Abstract
Our pioneering studies on the interplay between the small ubiquitin-like modifier (SUMO) and influenza A virus identified the nonstructural protein NS1 as the first known SUMO target of influenza virus and one of the most abundantly SUMOylated influenza virus proteins. Here, we further characterize the role of SUMOylation for the A/Puerto Rico/8/1934 (PR8) NS1 protein, demonstrating that NS1 is SUMOylated not only by SUMO1 but also by SUMO2/3 and mapping the main SUMOylation sites in NS1 to residues K219 and K70. Furthermore, by using SUMOylatable and non-SUMOylatable forms of NS1 and an NS1-specific artificial SUMO ligase (ASL) that increases NS1 SUMOylation ∼4-fold, we demonstrate that SUMOylation does not affect the stability or cellular localization of PR8 NS1. However, NS1's ability to be SUMOylated appears to affect virus multiplication, as indicated by the delayed growth of a virus expressing the non-SUMOylatable form of NS1 in the interferon (IFN)-competent MDCK cell line. Remarkably, while a non-SUMOylatable form of NS1 exhibited a substantially diminished ability to neutralize IFN production, increasing NS1 SUMOylation beyond its normal levels also exerted a negative effect on its IFN-blocking function. This observation indicates the existence of an optimal level of NS1 SUMOylation that allows NS1 to achieve maximal activity and suggests that the limited amount of SUMOylation normally observed for most SUMO targets may correspond to an optimal level that maximizes the contribution of SUMOylation to protein function. Finally, protein cross-linking data suggest that SUMOylation may affect NS1 function by regulating the abundance of NS1 dimers and trimers in the cell.
INTRODUCTION
Influenza A virus, a member of the Orthomyxoviridae family, is responsible for annual winter epidemics of respiratory illness and irregularly spaced pandemics usually associated with increased disease-related mortality (1). Despite substantial progress in our knowledge of the molecular roles played during infection by the different proteins encoded by the virus, the specific interactions established between viral proteins and host cell components are still being characterized. A better understanding of such virus-host interactions may lead to the identification of new potential targets for therapeutic intervention. Specifically, host-encoded proteins playing important roles as accessory factors needed for efficient viral replication, but whose inactivation exerts neutral or minimal effects on slowly proliferating cells, such as those of the respiratory epithelium (2), may constitute optimal new targets for the development of innovative antiviral therapies.
Out of the 10 to 11 viral proteins encoded by influenza A virus, one of the most functionally diverse is the nonstructural viral protein NS1, which has been associated with numerous roles during influenza infection, including the modulation of viral RNA (vRNA) replication (3–6), general inhibition of the nuclear export of mRNAs carrying polyadenylated tails (7, 8), inhibition of the transcriptional elongation of host genes (9), regulation of host and viral protein synthesis (recently reviewed by Yanguez and Nieto [10]), and the neutralization of the activity of some of the interferon (IFN)-induced antiviral proteins, such as 2′,5′-oligoadenylate synthetase (OAS)/RNase L (11) and the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) (12–18). However, the main function attributed to NS1 is the neutralization of the initial signaling pathway leading to the production of type I IFN (reviewed by Krug et al. and Hale et al. [19, 20]). Whereas the latter function is heavily dependent on NS1's RNA binding properties (21), the ability of this protein to interact with cellular and viral proteins also contributes to its IFN-blocking activity and constitutes the main determinant of its other numerous functions (19). For example, NS1's interactions with both the 30-kDa subunit of the cleavage and polyadenylation specificity factor protein (CPSF30) and polyadenylate binding protein 2 (PABP2) are thought to mediate NS1's ability to decrease the processing and maturation of cellular mRNAs, therefore leading to a substantial decrease in host protein synthesis (7, 22). Similarly, NS1's ability to inhibit the activation of the viral RNA sensor RIG-I is mediated by its ability to bind the tripartite motif protein TRIM25, a RING domain ubiquitin E3 ligase, therefore blocking its multimerization and in turn inhibiting its ability to ubiquitinate the CARD motif in RIG-I (23), a requirement to allow RIG-I interaction with its downstream effector MAVS/VISA/IPS-1/Cardif (24). Thus, NS1's ability to interact with numerous host and viral proteins is key to its multiple roles during infection.
Posttranslational modifications are well-known regulators of protein interactions. Phosphorylation was the first posttranslational modification described to affect NS1 (25), and although early reports established that NS1 phosphorylation is important for efficient viral replication (26, 27), a recent study provided strong evidence that, at least for the A/Udorn/72 virus strain, only one out of three identified phosphorylation events affects NS1's functions (28). Two additional posttranslational modifications affecting NS1 were almost simultaneously described, both belonging to the group of UBL proteins (proteins exhibiting a tertiary structure similar to that of ubiquitin). The first was SUMOylation, the conjugation of the small ubiquitin-like modifier (SUMO) to NS1 (29). The second was ISGylation, the conjugation of IFN-stimulated gene 15 (ISG15) to NS1 (30). The initial report identifying NS1 as an ISGylated protein indicated that ISGylation prevented the interaction of NS1 with importin-α, a karyopherin essential for its nuclear import (30). A subsequent report described three additional roles for NS1 ISGylation: disrupting NS1 dimerization, preventing its interaction with PKR, and exhibiting an overall negative effect on NS1's ability to neutralize the IFN system (31).
The negative effect mediated by NS1 ISGylation was somehow expected, considering that ISG15 is one of the cellular effectors whose synthesis is stimulated by IFN. We recently demonstrated that even though IFN stimulation does not trigger an increase in the activity of the cellular SUMOylation system, influenza infection produces a global increase in cellular SUMOylation (32), an effect contrary to the downregulation of cellular SUMOylation usually observed for most pathogenic microorganisms (33). The global increase in cellular SUMOylation suggested the possibility that SUMOylation could be providing a supportive function to the virus, potentially enhancing specific functions associated with the pool of viral proteins targeted by SUMOylation. This hypothetical model has been supported by our findings and those of other groups. First, we demonstrated that besides NS1, 4 additional viral proteins are SUMOylated in vivo, including M1, PB1, NP, and NEP (NS2), and many of the predicted SUMOylation sites in these proteins appear to be fairly well conserved (32). Second, large screenings looking for host proteins that are either required for viral multiplication or capable of interacting with specific viral proteins and required for optimal virus growth identified various members of the cellular SUMOylation system (34, 35). Finally, the last level of supporting information was obtained by other groups while studying the SUMOylation of specific influenza virus proteins: Xu et al. confirmed our initial identification of NS1 as a SUMOylation target, mapped one SUMOylation site in NS1, and assigned a potential role for NS1 SUMOylation, namely, extending NS1's half-life (36). Similarly, Wu et al. confirmed the SUMOylation of M1, identified the lysine residues in M1 that undergo SUMOylation, and demonstrated that this posttranslational modification exerts a regulatory function over viral morphogenesis, as a non-SUMOylatable form of M1 prevented normal virus budding at the plasma membrane (37).
Here, we expand our understanding of the interactions established between the cellular SUMOylation system and influenza A virus during infection by further characterizing the effects exerted by SUMOylation on the A/Puerto Rico/8/1934 influenza A virus (PR8) NS1 protein. Our data demonstrate that NS1 can be equally conjugated by SUMO1 and SUMO2/3 but appears to be preferentially modified by SUMO2/3 under the normal levels of SUMO components present in the cell. Furthermore, we identify residues K219 and K70 as the main SUMOylation sites in NS1 and demonstrate that mutations affecting NS1 SUMOylation decrease NS1's ability to neutralize the cellular IFN response. Interestingly, our data show that increasing NS1 SUMOylation through the use of an NS1-specific artificial SUMO ligase (ASL) also results in a diminished ability to counteract the IFN system, indicating the existence of an ideal proportion of SUMOylated versus non-SUMOylated NS1 that maximizes NS1's functionality during infection. Finally, our data indicate that, unlike previously reported for the NS1 protein of a highly pathogenic avian H5N1 virus (36), SUMOylation does not appear to regulate or affect the stability of PR8 NS1; instead, SUMOylation appears to regulate the abundance of NS1 dimers and trimers within the cell, a finding in agreement with SUMO's ability to regulate protein-protein interactions.
MATERIALS AND METHODS
Cells and viruses.
HEK293A cells (Invitrogen Corp., Carlsbad, CA), HEK293FT cells (Invitrogen Corp.), A549 cells (ATCC, Manassas, VA), and MDCK cells (ATCC) were maintained in complete medium consisting of 1× Dulbecco's modified essential medium (DMEM) supplemented with high glucose, l-glutamine, sodium pyruvate, and 10% fetal bovine serum. For HEK293FT cells, Geneticin (Invitrogen Corp.) was added to the complete medium at a final concentration of 500 μg/ml. All cell lines were maintained in a 37°C incubator at 5% CO2. Influenza A/Puerto Rico/8/1934 H1N1 (here referred to as PR8) was a gift from John M. Quarles (Department of Microbial and Molecular Pathogenesis, College of Medicine, Texas A&M Health Science Center) and was propagated in MDCK cells at a multiplicity of infection (MOI) of 0.001 by using 1× DMEM supplemented with 0.2% bovine serum albumin (BSA) and 2 μg/ml tosyl-phenylalanyl-chloromethyl-ketone (TPCK)-treated trypsin (Worthington Biochemical Corp., Lakewood, NJ). Vesicular stomatitis virus expressing enhanced green fluorescent protein (VSV-EGFP) was a gift from John Hiscott (Department of Molecular Oncology, Lady Davis Institute, Jewish General Hospital, McGill University, Montreal, Quebec, Canada) and was propagated in Vero cells at an MOI of 0.02 in complete medium.
Plasmids.
The A/WSN/1933 H1N1 (here referred to as WSN33) 12-plasmid reverse genetics system (38) was kindly provided by Yoshihiro Kawaoka (Department of Pathobiological Sciences, School of Veterinary Medicine, University of Wisconsin—Madison, Madison, WI). The expression plasmid for T7T7NS1 was developed by reverse transcription-PCR (RT-PCR) cloning using our laboratory strain of PR8 and the general methodology previously described by Hoffmann et al. (39). Briefly, viral RNA purified from MDCK infected-cell supernatants using the MagMAX viral RNA isolation kit (Applied Biosystems, Foster City, CA) was used as the template for a reverse transcription reaction using a UNI-12 primer targeting the 12 conserved nucleotides at the 3′ end of all viral RNA gene segments. The reverse transcription product was subsequently amplified by PCR using forward and reverse primers specific for the NS gene segment. The PCR product was cloned into the pcDNA3.1 mammalian expression vector (Invitrogen Corp., Carlsbad, CA) according to the In-Fusion Dry-Down PCR cloning method (Clontech, Mountain View, CA). Subsequently, a double T7 (T7T7) tag was inserted at the N terminus of the NS1 open reading frame (ORF) by using the Phusion site-directed mutagenesis kit (Finnzymes, Woburn, MA) according to the manufacturer's protocol. To map the SUMOylation site, specific mutations in the sequence of NS1 were subsequently inserted by using the same site-directed mutagenesis approach, resulting in the expression constructs for T7T7NS1K70A, T7T7NS1K219A, and the double mutant T7T7K70AK219A. To generate the pPolI/WSN/T7T7NS1 construct, the pPolI/WSN/NS construct derived from the WSN33 12-plasmid reverse genetics system (38) was PCR amplified by using primers complementary to the 5′ untranslated region (UTR) and the 3′ UTR of the WSN33 NS gene segment, producing a PCR product comprising the backbone of the original plasmid and the 5′ and 3′ UTRs of the NS gene segment but missing the totality of the ORF for NS1 and NS2. This PCR product was ligated with the PCR-amplified coding region of the NS gene segment from our PR8 strain, therefore generating a recombinant carrying the 5′ and 3′ UTRs of the WSN33 NS gene segment and the ORF from the PR8 NS gene segment. Similar overall approaches were employed to mutate the splicing acceptor site in the NS gene segment and generate the splicing-deficient pPolI/WSN/T7T7NS-derived mutants pPolI/WSN/T7T7NS1-SplAccptMut and pPolI/WSN/T7T7NS1K70AK219A-SplAccptMut and their NS2-shifted derivative constructs pPolI/WSN/T7T7NS1∼NS2 and pPolI/WSN/T7T7NS1K70AK219A∼NS2.
The dual expression plasmids Dual S1Q94P/I/U and Dual S3Q89P/I/U, containing mutations in SUMO1 and SUMO3, respectively, that are known to prevent their deconjugation from the target, were derived from the previously reported Dual S1/I/U and Dual S3/I/U constructs (29), using the Phusion site-directed mutagenesis kit (Finnzymes) according to the manufacturer's protocol.
ASL approach and ASL constructs.
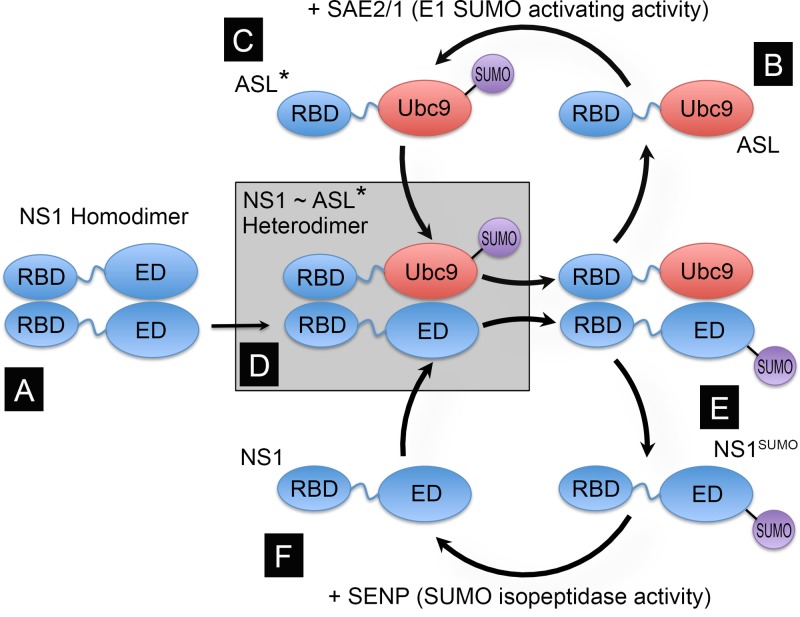
To enhance the SUMOylation of NS1, we engineered a construct expressing a fusion of the N-terminal RNA binding domain (RBD) of the PR8 NS1 protein with the SUMO-conjugating enzyme Ubc9. This fusion protein enhances the SUMOylation of full-length NS1 by means of the interaction established between the N-terminal region of NS1 and the N-terminal region of the NS1-Ubc9 fusion. This interaction positions Ubc9 in close proximity to the C-terminal domain of NS1, thus facilitating the SUMOylation of K219, the main SUMOylation acceptor site in NS1 (Fig. 1). To enable the free movement of Ubc9 in relation to the N-terminal region of NS1 in the fusion protein, something that we considered important to enhance the SUMOylation of NS1 by the NS1-Ubc9 fusion protein, the unstructured linker region of NS1 was also included in the fusion construct. Thus, the expression construct for the fusion protein coded for amino acid residues 1 to 87 of NS1 followed by the full-length amino acid sequence of Ubc9. Because it was shown previously that most of the IFN-blocking activity mediated by PR8 NS1 is associated with the RNA binding activity of its N-terminal domain (40), we also developed a mutant form of the NS1-Ubc9 construct containing two amino acid substitutions known to prevent RNA binding by its NS1 RBD portion, namely, an R-to-A substitution at position 38 and a K-to-A substitution at position 41 (R38AK41A) (41). Both the NS1-Ubc9 fusion carrying the wild-type (wt) NS1 sequence and the NS1-Ubc9 fusion carrying the amino acid substitutions preventing RNA binding were shown to enhance the SUMOylation of full-length wild-type NS1 when coexpressed by transfection in mammalian cells, specifically by enhancing SUMOylation at position K219 in PR8 NS1, and did not appear to exhibit self-SUMOylating activity. Therefore, these fusion proteins were designated artificial SUMO ligases (ASLs) and are referred to here simply as wt-ASL and mut-ASL (for the fusion proteins with and without RNA binding capabilities, respectively). The design and development of the ASL approach will be described in detail elsewhere (our unpublished data).
Fig 1.
Principle and design of the NS1-specific artificial SUMO ligase (ASL). NS1 contains two well-defined globular domains in its structure, the RNA binding domain (RBD) (amino acid residues 1 to 73) and the effector domain (ED) (amino acid residues 86 to 204), separated by an unstructured flexible linker (amino acid residues 74 to 85, represented by a wiggly blue line). Both domains are capable of forming dimers, thus allowing full-length NS1 to form dimers (A) and multimers. As the main SUMOylation site in NS1 is located near its C-terminal end, and the dimeric structures formed by the RBD alone are relatively unstable, we envisioned that a fusion protein consisting of NS1's RBD and linker region fused to the full-length SUMO-conjugating enzyme Ubc9 (RBD∼Ubc9) (B) would be “charged” by the E1 SUMO-activating enzyme (SAE2/1) present in the cell (C); form transient NS1:RBD∼Ubc9 heterodimers, thus positioning Ubc9 in close proximity to K219 in NS1 (D); and enhance NS1 SUMOylation (E). Thus, the RBD∼Ubc9 fusion would act as an ASL for NS1. Although it is likely that NS1 might subsequently be de-SUMOylated by the SUMO-specific isopeptidases present in the cell (referred to as SENPs) (F), the enhancing effect mediated by the RBD∼Ubc9 fusion would still increase the steady-state concentration of SUMOylated NS1 in the cell. ASL* represents the SUMO-charged form of the ASL, in which SUMO is attached to the active site in Ubc9, and ASL represents the uncharged form of the ASL.
In silico analyses and computer software.
To identify potential SUMOylation sites in PR8 NS1, two SUMO prediction analysis programs were used: SUMOsp2.0 (42) and SUMOplot (Abgent). For SUMOsp2.0, the cutoff value for predictions was set to medium. To estimate the conservation of the putative SUMOylation sites identified, multiple-sequence alignments were performed by using the tools freely available at the NIAID IRD (Influenza Research Database) website (http://www.fludb.org/) (43). The GenBank accession numbers of the sequences used in the alignment presented in Fig. 2 are as follows: J02150 for A/Puerto Rico/8/1934, GU135996 for A/Aichi/202/2009, CY026039 for A/Alabama/UR06_0482/2007, AF333238 for A/Brevig Mission/1/1918, FJ969514 for A/California/04/2009, CY008376 for A/Canterbury/204/2005, AB212059 for A/Hong Kong/213/2003, CY055160 for A/Hong Kong/3239/2008, CY050736 for A/New York/3309/2009, V01102 for A/Udorn/8/1972, and U13683 for A/WS/1933.
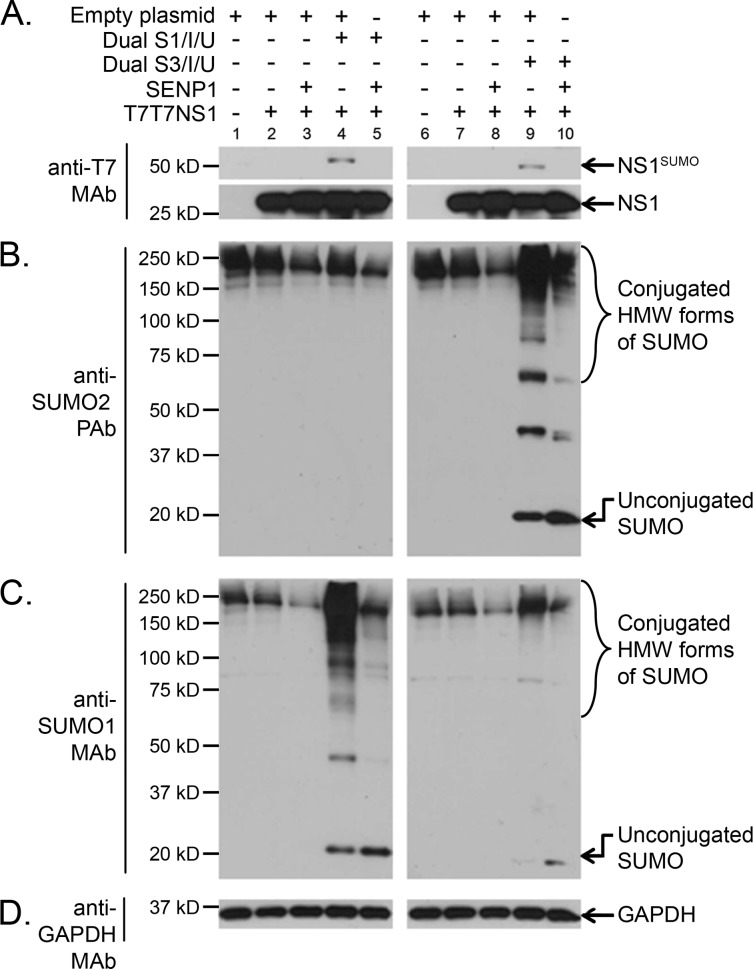
Fig 2.
NS1 is SUMOylated by both SUMO1 and SUMO2/3 in vivo. HEK293A cells cotransfected with (+) or without (−) the indicated combinations of mammalian expression plasmids, including T7T7NS1, the dicistronic expression constructs Dual S1/I/U and Dual S3/I/U, an expression construct for the SUMO-deconjugating enzyme SENP1, and an empty expression vector, were analyzed by SDS-PAGE and immunoblotting using anti-T7 MAb (A), anti-SUMO2 PAb (B), anti-SUMO1 MAb (C), and anti-GAPDH MAb (D). NS1SUMO, SUMOylated NS1; HMW, high molecular weight.
The confocal microscopy images used in this study were captured and analyzed by using ZEN 2009 software (Zeiss, New York, NY). The phosphordensitometry data were generated and analyzed by using Quantity One 1-D Analysis software (Bio-Rad Laboratories, Hercules, CA). All statistical analyses and graphics presented in this article were performed by using GraphPad Prism version 5.04 for Windows (GraphPad Software Inc., San Diego, CA). All figures generated for this report were created by using Adobe Photoshop CS5 extended version 12.0.3 ×64 (Adobe Systems Inc., San Jose, CA).
Transient transfections and generation of recombinant viruses by reverse genetics.
HEK293A and HEK293FT cells were seeded at a density of 1 × 105 cells/well into 24-well plates or 5 × 105 cells/well into 6-well plates. The following day, the cells were transfected by liposome-mediated transfection using the desired combination of CsCl-purified plasmids and TransIT-LT1 (Mirus Bio LLC, Madison, WI) according to the manufacturer's recommendations. For most transfections, either 2 to 3 μg of DNA and 4 to 6 μl of TransIT-LT1 reagent (for 24-well plates) or 12 to 15 μg of DNA and 24 to 30 μl of TransIT-LT1 reagent (for 6-well plates) were used. At the appropriate times posttransfection, cell extracts were collected by adding directly over the cells either boiling 2× sample buffer (25 mM Tris [pH 6.8], 5% glycerol, 2% SDS, 0.01% bromophenol blue) for SDS-PAGE and immunoblotting analyses or boiling 1× denaturing lysis buffer (1% SDS, 5 mM EDTA, 100 mM Tris [pH 7.8]) for immunoprecipitation analyses.
To generate recombinant viruses by reverse genetics, upon transfection, HEK293FT cells were incubated at 37°C in 5% CO2 for 24 h. The culture supernatant was then discarded and replaced with 1× DMEM supplemented with 0.2% BSA and 2 μg/ml TPCK-treated trypsin. The cells were further incubated for another 48 h at 37°C in 5% CO2, and the culture supernatants were subsequently collected and used for plaque assays.
Plaque assays.
MDCK cells were plated into 6-well plates at a density of 1 × 106 cells/well in complete medium and incubated at 37°C in 5% CO2 until the cells formed a confluent monolayer. The cells were then washed twice with 1× DMEM, and a virus dilution prepared in 1 ml of 1× DMEM supplemented with 0.2% BSA was added to the cells and incubated with the cells at 35°C for 30 min. The cells were washed twice again with 1× DMEM, and a 3-ml overlay of 1× DMEM plus 4 μg/ml TPCK-treated trypsin and 0.6% SeaKem ME agarose (Lonza, Rockland, ME), maintained at 40°C using a bench-top heater-shaker, was poured over the cells and allowed to solidify for ∼20 min at room temperature. The cells were subsequently incubated at 35°C in 5% CO2, and starting at ∼30 h postinfection (p.i.), the cells were visually screened for the presence of plaques. When the size of the plaques observed reached ∼1 to 2 mm in diameter, the overlay was removed, and the cells were fixed and stained with a solution containing 0.12% crystal violet in 20% ethanol. Alternatively, for plaque purification of recombinant viruses, the overlay was left in place, and well-isolated plaques were transferred into a vial containing 1× DMEM plus 0.2% BSA by using a sterile 1-ml pipette.
For the experiments aimed at measuring the viral growth of the recombinant viruses developed by reverse genetics, all of the viruses produced were purified by two consecutive rounds of plaque purification. Importantly, during the second round of plaque purification, plaques of identical diameter (as measured under a microscope) were selected as a way to initiate the experiment with similar virus numbers (i.e., using similar MOIs). The plaques selected were then incubated on 1 ml of 1× DMEM plus 0.2% BSA at 4°C for 4 h to allow the viruses to diffuse out of the agarose plug. Subsequently, the whole volume of 1× DMEM plus 0.2% BSA, including the agarose plug itself, was added to a confluent monolayer of either MDCK cells or Vero cells plated onto 10-cm2 petri dishes in 1× DMEM supplemented with 0.2% BSA and 2 μg/ml TPCK-treated trypsin. At 24 and 48 h p.i., the culture supernatants were removed, and their virus titers were analyzed by a plaque assay.
Immunoblot analyses.
Before SDS-PAGE analyses, all cell extracts generated were passed several times through a 29½-gauge needle to break down the genomic DNA released and decrease the viscosity of the samples. Subsequently, β-mercaptoethanol was added up to a final concentration of 10%, and the samples were boiled for 3 min. The samples were resolved by 10% SDS-PAGE gels, using either premade PAGEgel gels (PAGEgel Inc., San Diego, CA) or SDS-PAGE gels made in-house. Upon SDS-PAGE, the proteins were transferred onto either Immobilon-P for use with horseradish peroxidase (HRP)-conjugated secondary antibodies and chemiluminescence detection or Immobilon-FL for use with IRDye-conjugated secondary antibodies (LI-COR Biosciences Inc., Lincoln, NE) and infrared fluorescence imaging (both types of Immobilon membranes are from Millipore Corp., Bedford, MA). Upon transfer, the procedure used depended on the detection method to be used.
(i) Chemiluminescence detection.
Immobilon-P membranes were washed 3 times in 1× phosphate-buffered saline (PBS) supplemented with 0.05% Tween 20 (here referred to as 1× PBS-T), blocked with 1× PBS-T supplemented with 3% nonfat milk (here referred to as 1× Blotto) for a minimum of 30 min at room temperature, and incubated in 1× Blotto at 4°C overnight with the primary antibody at the indicated dilution. The membranes were then washed 3 times with 1× PBS-T and incubated for 1 h at room temperature in 1× Blotto with the appropriate horseradish peroxidase-conjugated secondary antibody at the indicated dilution. The membranes were then washed 3 times with 1× PBS-T and once again with 1× PBS and subsequently developed by using the Immobilon Western HRP Substrate system (Millipore), using Phenix Blue X-ray film (Phenix Research Products, Candler, NC) in a dark room.
(ii) Infrared fluorescence imaging.
Immobilon-FL membranes were washed 3 times in 1× PBS, blocked in Odyssey blocking buffer (OBB) (LI-COR Biosciences Inc.) for a minimum of 30 min at room temperature, and incubated in OBB supplemented with 0.1% Tween 20 (here referred to as OBB-T) at 4°C overnight with the primary antibody at the indicated dilution. The membranes were then washed 3 times with 1× PBS-T and incubated for 1 h at room temperature in OBB-T with the appropriate highly cross-absorbed IRDye 800 CW- and IRDye 680 LT-conjugated secondary antibodies (LI-COR Biosciences Inc.) at the indicated dilution. The membranes were then washed 3 times with 1× PBS-T and twice again with 1× PBS and scanned on an Odyssey CLx infrared imaging system (LI-COR Biosciences Inc.). Quantitative analyses of the images obtained was performed by using Odyssey Infrared Imaging System Application software version 3.0.29 (LI-COR Biosciences Inc.). Statistical analyses and graphics of the data generated were performed by using GraphPad Prism version 5.04 for Windows (GraphPad Software Inc.).
The following dilutions were employed for the various primary and secondary antibodies used for immunoblotting in this study: anti-T7 tag mouse monoclonal antibody (MAb) (Novagen, EMD Biosciences Inc., San Diego, CA) at a 1:5,000 dilution, anti-SUMO1 rabbit MAb Y299 (Epitomics Inc., Burlingame, CA) at a 1:5,000 dilution, anti-Ubc9 rabbit MAb EP2938Y (Epitomics Inc.) at a 1:5,000 dilution, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mouse MAb 2D4A7 (Santa Cruz Biotechnology Inc., Santa Cruz, CA) at a 1:5,000 dilution, anti-SUMO2 rabbit polyclonal antibody (PAb) (Invitrogen) at a 1:5,000 dilution, anti-C/EBP-β (C-19) rabbit PAb (Santa Cruz Biotechnology Inc.) at a 1:5,000 dilution, HRP-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology Inc.) at a 1:5,000 dilution, HRP-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology Inc.) at a 1:5,000 dilution, highly cross-absorbed IRDye 800 CW-conjugated goat anti-mouse IgG at a 1:20,000 dilution, highly cross-absorbed IRDye 680 LT-conjugated goat anti-mouse IgG at a 1:20,000 dilution, highly cross-absorbed IRDye 800 CW-conjugated goat anti-rabbit IgG at a 1:20,000 dilution, and highly cross-absorbed IRDye 680 LT-conjugated goat anti-rabbit IgG at a 1:20,000 dilution (all IRDye-conjugated secondary antibodies were obtained from LI-COR Biosciences Inc.).
In most immunoblotting analyses, the membranes were reused 3 to 5 times. Before they were reused, the membranes were stripped by incubation in boiling stripping buffer (1% SDS and 0.2% β-mercaptoethanol) for 10 min, washed 5 times with 1× PBS-T, and incubated with the appropriate blocking solution before being incubated with the new primary antibody.
Interferon inhibition assay.
Interferon inhibition assays were performed according to the general methodology established by Rehwinkel et al. to trigger IFN production using an active influenza virus RNA-dependent RNA polymerase (vRdRp) (44) and a bioassay established by Solorzano et al. (45), using the VSV-GFP developed by Stojdl et al. (46). Briefly, HEK293FT cells were transfected with various combinations of plasmids, including those needed to produce a functional vRdRp [here referred to as PolII(PB2+PB1+PA+NP)] and a construct transcribing the PB2 gene segment under an RNA polymerase I promoter in the reverse complementary direction [here referred to as PolI(PB2)]. At 48 h posttransfection, both culture supernatants and total cell extracts were collected (the latter were collected in 2× sample buffer). The supernatants collected were subsequently added undiluted to a monolayer of Vero cells plated 24 h earlier. At 24 h after exposure to the supernatant, the Vero cells were infected with VSV-GFP at an MOI of 2.0. At 8 h postinfection, the cells were trypsinized, fixed with 1× PBS plus 4% paraformaldehyde, and analyzed for expression of GFP by flow cytometry. Cells transfected with PolI(PB2) and PolII(PB2+PB1+PA+NP) in the absence of NS1 were assumed to produce the maximum amount of IFN achievable in this assay, and therefore, the percentage of Vero cells expressing GFP measured in cells treated with this supernatant was assumed to correspond to 0% inhibition of IFN production. Similarly, cells transfected with PolI(PB2) and PolII(PB2+PB1+PA+NP) in the presence of T7T7NS1 were arbitrarily assumed to produce the smallest amount of IFN due to the IFN inhibitory activity mediated by NS1, and therefore, the percentage of Vero cells expressing GFP measured in cells treated with this supernatant was assigned as 100% inhibition of IFN production. The IFN inhibitory activity of all other supernatants was calculated by comparison to the IFN inhibitory activity of the supernatants assigned as 0% inhibition and 100% inhibition.
Immunofluorescence analyses.
For immunofluorescence analyses, upon transfection with the plasmids indicated, the cells were incubated at 37°C in 5% CO2, and 12 h later, the cells were trypsinized and replated in a 96-well plate. Twelve hours later, the cells were then fixed with 1× PBS plus 4% paraformaldehyde for 30 min at 4°C, washed twice with 1× PBS, and permeabilized by incubation with 100% methanol for 10 min at 4°C. Upon permeabilization, the cells were incubated in blocking solution (1× PBS supplemented with 1% goat serum), followed by incubation overnight at 4°C with anti-T7 MAb (Novagen) and anti-Ubc9 MAb (Epitomics Inc.), both diluted 1:500 in blocking solution. Excess antibody was removed with three washes of 5 min each with 1× PBS. Fluorescently labeled secondary antibodies, namely, highly cross-absorbed Alexa Fluor 488 goat anti-mouse IgG and highly cross-absorbed Alexa Fluor 594 goat anti-rabbit IgG (both from Molecular Probes, Life Technologies Corp., Carlsbad, CA), were added to the cells at a 1:500 dilution in blocking solution and incubated for 2 h at room temperature. The cells were subsequently washed 3 times with 1× PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) for 5 min, and washed three additional times with 1× PBS. The images were captured by using an LSM 700 confocal microscope (Zeiss, New York, NY) with a 63× objective and three lasers, at 405 nm (DAPI), 488 nm (Alexa Fluor 488), and 555 nm (Alexa Fluor 594). Image acquisition was performed by using ZEN 2009 software (Zeiss, New York, NY). Since the microscope system is equipped with a monochromatic camera, ZEN 2009 software was used to add pseudocolor to the various images captured to produce the colored images presented.
Protein turnover analyses.
To estimate protein turnover, two approaches were used: cycloheximide treatment and pulse-chase analyses. For cycloheximide treatment, HEK293FT cells were plated and transfected in either 10-cm petri dishes or 6-well plates. At 6 h posttransfection, the cells were reseeded onto 24-well plates. This approach minimized sample-to-sample variation due to fluctuations in transfection efficiencies. At 24 h posttransfection, the cells were treated with either dimethyl sulfoxide (DMSO) as a vehicle control or 4 mM cycloheximide (Calbiochem, EMD Millipore, Billerica, MA). Samples were subsequently collected at the indicated times after cycloheximide treatment, using 2× sample buffer. Upon collection, the samples were processed and analyzed for SDS-PAGE and immunoblotting as described above. For pulse-chase analyses, HEK293FT cells were plated and transfected in 6-well plates. At 47 h posttransfection, the cells were starved in methionine- and cysteine-deficient medium for 1 h and pulsed for 1 h with medium containing 35S-labeled methionine and cysteine (200 μCi/well). Upon labeling, the cells were trypsinized, replated into 24-well plates, and incubated with complete medium containing 20% serum (chase) until collected. Cells were collected at 0, 4, 8, and 12 h postchase. For collection, the cells were treated with SDS lysis buffer (0.5% SDS, 50 mM Tris [pH 8.0], 1 mM dithiothreitol [DTT]), boiled for 3 min, and diluted with 4 volumes of radioimmunoprecipitation assay (RIPA) correction buffer (1.25% Nonidet P-40,1.25% sodium deoxycholate, 0.0125 M sodium phosphate [pH 7.2], 2 mM EDTA) supplemented with cOmplete Ultra protease inhibitor cocktail tablets (Roche Applied Science, Indianapolis, IN) and 20 mM N-ethylmaleimide. The resulting extract was passed through a 29½-gauge needle several times, spun down at 15,000 × g for 10 min at 4°C, and immunoprecipitated by incubation overnight at 4°C with 2 μg of anti-T7 MAb (Novagen), 2 μg of anti-GFP MAb (Santa Cruz Biotechnology Inc.), and 20 μl of a 50% bead slurry of protein A/G Plus-agarose (Santa Cruz Biotechnology Inc.). The beads were washed 4 times with 1× PBS-T and resuspended in 25 μl of 2× sample buffer. The immunoprecipitated proteins were resolved by SDS-PAGE, transferred onto Immobilon-P membranes, and analyzed by phosphordensitometry by using a personal molecular imager (Bio-Rad Laboratories). Quantitative analysis was performed by using Quantity One 1-D Analysis software (Bio-Rad Laboratories).
Chemical cross-linking to analyze NS1-containing protein complexes.
HEK293FT cells were transfected with the indicated constructs, and at 24 h posttransfection, the cells were treated for 30 min with either 1× PBS containing 4% DMSO alone (vehicle control) or 1× PBS containing 4% DMSO and 1 mM disuccinimidyl glutarate (DSG) (Thermo Fisher Scientific Inc., Waltham, MA). DSG is a short (7.7-Å spacer arm), amino-specific, homobifunctional, noncleavable, cell-permeable chemical cross-linker. Total cell extracts were collected in 300 μl of 2× sample buffer and processed for SDS-PAGE and immunoblotting as described above.
RESULTS
PR8 NS1 is SUMOylated by both SUMO1 and SUMO2/3 primarily through residues K70 and K219.
Previously, we identified nonstructural protein 1 of influenza A virus (NS1) as a bona fide target of the cellular SUMOylation system. Specifically, we demonstrated that NS1 is SUMOylated both when overexpressed by transient transfection in mammalian cells as well as during infection by SUMO1 conjugation (29, 32). However, the potential conjugation to NS1 of the other main SUMO modifiers in the cell (SUMO2/3), the main SUMOylation site in NS1, and the potential relevance of SUMOylation for the normal functions associated with NS1 during influenza infection were not assessed.
Proteomic analyses have demonstrated that the pool of cellular proteins SUMOylated with SUMO1 only partially overlaps the pool of proteins SUMOylated with SUMO2/3 (47–49). Furthermore, conjugation with SUMO2/3 is known to exert at least one potentially different effect on the target from that exerted by SUMO1 conjugation, namely, the formation of SUMO2/3 chains followed by the recognition of the poly-SUMOylated target by SUMO-dependent ubiquitin ligases that polyubiquitinylate it and direct it toward proteasomal degradation, as previously reviewed (50–52). Therefore, it was relevant to characterize whether NS1 could be equally SUMOylated by the two main types of SUMO proteins in the cell. To determine this, HEK293A cells were cotransfected with an expression construct for the A/Puerto Rico/8/1934 (here referred to simply as PR8) NS1 protein, our previously reported dual expression constructs coding for an S-tagged and His-tagged SUMO1 or SUMO3, and an expression construct for the de-SUMOylating enzyme SENP1. The cotransfected cells were collected in sample buffer, and the resulting cell extracts were analyzed by immunoblotting. The immunoblotting data obtained clearly demonstrated equivalent levels of SUMOylated NS1 in the presence of SUMO1 and SUMO3 (Fig. 2A, compare lanes 4 and 9), therefore indicating that, in addition to being SUMOylated in vivo by SUMO1 (29), NS1 can also be SUMOylated in vivo by SUMO2/3. The specificity of the global increases in SUMO1 or SUMO2/3 SUMOylation obtained when the cells were cotransfected with either of the dual expression constructs was demonstrated by immunoblotting performed with SUMO2- and SUMO1-specific antibodies (Fig. 2B and C, respectively).
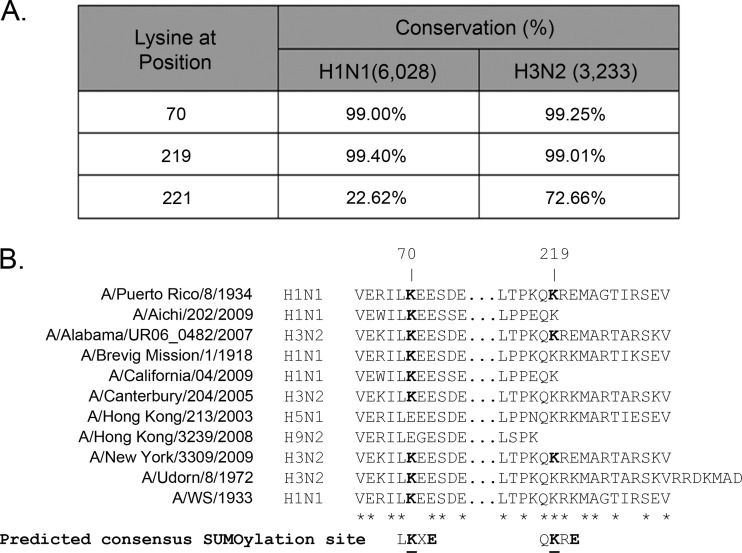
To map the SUMOylation sites in PR8 NS1, SUMOsp2.0 and SUMOplot software analyses were performed to identify likely SUMOylation sites. These analyses identified residues K70 and K219 as the primary potential SUMOylation sites in PR8 NS1. Sequence comparison with other reference laboratory influenza virus strains and a large array of human influenza virus isolates revealed that the first and second potential SUMOylation sites in NS1 were conserved in over 99% of all the H1N1 and H3N2 human influenza virus strains analyzed (Fig. 3A). However, K70 was not conserved among virus isolates representing sporadic bird-to-human transmission events (Fig. 3B, compare residue 70 in the H5N1 and H9N2 isolates with that in the other virus isolates presented), and although K219 was present in some virus strains representing sporadic bird-to-human transmission events (Fig. 3B, compare residue 219 in the H5N1 isolate with that in the other virus isolates presented), it constituted the last amino acid residue in some virus strains (e.g., A/Aichi/202/2009) (Fig. 3B), and in some others, NS1 did not even extend up to that residue (e.g., A/Hong Kong/3239/2008) (Fig. 3B). This indicated more de facto variability in the second putative SUMOylation site because lysine residues are not SUMOylated when located at the C-terminal end of a protein (see below). Importantly, K221, the main SUMOylation site previously identified in the NS1 protein derived from a highly pathogenic avian strain (36), showed much more limited conservation among the human H1N1 and H3N2 influenza virus strains analyzed (Fig. 3A).
Fig 3.
Conservation of likely SUMOylation sites in PR8 NS1 among human H1N1 and H3N2 influenza virus strains and influenza virus isolates representing sporadic bird-to-human transmission events. (A) Conservation of the two putative SUMOylation sites identified by SUMOsp2.0 and SUMOplot analyses of PR8 NS1 (K70 and K219) and a previously identified SUMOylation site in a highly pathogenic avian influenza virus (K221), among H1N1 and H3N2 human influenza virus strains. The numbers in parentheses indicate the total number of sequences analyzed for each type of human influenza virus. (B) Comparison of the primary sequence of the NS1 protein from various representative human and laboratory-adapted influenza virus strains and viral isolates representing sporadic bird-to-human transmission events. The sequence shown is the one around the two predicted SUMOylation sites in PR8 NS1 (K70 and K219). The predicted consensus SUMOylation sites identified in PR8 NS1 by in silico analyses are depicted underneath the sequence list, with the critical amino acids displayed in boldface type. Note that residue 221 in PR8 NS1 is glutamic acid.
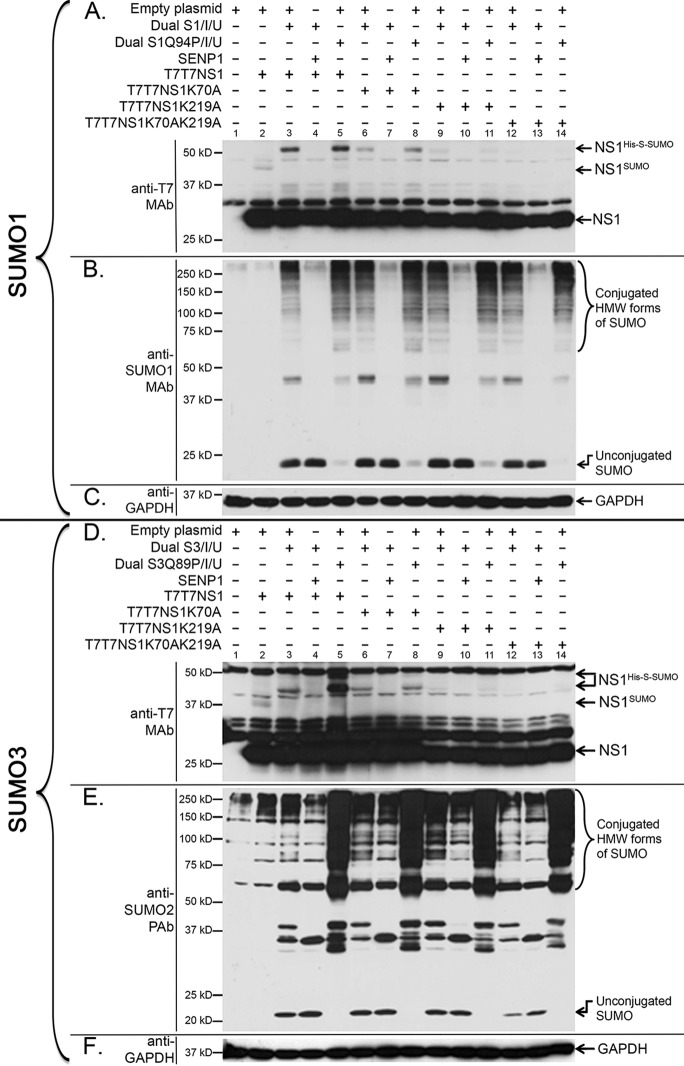
To determine whether K70 and K219 constituted the main SUMOylation sites in PR8 NS1, lysine-to-alanine (K-to-A) substitutions were introduced into each of those positions, both individually and simultaneously, in NS1 expression constructs carrying a dual T7 tag. The effect of the mutations was subsequently determined by immunoblot analysis of cells cotransfected with different combinations of (i) the wild-type or mutated NS1 expression constructs, (ii) dual expression plasmids coding for either deconjugatable or nondeconjugatable forms of His-tagged and S-tagged SUMO1 (Dual S1/I/U or Dual S1Q94P/I/U, respectively), and (iii) the de-SUMOylating enzyme SENP1.
While the K70A mutation produced a noticeable but slight decrease in NS1 SUMOylation (Fig. 4A, lanes 6 and 8), the K219A substitution produced a more dramatic decrease in NS1 SUMOylation (Fig. 4A, lanes 9 and 11), and the double mutant K70AK219A almost completely abolished NS1 SUMOylation (Fig. 4A, lanes 12 and 14). These data indicated that K219 and K70 constituted the primary and secondary SUMOylation sites in NS1, respectively. Longer exposures of the gel revealed, however, a residual amount of SUMOylated NS1 in the K70AK219A double mutant, therefore indicating the existence of other minor SUMOylation sites in NS1. Importantly, a mutant form of NS1 ending at position K219 exhibited SUMOylation levels comparable to those exhibited by the K219A mutant, therefore indicating that K219 is not SUMOylated when it constitutes the C-terminal end of NS1 (data not shown).
Fig 4.
K219 and K70 are the primary and secondary target sites in NS1, respectively, for both SUMO1 and SUMO2/3 SUMOylation. HEK293A cells were cotransfected with (+) or without (−) the indicated combinations of mammalian expression plasmids, including expression plasmids for T7T7-tagged NS1 (T7T7NS1), the single and double lysine mutant forms of NS1 (T7T7NS1K70A, T7T7NS1K219A, and T7T7NS1K70AK219A), an expression construct for the human de-SUMOylating enzyme SENP1, an empty expression vector used to equalize the total amount of DNA used to transfect the cells, and the dicistronic expression constructs Dual S1/I/Ubc9 and Dual S1Q94P/I/Ubc9 (S1Q94P is a mutant form of SUMO1 that prevents its de-SUMOylation upon its conjugation to a target) (A to C) or the dicistronic expression constructs Dual S3/I/Ubc9 and Dual S3Q89P/I/Ubc9 (S3Q89P is a mutant form of SUMO3 that prevents its de-SUMOylation upon its conjugation to a target) (D to F). Cell extracts were collected and analyzed by SDS-PAGE and immunoblotting using either anti-T7 MAb (A and D), anti-SUMO1 MAb (B), anti-SUMO2 PAb (E), or anti-GAPDH MAb (C and F). NS1His-S-SUMO, NS1 SUMOylated with His- and S-tagged SUMO; NS1SUMO, NS1 SUMOylated with endogenous SUMO; HMW, high molecular weight.
To determine whether SUMO2/3 SUMOylation of NS1 occurs through the same lysine residues as SUMO1 SUMOylation, an experiment similar to the one described above for SUMO1 was executed by using dual expression plasmids coding for deconjugatable or nondeconjugatable forms of His-tagged and S-tagged SUMO3 (Dual S3/I/U or Dual S3Q89P/I/U, respectively). The data obtained showed that, similarly to our previous observations for SUMO1 conjugation, SUMO3 conjugation to NS1 occurred primarily through residue K219, while residue K70 appeared to act as a secondary conjugation site (Fig. 4D). This finding indicated that SUMO2/3 conjugation of NS1 involved the same lysine residues associated with SUMO1 conjugation.
SUMOylation affects NS1's ability to neutralize the interferon response.
Having defined the SUMOylation sites in NS1 and the range of SUMO modifiers capable of affecting NS1, we aimed to address the potential effect(s) of SUMOylation on NS1 function. NS1 is a multifunctional viral protein, playing diverse roles during influenza infection. However, the main function attributed to NS1 is the neutralization of the host cell's IFN response (11, 19, 20, 53). To determine whether NS1 SUMOylation affected NS1's ability to neutralize the IFN response under conditions closely recapitulating the events taking place during influenza infection, we used a methodology previously established by Rehwinkel et al. (44). In this method, the replicase activity of the viral RNA-dependent RNA polymerase (vRdRp), in the presence of the PB2 vRNA template, produces PB2 cRNA and a large quantity of 5′-PPP PB2 vRNA. Such 5′-PPP vRNA acts as an agonist for RIG-I, thus activating it and stimulating IFN production in a way mirroring RIG-I activation during influenza infection (44).
HEK293FT cells were cotransfected with different combinations of (i) the expression plasmids for T7-tagged NS1 or NS1K70AK219A, (ii) a mix of RNA polymerase II (PolII)-driven expression plasmids carrying all the viral genes needed to reconstitute vRdRp activity (PB2, PB1, PA, and NP), and (iii) a plasmid carrying the PB2 gene segment under the transcriptional control of the human RNA polymerase I (PolI) promoter placed in the reverse complementary sense. Additionally, to better evaluate the effects mediated by NS1 SUMOylation, some samples were also cotransfected with a previously developed NS1-specific artificial SUMO ligase (ASL), whose development will be described in detail elsewhere (our unpublished data). The NS1-specific ASL (here referred to simply as ASL) consists of a fusion of the first N-terminal 87 amino acid residues in NS1 with the SUMO-conjugating enzyme Ubc9 (Fig. 1). Two different forms of the ASL were employed, one carrying the wild-type (wt) NS1 sequence (referred to as wt-ASL) and one carrying the R38AK41A double mutant form of NS1 (referred to as mut-ASL), which keeps the N-terminal region of NS1 from binding RNA. The production of IFN by the cells transfected with the different combinations of constructs described above was evaluated by a bioassay for type I IFN, as previously reported by Solorzano et al. (45). Briefly, Vero cells were incubated with the supernatant collected from the transfected HEK293FT cells and subsequently infected with a recombinant VSV expressing GFP (VSV-GFP) (54). The number of VSV-GFP-infected Vero cells (i.e., cells expressing GFP), determined 8 h after VSV infection by flow cytometry is proportional to the amount of IFN present in the supernatant, because IFN renders Vero cells resistant to VSV infection and therefore provides an accurate and sensitive assessment of IFN production by the transfected cells.
As expected, cotransfection with both the expression plasmids needed to reconstitute a fully functional vRdRp and the PolI(PB2) construct triggered the production of substantial amounts of IFN. The percentages of VSV-GFP-infected cells observed under these conditions in multiple experiments were consistently low, and their averages were arbitrarily defined as 0% IFN inhibitory activity. Also as expected, cotransfection with NS1 produced substantial decreases in IFN production, whose average was arbitrarily defined as 100% IFN inhibitory activity (Fig. 5A, bar 1). Cotransfection with NS1K70AK219A led to only an ∼40% decrease in IFN production (Fig. 5A, bar 2), indicating that NS1's ability to be SUMOylated is relevant for its ability to neutralize IFN production. Cotransfection with NS1 and wt-ASL led to a dramatic increase in IFN inhibitory activity, reaching values nearly 70% above the inhibitory activity of NS1 alone (Fig. 5A, bar 3). However, this increase was likely mediated by the RNA binding activity of wt-ASL and not by its ability to increase NS1 SUMOylation, because an identical increase in IFN inhibitory activity was observed when wt-ASL was transfected in the absence of NS1 (Fig. 5A, bar 5). In contrast, cotransfection with mut-ASL (which still increases NS1 SUMOylation but lacks RNA binding activity) and NS1 led to an ∼24% decrease in NS1's IFN inhibitory activity (Fig. 5A, bar 4). This decrease was likely mediated by the increased NS1 SUMOylation triggered by mut-ASL, because mut-ASL alone had no significant effect on IFN production (Fig. 5A, bar 6), and indicates that increasing NS1 SUMOylation beyond its normal levels is also deleterious for NS1's ability to neutralize the host's IFN response.
Fig 5.
SUMOylation affects NS1's ability to neutralize IFN production. HEK293FT cells were transfected with (+) or without (−) the indicated mammalian expression constructs, including (i) the T7T7NS1 and T7T7NS1K70AK219A constructs previously described; (ii) Pol II(PB2+PB1+PA+NP), a mix of four RNA polymerase II-driven constructs for the expression of the viral proteins PB2, PB1, PA, and NP, which together form the functional vRdRp; (iii) Pol I(PB2), an RNA polymerase I-driven construct for the production of the PB2 vRNA; (iv) wt-ASL, an expression construct coding for the wild-type form of the artificial SUMO ligase for NS1; and (v) mut-ASL, a mutated form of the artificial SUMO ligase for NS1 containing two amino acid substitutions, R38A and K41A. At 48 h posttransfection, culture supernatants and total cell extracts were collected. The IFN levels present in the supernatants were analyzed by using a bioassay. The total cell extracts were analyzed by SDS-PAGE and immunoblotting using anti-T7 MAb and anti-Ubc9 MAb or anti-SUMO1 MAb and anti-GAPDH MAb as primary antibodies and highly cross-absorbed IRDye 800 CW- and IRDye 680 LT-conjugated anti-mouse and anti-rabbit IgG antibodies as secondary antibodies. The immunoblots were read and analyzed by using an Odyssey CLx infrared imaging system. (A) Graphic representation of the percentage of inhibition of IFN production observed for cells transfected with the indicated constructs. The numbers in brackets indicate the corresponding sample in the gel presented in panel B. (B) Immunoblots of a representative group of transfected cells showing the expression of T7T7NS1 and SUMOylated T7T7NS1 (T7T7NS1SUMO), the overall SUMOylation profile obtained, and the levels of expression of the ASL. (C) Percentage of SUMOylated NS1 obtained in cells transfected with the indicated combinations of expression constructs. The numbers in brackets indicate the corresponding sample in the gel presented in panel B. (D) Relative quantification of the levels of ASL compared to the amount observed in cells transfected with wt-ASL. In all cases, the quantitative data presented represent the cumulative data obtained from four sets of samples obtained in independent experiments. Error bars indicate standard deviations.
To confirm that the effects observed in the IFN inhibition assays described above were associated with changes in NS1 SUMOylation, the percentage of SUMOylated NS1 present in every sample employed was determined by immunoblotting using IRDye-conjugated secondary antibodies and an Odyssey CLx infrared imaging system. A representative immunoblot exemplifying the data obtained is shown in Fig. 5B. NS1 expressed in the absence of the ASL exhibited limited but easily detectable SUMOylation (Fig. 5B, lane 3, and C, bar 1). The double mutant NS1K70AK219A exhibited SUMOylation levels barely above background levels (Fig. 5B, lane 4, and C, bar 2). Cotransfection with wt-ASL enhanced NS1 SUMOylation ∼5.5-fold (Fig. 5B, lane 5, and C, bar 3). Finally, cotransfection with mut-ASL enhanced NS1 SUMOylation by ∼4-fold (Fig. 5B, lane 6, and C, bar 4). Interestingly, in the samples cotransfected with wt-ASL and mut-ASL, enhanced imaging of the scanned data revealed the presence of higher-molecular-mass species of NS1 spaced by about ∼17 kDa, suggestive of the formation of poly-SUMO chains on NS1 (data not shown), which also indicates that NS1 is likely to be preferentially SUMOylated by SUMO2/3 in vivo. The difference observed in SUMO-enhancing ability between wt-ASL and mut-ASL appeared to be mediated by the differential accumulation of each ASL in the cell, as wt-ASL accumulated to levels about 40% higher than those of mut-ASL (Fig. 5B, anti-Ubc9 MAb panel, and D). Importantly, cotransfection with wt-ASL (but not with mut-ASL) led to increased SUMOylation of proteins other than NS1, as evidenced by an anti-SUMO1 immunoblot analysis (Fig. 5B, anti-SUMO1 MAb panel).
Altogether, the data obtained in these assays indicated that altering the levels of SUMOylated NS1 decreases NS1's ability to neutralize the IFN response, as both mutations precluding the efficient SUMOylation of NS1 and artificial increases in NS1 SUMOylation triggered by cotransfection with mut-ASL decreased NS1's IFN-neutralizing activity.
NS1 SUMOylation affects viral growth in IFN-competent and IFN-deficient host cells.
The decreased IFN-blocking activity exhibited by NS1K70AK219A led us to predict that a mutant virus carrying this double mutant form of NS1 should display growth defects in cell lines capable of producing type I IFN. To test this hypothesis, we aimed to develop recombinant viruses carrying the non-SUMOylatable form of NS1 using a previously reported influenza A/WSN/1933 (here referred to simply as WSN33) 12-plasmid reverse genetic system (38). However, since WSN33 NS1 lacks the second SUMOylation site (K219), and PR8 NS1 exhibits strong and well-characterized IFN-blocking activity (40, 55, 56), we replaced WSN33 NS with PR8 NS but maintained the original 5′ and 3′ UTRs of the WSN33 NS gene segment.
Introduction of the K219A mutation into NS1 resulted in an asparagine-to-histidine (N-to-H) substitution at amino acid residue 62 in NS2 (N62H). Recombinant viruses containing the NS1K70AK219A mutations could not be obtained despite numerous attempts. As a similar inability to generate K219R mutants was previously reported by others (30), we considered that this issue likely indicated that the N62H mutation in NS2 was lethal. Therefore, to allow us to test the relevance of NS1 SUMOylation in the absence of changes in the primary sequence of NS2, we followed a previously reported methodology to separate the ORF for NS1 from that for NS2 while still allowing NS2 to be produced as a splicing product of the primary NS mRNA transcript (57). Briefly, starting with PolI-driven constructs for either wt NS1 or NS1K70AK219A, the splicing acceptor site located in NS1 was inactivated by site-directed mutagenesis. A copy of the second exon for NS2, starting with a functional splicing acceptor site, was then placed downstream from the stop codon for NS1. The resulting NS2-shifted constructs were tested for their ability to produce NS1 and NS2 in transfection assays performed in the presence of a functional vRdRp. The data obtained demonstrated that although the amount of NS2 produced by the NS2-shifted constructs was decreased by half compared to that produced by the parental NS gene segments (Fig. 6A, compare the amounts of NS2 displayed by the different constructs, and B, compare bars 1 and 2 with bars 5 and 6), the amount of NS2 should still be sufficient to ensure virus survival because similar ratios of NS2 to NS1 are known to be produced by some human influenza viruses (58). Furthermore, no substantial difference in NS2 synthesis between the shifted constructs was observed (Fig. 6B, compare bars 5 and 6), indicating that the K70AK219A double mutation in the shifted construct did not affect the synthesis of NS2.
Fig 6.
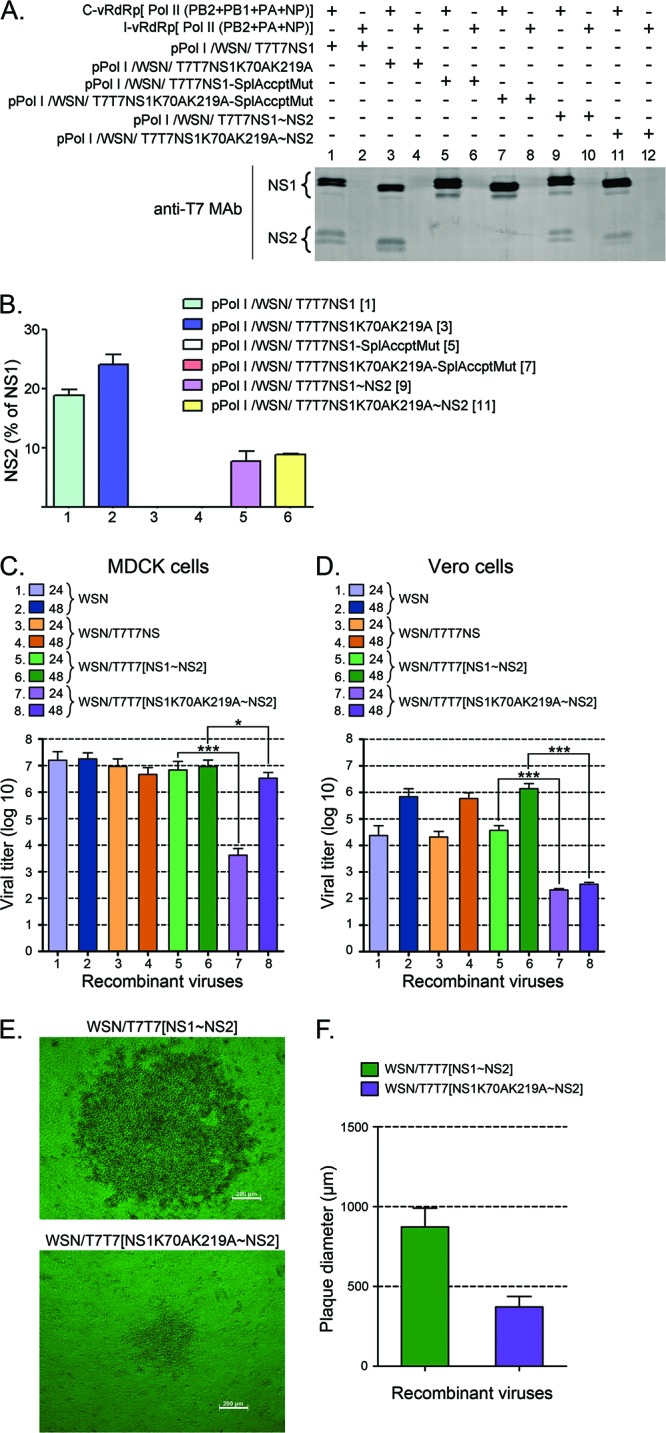
NS1 SUMOylation affects viral growth in MDCK and Vero cell lines. (A and B) Effect of shifting the second exon for NS2 downstream from the stop codon for NS1. HEK293FT cells transfected with (+) or without (−) the indicated plasmids, including a complete vRdRp mix (C-vRdRp), and an incomplete vRdRp mix (I-vRdRp) lacking PB1, were analyzed by immunoblotting with anti-T7 MAb and IRDye 800 CW-conjugated anti-mouse IgG antibodies to determine the levels of NS1 and NS2 expression. (A) Representative immunoblot displaying the data obtained. (B) Quantitative analysis of the percentage of NS2 observed in two independent experiments like the one presented in panel A. The numbers in brackets indicate the corresponding lane in the gel shown in panel A. (C and D) Viral growth of the recombinant viruses developed with the various pPolI/WSN/NS gene segments presented in panels A and B in either MDCK cells (C) or Vero cells (D). All recombinant viruses generated by reverse genetics, including WSN/T7T7[NS1∼NS2] and WSN/T7T7[NS1K70AK219A∼NS2], were purified by two consecutive rounds of plaque purification. Plaques of identical diameter produced during the second round of plaque purification were inoculated over a confluent monolayer of either MDCK cells or Vero cells plated onto 10-cm petri dishes. At 24 and 48 h p.i., the culture supernatants were removed, and their virus titers analyzed by plaque assays. (C) Virus titers obtained with the supernatants collected from MDCK cells. (D) Virus titers obtained with the supernatants collected from Vero cells. (E) Phase-contrast microscopy images of representative plaques for the WSN/T7T7[NS1∼NS2] and WSN/T7T7[NS1K70AK219A∼NS2] recombinant viruses. Bars, 200 μm. (F) Average diameter of the plaques obtained in MDCK cells for the WSN/T7T7[NS1∼NS2] and WSN/T7T7[NS1K70AK219A∼NS2] recombinant viruses by 36 h p.i. The values presented in panels C and D correspond to the data obtained in 3 independent experiments. The values presented in panel F correspond to the average diameters measured for 20 plaques chosen at random from the different plaque assays performed while quantifying virus titers for the different viruses. In all cases, error bars indicate standard deviations. Asterisks indicate P values of 0.05 (*) and 0.001 (***).
Using the indicated reverse genetics system and the different NS gene segments generated as indicated above, four different recombinant viruses were generated: WSN (carrying the original WSN NS gene segment), WSN/T7T7NS (carrying the PR8 T7T7-tagged NS gene segment), WSN/T7T7[NS1∼NS2] (carrying the NS2-shifted NS gene segment), and WSN/T7T7[NS1K70AK219A∼NS2] (carrying the NS2-shifted NS gene segment coding for the double mutant form of NS1, NS1K70AK219A). Note that the only difference between WSN/T7T7[NS1∼NS2] and WSN/T7T7[NS1K70AK219A∼NS2] is the presence of the two mutations that decrease NS1 SUMOylation in the latter. Each recombinant virus generated was purified by two rounds of plaque purification. To determine whether the different recombinant viruses generated exhibited differences in their growth rate in an IFN-competent cell line, plaques of identical diameter and morphology (as determined by light microscopy) produced during the second round of plaque purification were inoculated over a confluent monolayer of MDCK cells plated onto 10-cm petri dishes. At 24 and 48 h postinfection (p.i.), the culture supernatants were removed, and their virus titers were analyzed by a plaque assay. To provide statistical significance, three plaques were analyzed for every recombinant virus.
Three of the recombinant viruses generated, namely, WSN, WSN/T7T7NS, and WSN/T7T7[NS1∼NS2], exhibited very similar growth dynamics in MDCK cells, producing virus titers in the range of 1 × 107 to 3 × 107 PFU/ml by 24 h p.i. (Fig. 6C, bars 1, 3, and 5). In contrast, the virus carrying the double mutant form of NS1, WSN/T7T7[NS1K70AK219A∼NS2], exhibited a dramatic growth delay, producing virus titers in the range of 3.5 × 103 PFU/ml by 24 h p.i. Such titers are about 3.5 orders of magnitude lower than those of the other viruses, including the closely related recombinant virus WSN/T7T7[NS1∼NS2] (Fig. 6C, compare bars 5 and 7). Nevertheless, the double mutant virus, unlike the other viruses, continued to multiply, and by 48 h p.i., it reached titers closer to those reached by the other viruses, although they were still significantly lower (Fig. 6C, compare bars 6 and 8).
To determine whether the differences in viral growth observed between WSN/T7T7[NS1K70AK219∼NS2] and the other recombinant viruses were associated exclusively with differences in their ability to neutralize the host's IFN response, we repeated the experiment described above using Vero cells as a host for amplification of the viruses purified by plaque assays. Vero cells are unable to produce type I IFN (59), and therefore, viral growth in this host system is independent of the virus' ability to neutralize the IFN system. All recombinant viruses exhibited substantial decreases in growth speed and total virus yield in Vero cells compared to MDCK cells, with three out of the four recombinant viruses producing virus titers of 2 × 104 to 5 × 104 PFU/ml by 24 h p.i. (Fig. 6D, bars 1, 3, and 5) and 0.7 × 106 to 1.1 × 106 PFU/ml by 48 h p.i. (Fig. 6D, bars 2, 4, and 6). Importantly, WSN/T7T7[NS1K70AK219A∼NS2] still produced significantly lower virus titers than those produced by the other viruses, reaching titers of only ∼2.3 × 102 PFU/ml by 24 h p.i. (Fig. 6D, bar 7), and ceased to grow any further, as its titers appeared unchanged by 48 h p.i. (Fig. 6D, bar 8). This indicated that the growth deficiency related to the WSN/T7T7[NS1K70AK219A∼NS2] recombinant virus is not exclusively due to a limited ability to neutralize the IFN response but instead likely involves other activities related to NS1 that might be regulated by SUMOylation and appear more critical for growth in Vero cells.
Further evidence of the dramatic growth delay associated with the WSN/T7T7[NS1K70AK219A∼NS2] mutant virus was evidenced during the execution of the plaque assays performed to evaluate viral growth. In these assays, the plaques produced by WSN/T7T7[NS1K70AK219A∼NS2] were consistently half the diameter of those associated with the WSN/T7T7[NS1∼NS2] virus by 36 h p.i. (Fig. 6E and F).
SUMOylation does not affect either the cellular localization or the stability/turnover of PR8 NS1.
Knowing that NS1 SUMOylation affected NS1's ability to neutralize the cellular IFN response and other NS1-associated functions important for normal viral growth, we aimed to identify the molecular mechanism responsible for the observed SUMO-related effects on NS1.
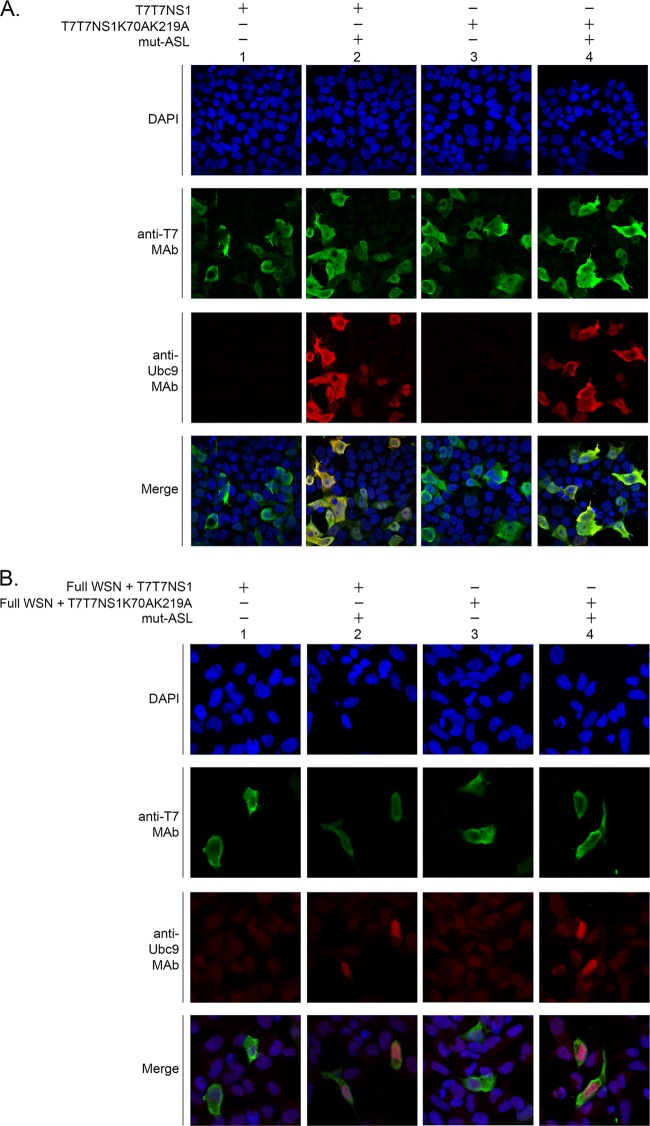
Different intracellular localization patterns, including nucleolar, nuclear, at promyelocytic leukemia nuclear bodies (PML-NBs), and cytoplasmic, have been assigned to NS1 (60–63), thus leading to the idea that NS1's cellular localization might be dependent on numerous factors, including the possible involvement of cellular factors (19). Since SUMOylation is known to affect the cellular localization of numerous SUMO targets, including that of several viral proteins (64–67), we decided to assess whether SUMOylation could be one of the cellular factors affecting NS1's cellular localization. To this end, we overexpressed either wt NS1 or the double mutant NS1K70AK219A by transient transfection. Furthermore, to enhance any potential effect mediated by SUMOylation, some cells were also cotransfected with mut-ASL. Additionally, to determine whether SUMOylation affected NS1's cellular localization under conditions more closely resembling those occurring during influenza infection, we also investigated the cellular distribution of NS1 in cells cotransfected with the full complement of reverse genetics plasmids.
Under all conditions tested, both wild-type NS1 and its double mutant form appeared to be mostly cytoplasmic, although most cells expressing NS1 also exhibited a weak diffuse nuclear NS1 signal (Fig. 7A and B, columns 1 and 3). Furthermore, coexpression of mut-ASL did not affect the cellular distribution of NS1 or its double mutant form, as most of the NS1 signal observed exhibited an identical profile as that in its absence (Fig. 7A and B, columns 2 and 4). Thus, these experiments indicated that SUMOylation did not appear to regulate the cellular localization of NS1 either when NS1 was expressed in the absence of other viral proteins or when NS1 was expressed in the context of all the viral components normally produced during influenza infection. Therefore, the effects exerted by SUMO modification on NS1 do not appear to involve regulation of NS1's cellular localization.
Fig 7.
SUMOylation does not affect the cellular localization of NS1. HEK293FT cells were transfected with either the T7T7NS1 or the T7T7NS1K70AK219A construct, alone (A) or together with the full complement of A/WSN/1933 gene segments and expression constructs required to generate recombinant viruses (Full WSN) (B). To increase any potential effects mediated by SUMOylation, some sets of cells were also cotransfected with mut-ASL. Immunofluorescence analyses were performed as described in the text, using anti-T7 MAb, anti-Ubc9 MAb, and DAPI stain. The images were captured by using an LSM 700 confocal microscope (Zeiss, New York, NY) with a 63× objective and three lasers, at 405 nm (DAPI), 488 nm, and 555 nm. ZEN 2009 software (Zeiss, New York, NY) was used for the acquisition and processing of the confocal images.
SUMOylation is also known to affect the turnover of some of its targets, including that of several viral proteins (68–70). Importantly, for certain substrates, SUMOylation with SUMO2/3 is known to trigger the formation of long poly-SUMO chains that are recognized by ubiquitin ligases and lead to the polyubiquitinylation of the SUMOylated substrate, which in turns leads to their proteasomal degradation (71–74). Data obtained during the execution of these studies revealed the formation of poly-SUMOylated NS1 in the presence of wt-ASL and mut-ASL (see, for example, Fig. 5B, lanes 5 and 6). As these occurred in the absence of exogenously added SUMO, the data indicated that NS1 was likely to be preferentially modified by SUMO2/3 in vivo and therefore suggested a potential role for SUMOylation in regulating the stability of NS1. Furthermore, a previous report indicated that SUMOylation affects the function of the NS1 protein of a highly pathogenic avian influenza A virus strain by increasing its half-life (36). Therefore, we considered it essential to determine whether SUMOylation affected PR8 NS1's stability.
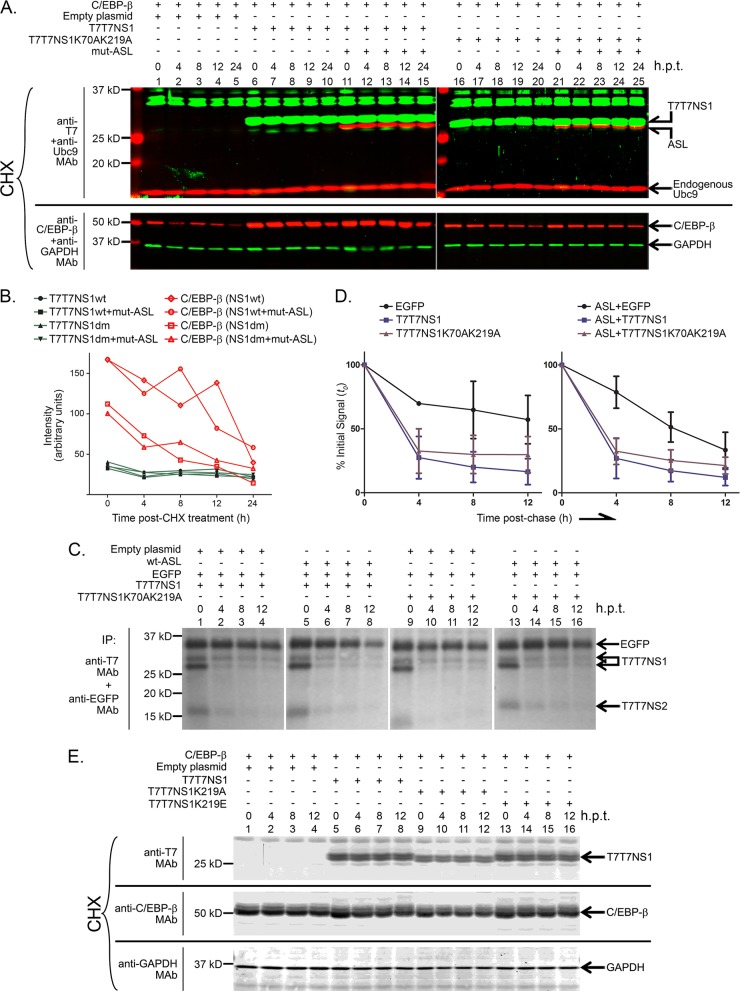
To measure the effect of SUMOylation on PR8 NS1's stability, we cotransfected HEK293FT cells with expression constructs for wt NS1 and NS1K70AK219A. To potentiate the effects mediated by SUMOylation, some cells were also transfected with mut-ASL. Additionally, the cells were also cotransfected with C/EBP-β1, a transcriptional regulator that exhibits an intermediate half-life in the cell. Next, at 24 h posttransfection, the cells were treated with cycloheximide, an inhibitor of protein synthesis, and samples were collected at 4-h intervals after cycloheximide treatment. Under these experimental conditions, both wt NS1 and NS1K70AK219A accumulated to similar levels in the cell and appeared very stable, not displaying any significant decrease in cellular levels during the time span analyzed (Fig. 8A, compare, for example, lanes 6 and 10 to lanes 16 and 20). Furthermore, the enhanced SUMOylation observed upon addition of mut-ASL did not seem to affect the stability of NS1, as very similar profiles were observed in the presence and absence of mut-ASL (Fig. 8A, compare lanes 11 and 15 to lanes 21 and 25). Quantitative analyses performed by using an Odyssey CLx infrared imaging system in combination with IRDye-conjugated secondary antibodies demonstrated little fluctuation in the cellular quantities of NS1 throughout the experiment (Fig. 8B). Similar analyses performed with the coexpressed protein C/EBP-β1 showed substantial changes in the cellular concentration of C/EBP-β1 throughout the time frame analyzed, therefore demonstrating that the cycloheximide treatment had been effective (Fig. 8B).
Fig 8.
SUMOylation does not affect the stability of NS1. (A, C, and E) HEK293FT cells were cotransfected with (+) or without (−) the indicted expression constructs, including expression constructs for C/EBP-β1 and EGFP, which were used as controls. (A and E) At 24 h posttransfection (h.p.t.), cycloheximide (CHX) was added to the cells at a final concentration of 4 mM, and the cells were collected 0, 4, 8, 12, and 24 h later in 2× sample buffer (the sample at 24 h posttransfection is not shown in panel E). The cell extracts generated were analyzed by SDS-PAGE and successive rounds of immunoblotting using anti-T7 MAb and anti-Ubc9 MAb, and anti-C/EBP-β MAb and anti-GAPDH MAb, as primary antibodies. Highly cross-absorbed IRDye 800 CW- and IRDye 680 LT-conjugated anti-mouse and anti-rabbit IgG antibodies were used as secondary antibodies. The immunoblots were read and analyzed by using an Odyssey CLx infrared imaging system. (B) Quantitative analysis of the data presented in panel A. The signals for C/EBP-β shown are those measured in samples cotransfected with the constructs indicated in parentheses. (C) At 24 h posttransfection, the cells were trypsinized and replated into a 24-well plate. Next, at 47 h posttransfection, the cells were starved in methionine- and cysteine-deficient medium for 1 h, pulsed for 1 h with medium containing 35S-labeled methionine and cysteine, and chased with complete medium containing 20% serum until collected. Cells were collected at 0, 4, 8, and 12 h postchase and processed for immunoprecipitation as described above. The immunoprecipitated (IP) samples were resolved by SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and analyzed by phosphordensitometry using a personal molecular imager. (D) Quantitative analysis of the phosphordensitometry data obtained in two independent experiments like the one presented in panel C.
As an alternative approach to determine whether SUMOylation affected the stability of NS1, we performed pulse-chase analyses of cells transfected with various combinations of wt NS1, NS1K70AK219A, wt-ASL, and EGFP. In these experiments, the cells were collected and lysed at different times postchase, using the same 4-h intervals used in the cycloheximide experiments, and the resulting extracts were immunoprecipitated with antibodies directed against the T7 tag and EGFP. Interestingly, in these analyses, a substantial decrease in the amount of NS1 that could be immunoprecipitated from the soluble fraction was observed soon after the chase, with a decrease of more than 50% of the initial signal between the time-zero and the 4-h-postchase samples (Fig. 8C, compare lanes 1 and 2, 5 and 6, 9 and 10, and 13 and 14, and Fig. 8D, compare initial values with those observed at 4 h postchase). As our previous cycloheximide analyses had not shown substantial decreases in the total amount of NS1 present in whole-cell extracts, these differences most likely represented the incorporation of a substantial amount of NS1 into an insoluble fraction that was not solubilized under the conditions used for our immunoprecipitation assay. Nevertheless, similar profiles were observed for both wt NS1 and NS1K70AK219A, and no substantial changes were observed in the presence of wt-ASL, therefore indicating that altering the SUMOylation of NS1 did not affect its stability.
To further confirm our findings related to the lack of effects mediated by SUMOylation on the cellular stability of NS1, we decided to also compare the stability of wt NS1 to that of two single mutant forms of NS1: NS1K219A and NS1K219E. These mutants introduce different substitutions at residue K219, one of which introduces an opposite charge to the one normally present in NS1; however, both mutations led to a similar substantial decrease in NS1 SUMOylation (data not shown). The stability of these single mutant forms of NS1 was assessed by cycloheximide treatment, as described above. Similarly, as observed for the double mutant NS1K70AK219A, the single mutants NS1K219A and NS1K219E appeared to be as stable as wt NS1, therefore confirming that SUMOylation did not appear to affect the cellular stability of NS1 (Fig. 8E). Furthermore, similar cycloheximide experiments performed with NS1, its non-SUMOylatable double mutant NS1K70AK219A, and the single mutant NS1K219E, all in the presence of the full complement of reverse genetics plasmids (as a way to more closely reproduce the predominant conditions during viral infection), produced similar results (data not shown).
Altogether, our stability analysis performed by using two different but complementary approaches, pulse-chase analysis and cycloheximide treatment, indicated that the non-SUMOylatable form of PR8 NS1 exhibited stability similar to that of its wt form and that coexpression of mut-ASL did not affect their stability, therefore strongly indicating that SUMOylation does not affect the stability of PR8 NS1.
NS1 SUMOylation affects the abundance of NS1 dimers and trimers.
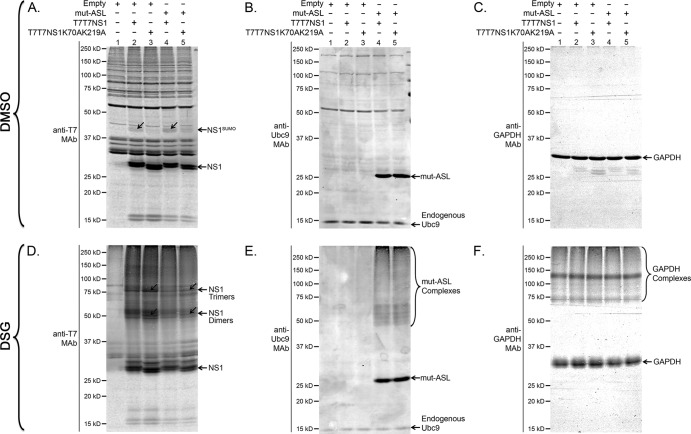
Most of the activities associated with SUMO conjugation are thought to be mediated by SUMO's ability to enhance and regulate protein-protein interactions. Hence, SUMO has been dubbed a “molecular matchmaker.” To determine whether NS1 SUMOylation affected some of the molecular interactions established by NS1, we overexpressed wt NS1 and its non-SUMOylatable double mutant by transfection, either alone or in the presence of mut-ASL. At 24 h posttransfection, the cells were incubated for 30 min in the presence of DSG, a membrane-permeable chemical cross-linker, to stabilize the protein complexes formed by NS1 under the different experimental conditions used. The cells were then lysed, and the resulting cell extracts were analyzed by immunoblotting. The images obtained revealed the presence of substantially larger amounts of NS1 complexes exhibiting molecular masses of approximately ∼54 kDa and ∼81 kDa, consistent with the expected size of NS1 dimers and trimers, in cells overexpressing the non-SUMOylatable NS1K70AK219A, compared to cells overexpressing wt NS1 (Fig. 9D, compare lanes 2 and 3). Importantly, cotransfection with mut-ASL, which increased the concentration of SUMOylated NS1 for both wt NS1 and NS1K70AK219A (Fig. 9A, compare lanes 2 and 4 and lanes 3 and 5), led to a noticeable decrease in the apparent abundance of the ∼54-kDa and ∼81-kDa bands. In contrast, a subsequent immunoblot analysis using anti-GAPDH antibodies revealed a very similar pattern among all different samples (Fig. 9C and F, compares lanes 1 to 5), therefore providing strong support to the specificity and relevance of the large differences observed for the NS1 complexes. These data suggested that SUMOylation decreased the abundance of NS1 dimers and trimers, therefore indicating a role for SUMOylation in preventing or disrupting the homomeric interactions normally established by NS1 in the cell.
Fig 9.
NS1 SUMOylation affects the abundance of NS1 dimers and trimers. HEK293FT cells were cotransfected with (+) or without (−) the indicated mammalian expression constructs. At 24 h posttransfection, the cells were treated with either 1 mM disuccinimidyl glutarate (DSG) or vehicle only. Half an hour later, the cells were collected and analyzed by SDS-PAGE and immunoblotting using anti-T7 MAb (A and D), anti-Ubc9 MAb (B and E), and anti-GAPDH MAb (C and F). NS1SUMO, SUMOylated NS1. The positions of NS1 dimers and trimers are indicated and emphasized by diagonal arrows.
DISCUSSION
The nonstructural protein of influenza A virus NS1 plays a critical role during viral infection, neutralizing the innate antiviral responses of the host cell and enhancing viral protein expression. Despite substantial knowledge related to NS1's basic functional properties and recent advances in the characterization of its molecular architecture, our knowledge of the mechanisms regulating its function is still limited. Posttranslational modifications are likely to play an important role as critical regulators of NS1 function. Here, we map the SUMOylation sites in the NS1 protein derived from the PR8 virus strain to residues K70 and K219 and present evidence indicating that, besides being SUMOylated by SUMO1 as previously reported, NS1 can also be SUMOylated by SUMO2/3 and is likely to be preferentially SUMOylated by SUMO2/3 with normal cellular levels of SUMOylation. Furthermore, we also show that SUMOylation appears to affect NS1's ability to neutralize the IFN system and probably other functional properties of NS1, as indicated by the finding that a recombinant virus carrying a non-SUMOylatable NS1 exhibited substantial growth defects even in IFN-deficient cells (Vero cells). Additionally, our data show that SUMOylation does not affect NS1's stability or cellular localization. Instead, SUMOylation appears to decrease the abundance of protein complexes exhibiting molecular weights consistent with their identity corresponding to NS1 dimers and trimers. Finally, our data show that both increases and decreases in the proportion of SUMOylated NS1 have negative effects on NS1 function. This observation suggests that the limited levels of SUMOylated NS1 normally present in the cell during infection may correspond to an optimal level that maximizes the contribution of SUMOylation to protein function. As most SUMOylated proteins in the cell exhibit similar limited levels of SUMOylation, the latter hypothetical scenario may be applicable to most known SUMO targets.
While our data indicate that SUMOylation plays an essential role for the NS1 protein derived from the PR8 virus strain, the key amino acid residues responsible for the SUMOylation of this protein exhibit limited conservation: residue K70, the secondary SUMOylation site in PR8 NS1, is very well conserved in all strains normally transmitted in humans but does not appear to be conserved in virus strains circulating in birds or other animals. Similarly, residue K219, the primary SUMOylation site in PR8 NS1, while widely conserved among human influenza virus strains, constitutes the last amino acid residue in the sequence of several of them, most notably in those derived from the 2009 pandemic strain. Our data showed that K219 is not SUMOylated when it constitutes the C-terminal end of the protein, an observation consistent with data reported previously by Xu et al. (36). Importantly, in that study, Xu et al. mapped the SUMOylation site in the NS1 protein derived from the A/Duck/Hubei/L-1/2004 (H5N1) virus strain to residues K221 (primary SUMOylation site) and K219 (secondary SUMOylation site) and showed that NS1 proteins derived from several virus strains were also SUMOylated. Thus, it appears that while most NS1 proteins share the ability to be SUMOylated, with the NS1 protein from the pandemic viruses being a remarkable exception, it is likely that different lysine residues are used as the main SUMOylation site in NS1 in a strain-specific manner. The ability of other lysine residues within NS1 to become SUMOylated is supported by our observation that some residual SUMOylation was observed in the NS1K70AK219A mutant. Previous studies considered NS1's ability to bind to specific cellular factors as a critical contributor to viral pathogenicity. However, some of those interactions have failed to correlate with increased pathogenicity when empirically tested (75, 76). It will be exciting to investigate whether NS1's ability to become SUMOylated may constitute a better predictor of the potential contribution of the NS1 gene to viral pathogenicity. To this end, it is worth mentioning that the NS1 protein derived from the Spanish flu pandemic virus, A/Brevig Mission/1/1918 (H1N1), exhibits the sequence IKSEV at its C terminus, which is a perfect match to the ΨKXE consensus SUMOylation site (in which Ψ is a bulky hydrophobic amino acid residue and X is any amino acid residue).
Our analyses performed in the presence of mut-ASL provide a new approach toward studying the functional effects mediated by SUMOylation. The classical approach of mutating the SUMOylated residue and studying the effects of that mutation on the target protein has the disadvantage of providing a unidimensional picture of the potential role of SUMOylation. A more complete picture of the role mediated by SUMOylation becomes available when the SUMOylation of the protein can be increased severalfold beyond the SUMOylation levels normally observed in the cell. This is particularly true when no changes in the structure of the protein are needed to achieve this increase, as it is allowed by the use of an ASL. In this study, the use of the ASL provided four important insights into the effects of SUMOylation on NS1 function.
First, the NS1 SUMOylation profile observed upon addition of the ASL revealed the formation of a ladder (Fig. 4 and data not shown), suggestive of the formation of SUMO chains on PR8 NS1. As SUMO chains are associated exclusively with SUMO2/3 conjugation, this observation indicates that NS1 is likely preferentially SUMOylated by SUMO2/3. SUMO2/3 chains are known to stimulate the polyubiquitination of some SUMOylated proteins, therefore triggering their proteasomal degradation (71–74). However, our data indicate that SUMOylation does not affect the stability of NS1 (see below). Therefore, both the actual preferential modification of NS1 by SUMO2/3 and the potential functional consequences of this preference remain to be studied further.
Second, the ASL also allowed us to demonstrate that SUMOylation does not affect the cellular localization of PR8 NS1. The cellular localization of NS1 observed in our analyses appeared to be preferentially cytoplasmic, in contrast with previous studies that have shown a preferentially nuclear localization for NS1 (60–63). Nevertheless, we observed the same cellular distribution of NS1 in cells expressing the non-SUMOylatable form of NS1 as well as in cells coexpressing the ASL, therefore conclusively indicating that dramatic changes in the percentage of SUMOylated NS1 in the cell do not translate into changes in its cellular distribution.
Third, the ASL also allowed us to demonstrate that SUMOylation does not affect the stability of PR8 NS1. Our data clearly show that NS1 is a very stable protein, exhibiting a half-life longer than 12 h. Such stability is not altered when the cells are cotransfected with the ASL or by the introduction of the two amino acid substitutions that prevent NS1 SUMOylation. As similar data were obtained by using two different experimental approaches (pulse-chase analysis and cycloheximide treatment) and two different types of amino acid substitutions in the primary SUMOylation site in NS1 (K219A and K219E, the second reflecting a naturally occurring mutation in some avian influenza virus strains), we consider that these results support the conclusion that drastic changes in the abundance of SUMOylated PR8 NS1 do not result in changes in its turnover. This finding is radically different from those of Xu et al. (36), who reported that a non-SUMOylatable K219RK221R mutant form of the NS1 protein derived from influenza A/Duck/Hubei/L-1/2004 [H5N1] showed a dramatically decreased stability compared to that of its wild-type SUMOylatable form, therefore concluding that SUMOylation regulates the stability of NS1. This raises the possibility that SUMOylation may exert different effects on NS1 proteins derived from different influenza virus strains and indicates the need to empirically determine the effects of SUMOylation on a wide range of NS1 proteins derived from both avian and human influenza A virus strains.
Finally, the ASL also allowed us to demonstrate that the abundance of the SUMOylated form of NS1 in the cell is a major determinant of NS1's ability to neutralize the IFN response. Interestingly, our data indicate that while SUMOylation is important to enhance NS1's ability to neutralize the IFN response, increasing the relative abundance of SUMOylated NS1 beyond a certain threshold also decreases NS1's ability to neutralize the IFN response. This observation led us to conclude that there is an optimal proportion of SUMOylated NS1 that must be present in the cell to maximize NS1's ability to neutralize the IFN response. As most SUMO substrates known to date also exhibit low steady-state SUMOylation levels, it is possible that this conclusion may be true for most SUMOylated proteins. The intriguing inference that little SUMOylation might represent optimal SUMOylation for most SUMO targets will require experimental confirmation, likely achievable by developing ASLs specific for other SUMO targets. Meanwhile, an important consequence of this observation as it relates to the role played by NS1 during influenza infection is that the SUMOylation of NS1 potentially constitutes an optimal target for innovative anti-influenza therapies, as interventions leading to either increases or decreases in the percentage of SUMOylated NS1 are likely to diminish viral growth by affecting NS1 function.
Besides affecting NS1's ability to neutralize the host's IFN response, the data obtained with the recombinant viruses carrying the NS2-shifted NS gene segment indicate that SUMOylation also regulates other activities associated with NS1. This is supported by the finding that the viral mutant carrying the non-SUMOylatable form of NS1 exhibited an ∼2-log growth defect compared to its SUMOylatable counterpart in a cell line defective for IFN production. A previous study indicated a requirement for specific nucleotide sequences located at both the 5′ and 3′ ends of the NS gene segment for proper viral packaging (77). Thus, to minimize the potential interference on viral packaging of the changes intentionally introduced into the NS gene segment, the methodology used to assess the relevance of NS1 SUMOylation for viral growth avoided the need for any viral amplification. The success of this approach was evidenced by the lack of substantial differences in the virus titers produced by the WSN, WSN/T7T7NS, and WSN/T7T7[NS1∼NS2] viruses, a finding probably associated with the recently reported lesser relevance of the 5′ and 3′ UTR sequences of the NS gene segment for viral packaging (78). Nevertheless, some of the growth defects observed for the double mutant virus could be partially associated with the cumulative effect of the various mutations introduced and not exclusively due to SUMO-specific effects. Full confirmation of the relevance of SUMOylation for the data observed with the recombinant viruses will require the development of other recombinant viruses carrying alternative amino acid substitutions in the predicted SUMOylation sites.
Most of the effects attributed to SUMO conjugation are related to SUMO's ability to modulate protein-protein interactions. While our analyses showed that SUMOylation did not affect the cellular localization or the stability of NS1, our cross-linking data suggest a role for SUMO as a modulator of the abundance of NS1 dimers and trimers in the cell. NS1 dimerization plays a critical role in RNA binding (41), and RNA binding is essential to neutralize IFN production (21). Recent models derived from the crystal structure of full-length NS1 indicate that, besides dimers, NS1 is also capable of forming larger homopolymeric structures, which might enhance NS1's anti-IFN function (79). However, dimeric NS1 might be the predominant form of NS1 involved in some of the protein-protein interactions established between NS1 and some cellular factors (80, 81). Therefore, SUMOylation may exert a broad regulatory role on NS1 function simply by helping modulate the proportion of NS1 monomers, dimers, trimers, and polymers present in the cell. We are currently assessing whether NS1 SUMOylation affects each of all currently known protein interactions established by NS1 during infection. Similarly, we are also exploring whether NS1 SUMOylation mediates interactions with other currently unknown cellular and viral factors. Further characterization of the effects of SUMOylation on NS1 may lead to the identification of new molecular targets amenable for the future development of innovative anti-influenza therapies.
ACKNOWLEDGMENTS
This research was supported by grant number 1SC2AI081377 from the National Institute of Allergy and Infectious Diseases (NIAID) and the National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH), and grant number 1SC1AI098976 from the NIAID, NIH, both to Germán Rosas-Acosta. Andres Santos was supported by RISE grant number R25GM069621-09 from the NIGMS, NIH. The Border Biomedical Research Center (BBRC) and its associated core facilities, namely, the DNA Sequencing, Biomolecule Characterization, and Cell Culture Core Facilities, provided essential technical support to this research. Support to the BBRC is provided by grant number G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD), NIH.
We are immensely grateful to Van G. Wilson (Texas A&M Health Science Center College of Medicine, College Station, TX), Daniel R. Perez (Department of Veterinary Medicine, University of Maryland, College Park, MD), Adolfo Garcia-Sastre and Megan L. Shaw (both at the Department of Microbiology, Mount Sinai School of Medicine, New York, NY), and Michael J. Matunis (Bloomberg School of Public Health, Department of Biochemistry and Molecular Biology, Johns Hopkins University, Baltimore, MD) for providing insightful discussions and suggestions related to this work. We are also immensely grateful to Yoshihiro Kawaoka (Department of Pathobiological Sciences, School of Veterinary Medicine, University of Wisconsin—Madison, Madison, WI) for providing us with the 12-plasmid reverse genetics system for the A/WSN/1933 H1N1 virus.
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. Taubenberger JK, Morens DM. 2008. The pathology of influenza virus infections. Annu. Rev. Pathol. 3:499–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rock JR, Hogan BL. 2011. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27:493–512 [DOI] [PubMed] [Google Scholar]
- 3. Falcon AM, Marion RM, Zurcher T, Gomez P, Portela A, Nieto A, Ortin J. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78:3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marion RM, Zurcher T, de la Luna S, Ortin J. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78(Part 10):2447–2451 [DOI] [PubMed] [Google Scholar]
- 5. Min JY, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Twu KY, Kuo RL, Marklund J, Krug RM. 2007. The H5N1 influenza virus NS genes selected after 1998 enhance virus replication in mammalian cells. J. Virol. 81:8112–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000 [DOI] [PubMed] [Google Scholar]
- 8. Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 104:1853–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, Garcia-Sastre A, Roeder RG, Tarakhovsky A. 2012. Suppression of the antiviral response by an influenza histone mimic. Nature 483:428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yanguez E, Nieto A. 2011. So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res. 156:1–12 [DOI] [PubMed] [Google Scholar]
- 11. Lin D, Lan J, Zhang Z. 2007. Structure and function of the NS1 protein of influenza A virus. Acta Biochim. Biophys. Sin. (Shanghai) 39:155–162 [DOI] [PubMed] [Google Scholar]
- 12. Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324–328 [DOI] [PubMed] [Google Scholar]
- 14. Hatada E, Saito S, Fukuda R. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21 [DOI] [PubMed] [Google Scholar]
- 16. Lu Y, Wambach M, Katze MG, Krug RM. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222–228 [DOI] [PubMed] [Google Scholar]
- 17. Min JY, Li S, Sen GC, Krug RM. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236–243 [DOI] [PubMed] [Google Scholar]
- 18. Tan SL, Katze MG. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757–766 [DOI] [PubMed] [Google Scholar]
- 19. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 20. Krug RM, Yuan W, Noah DL, Latham AG. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181–189 [DOI] [PubMed] [Google Scholar]
- 21. Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257–13266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Privalsky ML, Penhoet EE. 1981. The structure and synthesis of influenza virus phosphoproteins. J. Biol. Chem. 256:5368–5376 [PubMed] [Google Scholar]
- 26. Hale BG, Knebel A, Botting CH, Galloway CS, Precious BL, Jackson D, Elliott RM, Randall RE. 2009. CDK/ERK-mediated phosphorylation of the human influenza A virus NS1 protein at threonine-215. Virology 383:6–11 [DOI] [PubMed] [Google Scholar]
- 27. Mahmoudian S, Auerochs S, Grone M, Marschall M. 2009. Influenza A virus proteins PB1 and NS1 are subject to functionally important phosphorylation by protein kinase C. J. Gen. Virol. 90:1392–1397 [DOI] [PubMed] [Google Scholar]
- 28. Hsiang TY, Zhou L, Krug RM. 2012. Roles of the phosphorylation of specific serines and threonines in the NS1 protein of human influenza A viruses. J. Virol. 86:10370–10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pal S, Rosas JM, Rosas-Acosta G. 2010. Identification of the non-structural influenza A viral protein NS1A as a bona fide target of the small ubiquitin-like modifier by the use of dicistronic expression constructs. J. Virol. Methods 163:498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao C, Hsiang TY, Kuo RL, Krug RM. 2010. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 107:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang Y, Zhong G, Zhu L, Liu X, Shan Y, Feng H, Bu Z, Chen H, Wang C. 2010. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J. Immunol. 184:5777–5790 [DOI] [PubMed] [Google Scholar]
- 32. Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. 2011. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 158:12–27 [DOI] [PubMed] [Google Scholar]
- 33. Bekes M, Drag M. 2012. Trojan horse strategies used by pathogens to influence the small ubiquitin-like modifier (SUMO) system of host eukaryotic cells. J. Innate Immun. 4:159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Konig R, Stertz S, Zhou Y, Inoue A, Heinrich Hoffmann H, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu K, Klenk C, Liu B, Keiner B, Cheng J, Zheng BJ, Li L, Han Q, Wang C, Li T, Chen Z, Shu Y, Liu J, Klenk HD, Sun B. 2011. Modification of nonstructural protein 1 of influenza A virus by SUMO1. J. Virol. 85:1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu CY, Jeng KS, Lai MM. 2011. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J. Virol. 85:6618–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 40. Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. 2009. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics 9:3409–3412 [DOI] [PubMed] [Google Scholar]
- 43. Squires RB, Noronha J, Hunt V, Garcia-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. 2012. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi. Viruses 6:404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408 [DOI] [PubMed] [Google Scholar]
- 45. Solorzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535–7543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275 [DOI] [PubMed] [Google Scholar]
- 47. Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell. Proteomics 4:56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. 2006. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics 5:2298–2310 [DOI] [PubMed] [Google Scholar]
- 49. Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. 2004. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 279:33791–33798 [DOI] [PubMed] [Google Scholar]
- 50. Hay RT. 2007. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17:370–376 [DOI] [PubMed] [Google Scholar]
- 51. Ulrich HD. 2008. The fast-growing business of SUMO chains. Mol. Cell 32:301–305 [DOI] [PubMed] [Google Scholar]
- 52. Vertegaal AC. 2010. SUMO chains: polymeric signals. Biochem. Soc. Trans. 38:46–49 [DOI] [PubMed] [Google Scholar]
- 53. Schneider J, Wolff T. 2009. Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine 27:6312–6316 [DOI] [PubMed] [Google Scholar]
- 54. Cesaire R, Oliere S, Sharif-Askari E, Loignon M, Lezin A, Olindo S, Panelatti G, Kazanji M, Aloyz R, Panasci L, Bell JC, Hiscott J. 2006. Oncolytic activity of vesicular stomatitis virus in primary adult T-cell leukemia. Oncogene 25:349–358 [DOI] [PubMed] [Google Scholar]
- 55. Hayman A, Comely S, Lackenby A, Murphy S, McCauley J, Goodbourn S, Barclay W. 2006. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology 347:52–64 [DOI] [PubMed] [Google Scholar]
- 56. Kuo RL, Zhao C, Malur M, Krug RM. 2010. Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-beta transcription. Virology 408:146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. 2010. Engineered RNA viral synthesis of microRNAs. Proc. Natl. Acad. Sci. U. S. A. 107:11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Backstrom Winquist E, Abdurahman S, Tranell A, Lindstrom S, Tingsborg S, Schwartz S. 2012. Inefficient splicing of segment 7 and 8 mRNAs is an inherent property of influenza virus A/Brevig Mission/1918/1 (H1N1) that causes elevated expression of NS1 protein. Virology 422:46–58 [DOI] [PubMed] [Google Scholar]
- 59. Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 85:5259–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Greenspan D, Palese P, Krystal M. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li Y, Yamakita Y, Krug RM. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 95:4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Melen K, Kinnunen L, Fagerlund R, Ikonen N, Twu KY, Krug RM, Julkunen I. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 81:5995–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sato Y, Yoshioka K, Suzuki C, Awashima S, Hosaka Y, Yewdell J, Kuroda K. 2003. Localization of influenza virus proteins to nuclear dot 10 structures in influenza virus-infected cells. Virology 310:29–40 [DOI] [PubMed] [Google Scholar]
- 64. Endter C, Kzhyshkowska J, Stauber R, Dobner T. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 98:11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Janssen K, Hofmann TG, Jans DA, Hay RT, Schulze-Osthoff K, Fischer U. 2007. Apoptin is modified by SUMO conjugation and targeted to promyelocytic leukemia protein nuclear bodies. Oncogene 26:1557–1566 [DOI] [PubMed] [Google Scholar]
- 66. Lethbridge KJ, Scott GE, Leppard KN. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259–268 [DOI] [PubMed] [Google Scholar]
- 67. Palacios S, Perez LH, Welsch S, Schleich S, Chmielarska K, Melchior F, Locker JK. 2005. Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell 16:2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen SC, Chang LY, Wang YW, Chen YC, Weng KF, Shih SR, Shih HM. 2011. Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J. Biol. Chem. 286:31373–31384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marusic MB, Mencin N, Licen M, Banks L, Grm HS. 2010. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J. Virol. 84:11585–11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weger S, Hammer E, Heilbronn R. 2004. SUMO-1 modification regulates the protein stability of the large regulatory protein Rep78 of adeno associated virus type 2 (AAV-2). Virology 330:284–294 [DOI] [PubMed] [Google Scholar]
- 71. Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10:538–546 [DOI] [PubMed] [Google Scholar]
- 72. Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282:34167–34175 [DOI] [PubMed] [Google Scholar]
- 73. van Hagen M, Overmeer RM, Abolvardi SS, Vertegaal AC. 2010. RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated hypoxia-inducible factor-2alpha. Nucleic Acids Res. 38:1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weisshaar SR, Keusekotten K, Krause A, Horst C, Springer HM, Gottsche K, Dohmen RJ, Praefcke GJ. 2008. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582:3174–3178 [DOI] [PubMed] [Google Scholar]
- 75. Hale BG, Steel J, Manicassamy B, Medina RA, Ye J, Hickman D, Lowen AC, Perez DR, Garcia-Sastre A. 2010. Mutations in the NS1 C-terminal tail do not enhance replication or virulence of the 2009 pandemic H1N1 influenza A virus. J. Gen. Virol. 91:1737–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shelton H, Smith M, Hartgroves L, Stilwell P, Roberts K, Johnson B, Barclay W. 2012. An influenza reassortant with polymerase of pH1N1 and NS gene of H3N2 influenza A virus is attenuated in vivo. J. Gen. Virol. 93:998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 79:3766–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gao Q, Chou YY, Doganay S, Vafabakhsh R, Ha T, Palese P. 2012. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J. Virol. 86:7043–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bornholdt ZA, Prasad BV. 2008. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456:985–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, Krug RM, Montelione GT. 2008. Structural basis for suppression of a host antiviral response by influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 105:13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hale BG, Kerry PS, Jackson D, Precious BL, Gray A, Killip MJ, Randall RE, Russell RJ. 2010. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 107:1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]