result from infection of rhesus macaques by SIVsmE660 or SIVmac251 or by humans infected by HIV-2. The exquisite neutralization sensitivity of SIVsmE660 was evident regardless of the cell derivation of the virus or viral Envs, including pseudotyped Envs and full-length IMCs, whether viruses were derived from 293T or primary rhesus CD4+ T cells or tested in TZM-bl or primary rhesus CD4+ T lymphocytes.
Compared with primary HIV-1 strains, SIVsmE660 differs substantially in neutralization sensitivities to polyclonal and monoclonal antibodies. The SIVsmE660 isolate was susceptible to high-titer neutralization by homologous and heterologous SIV+ plasmas: SIVsmE660-infected macaque plasma samples generated median NAb titers greater than 1:100,000, while SIVmac251-infected plasma samples induced median titers greater than 1:10,000. Primary HIV-1 isolates, in contrast, are generally resistant to neutralization by the majority of heterologous plasmas, with most having tier 2 or tier 3 neutralization phenotypes that generally correspond to NAb titers of <1:100 (36, 85–87). Cross-reactive neutralizing antibodies capable of generating substantial neutralization titers against diverse HIV-1 strains arise in only a small fraction (∼10%) of HIV-1-infected individuals after years of infection (88–91).
Surprisingly, we found SIVsmE660 to be highly sensitive to neutralization by V3-, CD4i-, CD4bs-, and V4-specific MAbs. The accessibility of V3, CD4i, CD4bs, and V4 epitopes to neutralizing antibodies is quite different from the accessibility of these regions to antibody binding in functional HIV-1 Env trimers. In natural HIV-1 infection, V3 and CD4i regions are highly immunogenic and elicit early potentially neutralizing antibody responses (70, 75, 92); however, both V3 and CD4i epitope regions are well shielded on HIV-1 Env trimers. Studies of primary HIV-1 viruses, including T/F viruses, demonstrate resistance to V3 and CD4i MAbs (30, 36, 75, 93), corroborating structural data indicating that CD4i epitopes and the V3 loop region are not well exposed on the HIV-1 glycoprotein (93, 94). It is interesting, though not entirely unexpected given their common origin and degree of genetic homology (9, 13, 14, 16), to note the extent to which the SIVsmE660 and HIV-2 neutralization profiles overlap. Three recent studies of the antibody response to HIV-2 infection independently demonstrated that HIV-2-infected individuals mount high-titer, broadly reactive NAb responses (39, 95, 96). Similar to the uncloned SIVsmE660 isolate and Envs described here, the majority of primary HIV-2 strains, including SGA-derived Envs, were highly sensitive to antibody-mediated neutralization through multiple exposed epitopes (V3, CD4i, CD4bs, and V4), suggesting that SIVsmE660 and HIV-2 have more open, flexible Env conformations, akin to those of lab-adapted HIV-1 strains and distinct from those of primary HIV-1 isolates and T/F viruses (39). Thus, high-titer V3 and CD4i NAbs are present in SIV infection of macaques and HIV-1 and HIV-2 infection of humans, but such antibodies are able to neutralize virus only in the SIVsmE660/macaque model and HIV-2-infected humans. These data raise the possibility that misleading correlates of antibody-mediated protection could be derived from SIVsmE660 challenge studies, especially following low-dose mucosal challenge. For example, a vaccine strategy that elicits neutralizing antibodies targeting V3 or CD4i epitopes could have a substantial protective effect against most transmitted viruses in the SIVsmE660/macaque model but would be expected to be largely ineffective in HIV-1 infection. The exquisite neutralization sensitivity and lack of conformational masking of most viruses comprising the uncloned SIVsmE660 isolate may thus hamper the ability of SIVsmE660 challenge models to predict antibody responses to HIV-1 vaccine candidates.
A second principal finding from this study is the demonstration of striking heterogeneity in neutralization sensitivity within the SIVsmE660 isolate. Of the 21 Env clones we tested, the majority were exquisitely neutralization sensitive. Approximately 10%, however, were outright neutralization resistant, and an additional 10 to 25% exhibited an intermediate phenotype. These resistant and intermediate Envs did not cluster within the phylogeny of the overall SIVsmE660 tree (Fig. 2), and we could not identify within the sequences characteristic genetic signatures that were necessary and sufficient for neutralization resistance. We speculate that conformational changes in structure resulting from multiple or different sequence polymorphisms are responsible for the substantial differences that we observed in neutralization sensitivities. The 10 SIVsmE660 isolate-derived Env clones and the 11 T/F Env clones demonstrated a similar degree of neutralization heterogeneity, with comparable ratios of sensitive, intermediate, and resistant Envs (Fig. 6). The similar proportions further support the existence of a small population of resistant virus among a largely neutralization-sensitive virus quasispecies. These data also suggest that there is no significant transmission bottleneck based on neutralization sensitivity in the nonimmunized low-dose mucosal transmission model. The minority population of more neutralization-resistant viruses could have greater significance, however, depending on whether animals are inoculated with large or small (limiting-dilution) amounts of virus, where a minor population of NAb-resistant variants could prevail over sensitive variants in animals vaccinated with Env-based immunogens.
Within the minority population of more resistant SIVsmE660 viruses, we identified 5 of 21 (24%) with an intermediate neutralization phenotype. The neutralization curves from these viruses all displayed plateaus of retained infectivity at high antibody concentrations (Fig. 6, center column), with a 100-fold range of IC50s for plasma and MAbs. The plateau phenomenon has been seen in other studies of SIV and HIV-2 neutralization by several research groups (39, 97) and rarely if ever occurs with HIV-1 viruses tested with the same TZM-bl methodology. Given the range of IC50s and the potential that this phenomenon may be unique to SIV/HIV-2 viruses, it is unclear whether these viruses act in vivo as intermediate, tier 2-like viruses or as neutralization-sensitive viruses. This question merits further investigation in relation to SIVsmm viruses as vaccine challenge stocks and mechanistic explanations of virus neutralization. Depending on the in vivo properties of these plateau viruses, SIVsmE660 is comprised of 65 to 90% highly neutralization-sensitive viruses with a minority population of more resistant viruses. Of note, two of the HIV-2 studies described above also identified heterogeneity within the HIV-2 Env clones they studied; the vast majority were highly sensitive tier 1-like Envs, while a minority were highly resistant tier-3 like Envs (39, 95). This dichotomy of neutralization sensitivity, without representation of the intermediate-sensitivity tier 2-like Envs that predominate in HIV-1 infection, highlights still-unexplained differences in Env biology and Env-antibody interactions between HIV-2 and HIV-1.
To assess the neutralization sensitivity of SIVsmE660 as it relates to primary HIV-1 strains and other challenge viruses, we utilized pseudotyped Envs and the sensitive and standardized TZM-bl single-round infectivity assay (76, 98) to generate neutralization titers that translate directly to clinical and preclinical studies of HIV-1 and other SIV viruses. Neutralization assays using multiple rounds of virus infection in CD4+ T target cells may better reflect certain biologically relevant virus-cell interactions in vivo, but they are still only an approximation and are less standardized, less precise, and less widely implemented. Of relevance to the present study, an earlier report by Letvin et al. demonstrated incomplete neutralization of the SIVsmE660 isolate by immune plasma samples when tested in a PBMC target cell assay; these data suggested an intermediate or tier 2-like neutralization profile for the SIVsmE660 virus swarm (24). Our findings were different: we found that both the SIVsmE660 isolate and the T/F IMC pE660.CG7V.ir1 IMC-derived virus were highly neutralization sensitive to dilute immune plasma (1 × 10−4 dilution) and low concentrations (0.1 μg/ml) of V3-targeting MAbs in the primary rhesus CD4+ T cell assay. Our results also corroborate other published comparisons of neutralization differences between the TZM-bl and PBMC target cell assays (39), which demonstrated only modest differences in neutralization sensitivities of primary viruses between the two assay systems. To assess the influence of pseudotyped Envs versus that of full-length viruses and the role of the cellular source of these viruses, we compared isogenic Envs and IMCs generated in 293T cells and primary rhesus CD4+ T cells. Utilizing one highly neutralization-sensitive virus (CG7G) and one intermediate virus (CG7V), we found only modest differences in the neutralization profiles of the viruses for MAb and polyclonal plasma antibodies based on the cell of derivation and the Env pseudotype versus IMC. The general phenotypic properties and order of neutralization sensitivities were relatively consistent (Fig. 7 and data not shown). The changes in neutralization sensitivity between pseudoviruses and IMCs and by the cell of derivation compare favorably with previous reports of similar studies conducted with HIV-1 and HIV-2 (39, 82–84) and likely are due to the same proposed mechanisms of differential Env incorporation, glycosylation pattern, or host cell protein incorporation. It remains unclear, however, how in vitro NAb titers assayed by the TZM-bl system compare with in vivo effective NAb titers. Recent studies suggest that low titers of NAbs in vivo can substantially affect HIV-1 infectivity and replication efficiency in vivo (99).
In summary, we have demonstrated that the SIVsmE660 isolate quasispecies is highly sensitive to neutralization by a broad range of human (HIV-2), homologous rhesus (SIVsmE660), and heterologous rhesus (SIVmac251) antibodies, by a mechanism that involves antibody access to V3, CD4i, CD4bs, and V4 epitopes. We also demonstrated that the uncloned isolate possesses a minor fraction of substantially more resistant viruses, and this dichotomy could have unanticipated effects on the outcome of low-dose SIVsmE660 challenge studies in terms of both disease natural history and vaccine-elicited immune protection by Env antibodies. While these features of the SIVsmE660-rhesus infection model distinguish it from HIV-1 infection in humans, they have interesting and still-unexplained parallels with HIV-2 infection of humans (39, 95, 96). The converse of these features of SIVsmE660 and HIV-2 neutralization sensitivity is the property of pan-neutralization resistance of SIVmac251 and SIVmac239, which mechanistically remains largely unexplained (17–19, 100). Thus, much more needs to be understood about structure-function-antigen relationships of HIV-1, HIV-2, and the SIV strains that are used to model these important human pathogens. In the meantime, the exquisite neutralization sensitivity and heterogeneity in the SIVsmE660 isolate are relevant to the interpretation of recent NHP vaccine challenge studies demonstrating significant, but incomplete, vaccine-induced protection from repeated low-dose mucosal challenge with SIVsmE660 (24, 101, 102). Indeed, our data suggest the possibility that the variable protection afforded by heterologous immunization in animals challenged mucosally with SIVsmE660 may be due to vaccine-induced protection only against the majority population of highly sensitive viruses, with breakthrough infections arising from the minority population of resistant viruses. Characterization of the neutralization sensitivities of these breakthrough T/F viruses to test this hypothesis may be informative. In contrast, and despite its widespread use in vaccine challenge studies, we are aware of only a single report of an 80% reduction in per-challenge risk against mucosal challenge with SIVmac251 (35). For these and other reports of vaccine-induced protection from heterologous challenge, the improved understanding of the relative neutralization sensitivities of SIVsmE660 and SIVmac251 in comparison with that of HIV-1 will facilitate interpretation of NHP vaccine challenge trials, as well as the design of future vaccine challenge studies using the NHP model.
ACKNOWLEDGMENTS
We thank D. Watkins (University of Miami) for rhesus macaque plasma specimens, the core research facilities at the Center for HIV/AIDS Research at UAB and the University of Pennsylvania, and Patricia Crystal and Shivani Sethi for manuscript preparation.
This work was supported by an early-stage investigator grant through the Center for HIV/AIDS Vaccine Immunology and the HIV Vaccines Trials Network (U19AI067854) and by grants from the National Institutes of Health (AI087383, AI088564, and AI095985).
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. Benveniste RE, Arthur LO, Tsai CC, Sowder R, Copeland TD, Henderson LE, Oroszlan S. 1986. Isolation of a lentivirus from a macaque with lymphoma: comparison with HTLV-III/LAV and other lentiviruses. J. Virol. 60:483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daniel MD, Letvin NL, King NW, Kannagi M, Sehgal PK, Hunt RD, Kanki PJ, Essex M, Desrosiers RC. 1985. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228:1201–1204 [DOI] [PubMed] [Google Scholar]
- 3. Letvin NL, Daniel MD, Sehgal PK, Desrosiers RC, Hunt RD, Waldron LM, MacKey JJ, Schmidt DK, Chalifoux LV, King NW. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71–73 [DOI] [PubMed] [Google Scholar]
- 4. Murphey-Corb M, Martin LN, Rangan SR, Baskin GB, Gormus BJ, Wolf RH, Andes WA, West M, Montelaro RC. 1986. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature 321:435–437 [DOI] [PubMed] [Google Scholar]
- 5. Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389–392 [DOI] [PubMed] [Google Scholar]
- 6. Fultz PN, McClure HM, Anderson DC, Swenson RB, Anand R, Srinivasan A. 1986. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys). Proc. Natl. Acad. Sci. U. S. A. 83:5286–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. 2007. Understanding the benign nature of SIV infection in natural hosts. J. Clin. Invest. 117:3148–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427–431 [DOI] [PubMed] [Google Scholar]
- 9. Hirsch VM, Zack PM, Vogel AP, Johnson PR. 1991. Simian immunodeficiency virus infection of macaques: end-stage disease is characterized by widespread distribution of proviral DNA in tissues. J. Infect. Dis. 163:976–988 [DOI] [PubMed] [Google Scholar]
- 10. Hirsch VM, Lifson JD. 2000. Simian immunodeficiency virus infection of monkeys as a model system for the study of AIDS pathogenesis, treatment, and prevention. Adv. Pharmacol. 49:437–477 [DOI] [PubMed] [Google Scholar]
- 11. Goldstein S, Elkins WR, London WT, Hahn A, Goeken R, Martin JE, Hirsch VM. 1994. Immunization with whole inactivated vaccine protects from infection by SIV grown in human but not macaque cells. J. Med. Primatol. 23:75–82 [DOI] [PubMed] [Google Scholar]
- 12. Lewis MG, Bellah S, McKinnon K, Yalley-Ogunro J, Zack PM, Elkins WR, Desrosiers RC, Eddy GA. 1994. Titration and characterization of two rhesus-derived SIVmac challenge stocks. AIDS Res. Hum. Retroviruses 10:213–220 [DOI] [PubMed] [Google Scholar]
- 13. Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Greene BM, Sharp PM, Shaw GM, Hahn BH. 1992. Human infection by genetically diverse SIVsm-related HIV-2 in west Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 14. Marx PA, Li Y, Lerche NW, Sutjipto S, Gettie A, Yee JA, Brotman BH, Prince AM, Hanson A, Webster RG, et al. 1991. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type 2 from a west African pet sooty mangabey. J. Virol. 65:4480–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 16. Shedlock DJ, Silvestri G, Weiner DB. 2009. Monkeying around with HIV vaccines: using rhesus macaques to define ‘gatekeepers’ for clinical trials. Nat. Rev. Immunol. 9:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson WE, Lifson JD, Lang SM, Johnson RP, Desrosiers RC. 2003. Importance of B-cell responses for immunological control of variant strains of simian immunodeficiency virus. J. Virol. 77:375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson WE, Sanford H, Schwall L, Burton DR, Parren PW, Robinson JE, Desrosiers RC. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Means RE, Greenough T, Desrosiers RC. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeh WW, Rahman I, Hraber P, Coffey RT, Nevidomskyte D, Giri A, Asmal M, Miljkovic S, Daniels M, Whitney JB, Keele BF, Hahn BH, Korber BT, Shaw GM, Seaman MS, Letvin NL. 2010. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J. Virol. 84:6018–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu F, Ourmanov I, Kuwata T, Goeken R, Brown CR, Buckler-White A, Iyengar R, Plishka R, Aoki ST, Hirsch VM. 2012. Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J. Virol. 86:8835–8847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ourmanov I, Bilska M, Hirsch VM, Montefiori DC. 2000. Recombinant modified vaccinia virus Ankara expressing the surface gp120 of simian immunodeficiency virus (SIV) primes for a rapid neutralizing antibody response to SIV infection in macaques. J. Virol. 74:2960–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ourmanov I, Kuwata T, Goeken R, Goldstein S, Iyengar R, Buckler-White A, Lafont B, Hirsch VM. 2009. Improved survival in rhesus macaques immunized with modified vaccinia virus Ankara recombinants expressing simian immunodeficiency virus envelope correlates with reduction in memory CD4+ T-cell loss and higher titers of neutralizing antibody. J. Virol. 83:5388–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, Yeh WW, Asmal M, Gelman RS, Shen L, Whitney JB, Seoighe C, Lacerda M, Keating S, Norris PJ, Hudgens MG, Gilbert PB, Buzby AP, Mach LV, Zhang J, Balachandran H, Shaw GM, Schmidt SD, Todd JP, Dodson A, Mascola JR, Nabel GJ. 2011. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci. Transl. Med. 3:81ra36 doi:10.1126/scitranslmed.3002351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reynolds MR, Sacha JB, Weiler AM, Borchardt GJ, Glidden CE, Sheppard NC, Norante FA, Castrovinci PA, Harris JJ, Robertson HT, Friedrich TC, McDermott AB, Wilson NA, Allison DB, Koff WC, Johnson WE, Watkins DI. 2011. The TRIM5α genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J. Virol. 85:9637–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yeh WW, Rao SS, Lim SY, Zhang J, Hraber PT, Brassard LM, Luedemann C, Todd JP, Dodson A, Shen L, Buzby AP, Whitney JB, Korber BT, Nabel GJ, Mascola JR, Letvin NL. 2011. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J. Virol. 85:10389–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274 doi:10.1371/journal.ppat.1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890 doi:10.1371/journal.ppat.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J, Keele BF, Li H, Keating S, Norris PJ, Carville A, Mansfield KG, Tomaras GD, Haynes BF, Kolodkin-Gal D, Letvin NL, Hahn BH, Shaw GM, Barouch DH. 2010. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J. Virol. 84:10406–10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim SY, Rogers T, Chan T, Whitney JB, Kim J, Sodroski J, Letvin NL. 2010. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6:e1000738 doi:10.1371/journal.ppat.1000738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, Leon EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J. Virol. 84:9190–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kong R, Li H, Bibollet-Ruche F, Decker JM, Zheng NN, Gottlieb GS, Kiviat NB, Sow PS, Georgiev I, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2012. Broad and potent neutralizing antibody responses elicited in natural HIV-2 infection. J. Virol. 86:947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kong R, Li H, Georgiev I, Changela A, Bibollet-Ruche F, Decker JM, Rowland-Jones SL, Jaye A, Guan Y, Lewis GK, Langedijk JPM, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2012. Epitope mapping of broadly neutralizing human HIV-2 monoclonal antibodies. J. Virol. 86:12115–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Letvin NL, Rao SS, Dang V, Buzby AP, Korioth-Schmitz B, Dombagoda D, Parvani JG, Clarke RH, Bar L, Carlson KR, Kozlowski PA, Hirsch VM, Mascola JR, Nabel GJ. 2007. No evidence for consistent virus-specific immunity in simian immunodeficiency virus-exposed, uninfected rhesus monkeys. J. Virol. 81:12368–12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 44. Maddison WP, Maddison DR. 1992. MacClade—analysis of phylogeny and character evolution—version 3. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 45. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 46. Swofford DL. 1999. PAUP* 4.0: phylogenetic analysis using parsimony (*and other methods), 4.0b2a ed. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 47. Leitner T, Korber B, Daniels M, Calef C, Foley B. 2006. HIV-1 subtype and circulating recombinant form (CRF) reference sequences, 2005, p 41–48 In Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B. (ed), HIV sequence compendium 2005. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 48. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 49. Nickle DC, Heath L, Jensen MA, Gilbert PB, Mullins JI, Kosakovsky Pond SL. 2007. HIV-specific probabilistic models of protein evolution. PLoS One 2:e503 doi:10.1371/journal.pone.0000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 51. Akaike H. 1974. A new look at statistical model identification. IEEE Trans. Automat. Contr. 19:716–723 [Google Scholar]
- 52. Stone M, Keele BF, Ma ZM, Bailes E, Dutra J, Hahn BH, Shaw GM, Miller CJ. 2010. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J. Virol. 84:7083–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiao P, Patterson LJ, Kuate S, Brocca-Cofano E, Thomas MA, Venzon D, Zhao J, DiPasquale J, Fenizia C, Lee EM, Kalisz I, Kalyanaraman VS, Pal R, Montefiori D, Keele BF, Robert-Guroff M. 2012. Replicating adenovirus-simian immunodeficiency virus (SIV) recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low-dose rectal SIV(mac251) challenge. J. Virol. 86:4644–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fischer W, Apetrei C, Santiago ML, Li Y, Gautam R, Pandrea I, Shaw GM, Hahn BH, Letvin NL, Nabel GJ, Korber BT. 2012. Distinct evolutionary pressures underlie diversity in simian and human immunodeficiency virus lineages. J. Virol. 86:13217–13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho DD, Marx PA. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Courgnaud V, Laure F, Fultz PN, Montagnier L, Brechot C, Sonigo P. 1992. Genetic differences accounting for evolution and pathogenicity of simian immunodeficiency virus from a sooty mangabey monkey after cross-species transmission to a pig-tailed macaque. J. Virol. 66:414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dewhurst S, Embretson JE, Anderson DC, Mullins JI, Fultz PN. 1990. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature 345:636–640 [DOI] [PubMed] [Google Scholar]
- 58. Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins WR, Montefiori DC. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, et al. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109–1112 [DOI] [PubMed] [Google Scholar]
- 60. Marthas ML, Banapour B, Sutjipto S, Siegel ME, Marx PA, Gardner MB, Pedersen NC, Luciw PA. 1989. Rhesus macaques inoculated with molecularly cloned simian immunodeficiency virus. J. Med. Primatol. 18:311–319 [PubMed] [Google Scholar]
- 61. Novembre FJ, De Rosayro J, O'Neil SP, Anderson DC, Klumpp SA, McClure HM. 1998. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J. Virol. 72:8841–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Novembre FJ, Hirsch VM, McClure HM, Fultz PN, Johnson PR. 1992. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology 186:783–787 [DOI] [PubMed] [Google Scholar]
- 63. Novembre FJ, Johnson PR, Lewis MG, Anderson DC, Klumpp S, McClure HM, Hirsch VM. 1993. Multiple viral determinants contribute to pathogenicity of the acutely lethal simian immunodeficiency virus SIVsmmPBj variant. J. Virol. 67:2466–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. O'Connor DH, McDermott AB, Krebs KC, Dodds EJ, Miller JE, Gonzalez EJ, Jacoby TJ, Yant L, Piontkivska H, Pantophlet R, Burton DR, Rehrauer WM, Wilson N, Hughes AL, Watkins DI. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 78:14012–14022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Anderson JP, Rodrigo AG, Learn GH, Madan A, Delahunty C, Coon M, Girard M, Osmanov S, Hood L, Mullins JI. 2000. Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J. Virol. 74:10752–10765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gao F, Robertson DL, Morrison SG, Hui H, Craig S, Decker J, Fultz PN, Girard M, Shaw GM, Hahn BH, Sharp PM. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 68. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 69. Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C, Ngole EM, Shaw GM, Peeters M, Sharp PM, Hahn BH. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. 2009. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387:414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pilgrim AK, Pantaleo G, Cohen OJ, Fink LM, Zhou JY, Zhou JT, Bolognesi DP, Fauci AS, Montefiori DC. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924–932 [DOI] [PubMed] [Google Scholar]
- 72. Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, Caudrelier P, El Habib R, Klein M, Lazzarin A, Stablein DM, Deers M, Corey L, Greenberg ML, Schwartz DH, Montefiori DC. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:2019–2035 [DOI] [PubMed] [Google Scholar]
- 73. Park SY, Love TM, Nelson J, Thurston SW, Perelson AS, Lee HY. 2011. Designing a genome-based HIV incidence assay with high sensitivity and specificity. AIDS 25:F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485:395–405 [DOI] [PubMed] [Google Scholar]
- 77. Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Path. 8:e1002686 doi:10.1371/journal.ppat.1002686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kraus MH, Parrish NF, Shaw KS, Decker JM, Keele BF, Salazar-Gonzalez JF, Grayson T, McPherson DT, Ping LH, Anderson JA, Swanstrom R, Williamson C, Shaw GM, Hahn BH. 2010. A rev1-vpu polymorphism unique to HIV-1 subtype A and C strains impairs envelope glycoprotein expression from rev-vpu-env cassettes and reduces virion infectivity in pseudotyping assays. Virology 397:346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, Hahn BH, Kappes JC. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T-lymphocytes and monocyte-derived macrophages. J. Virol. 86:2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cole KS, Alvarez M, Elliott DH, Lam H, Martin E, Chau T, Micken K, Rowles JL, Clements JE, Murphey-Corb M, Montelaro RC, Robinson JE. 2001. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology 290:59–73 [DOI] [PubMed] [Google Scholar]
- 81. Robinson JE, Cole KS, Elliott DH, Lam H, Amedee AM, Means R, Desrosiers RC, Clements J, Montelaro RC, Murphey-Corb M. 1998. Production and characterization of SIV envelope-specific rhesus monoclonal antibodies from a macaque asymptomatically infected with a live SIV vaccine. AIDS Res. Hum. Retroviruses 14:1253–1262 [DOI] [PubMed] [Google Scholar]
- 82. Louder MK, Sambor A, Chertova E, Hunte T, Barrett S, Ojong F, Sanders-Buell E, Zolla-Pazner S, McCutchan FE, Roser JD, Gabuzda D, Lifson JD, Mascola JR. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339:226–238 [DOI] [PubMed] [Google Scholar]
- 83. Provine NM, Cortez V, Chohan V, Overbaugh J. 2012. The neutralization sensitivity of viruses representing human immunodeficiency virus type 1 variants of diverse subtypes from early in infection is dependent on producer cell, as well as characteristics of the specific antibody and envelope variant. Virology 427:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Provine NM, Puryear WB, Wu X, Overbaugh J, Haigwood NL. 2009. The infectious molecular clone and pseudotyped virus models of human immunodeficiency virus type 1 exhibit significant differences in virion composition with only moderate differences in infectivity and inhibition sensitivity. J. Virol. 83:9002–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 173:340–348 [DOI] [PubMed] [Google Scholar]
- 86. Wrin T, Nunberg JH. 1994. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS 8:1622–1623 [DOI] [PubMed] [Google Scholar]
- 87. Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, Montefiori DC, Binley JM, Shaw GM, Haynes BF, Mascola JR, Morris L. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 94. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. de Silva TI, Aasa-Chapman M, Cotten M, Hue S, Robinson J, Bibollet-Ruche F, Sarge-Njie R, Berry N, Jaye A, Aaby P, Whittle H, Rowland-Jones S, Weiss R. 2012. Potent autologous and heterologous neutralizing antibody responses occur in HIV-2 infection across a broad range of infection outcomes. J. Virol. 86:930–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ozkaya Sahin G, Holmgren B, da Silva Z, Nielsen J, Nowroozalizadeh S, Esbjornsson J, Mansson F, Andersson S, Norrgren H, Aaby P, Jansson M, Fenyo EM. 2012. Potent intratype neutralizing activity distinguishes human immunodeficiency virus type 2 (HIV-2) from HIV-1. J. Virol. 86:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bixby JG, Laur O, Johnson WE, Desrosiers RC. 2010. Diversity of envelope genes from an uncloned stock of SIVmac251. AIDS Res. Hum. Retroviruses 26:1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kirchherr JL, Hamilton J, Lu X, Gnanakaran S, Muldoon M, Daniels M, Kasongo W, Chalwe V, Mulenga C, Mwananyanda L, Musonda RM, Yuan X, Montefiori DC, Korber BT, Haynes BF, Gao F. 2011. Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology 409:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM. 2012. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 8:e1002721 doi:10.1371/journal.ppat.1002721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Johnson WE, Morgan J, Reitter J, Puffer BA, Czajak S, Doms RW, Desrosiers RC. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Flatz L, Cheng C, Wang L, Foulds K, Ko SY, Kong WP, Roychoudhuri R, Shi W, Bao S, Todd JP, Asmal M, Shen L, Donaldson M, Schmidt SD, Gall J, Pinschewer DD, Letvin NL, Rao S, Mascola JR, Roederer M, Nabel GJ. 2012. Gene-based vaccination with a mis-matched envelope protects against simian immunodeficiency virus infection in non-human primates. J. Virol. 86:7760–7770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, Johnson WE, Ferrari G, Hirsch VM, Felber BK, Pavlakis GN, Earl PL, Moss B, Amara RR, Robinson HL. 2012. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine 30:1737–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 79:8991–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236 [DOI] [PubMed] [Google Scholar]