Abstract
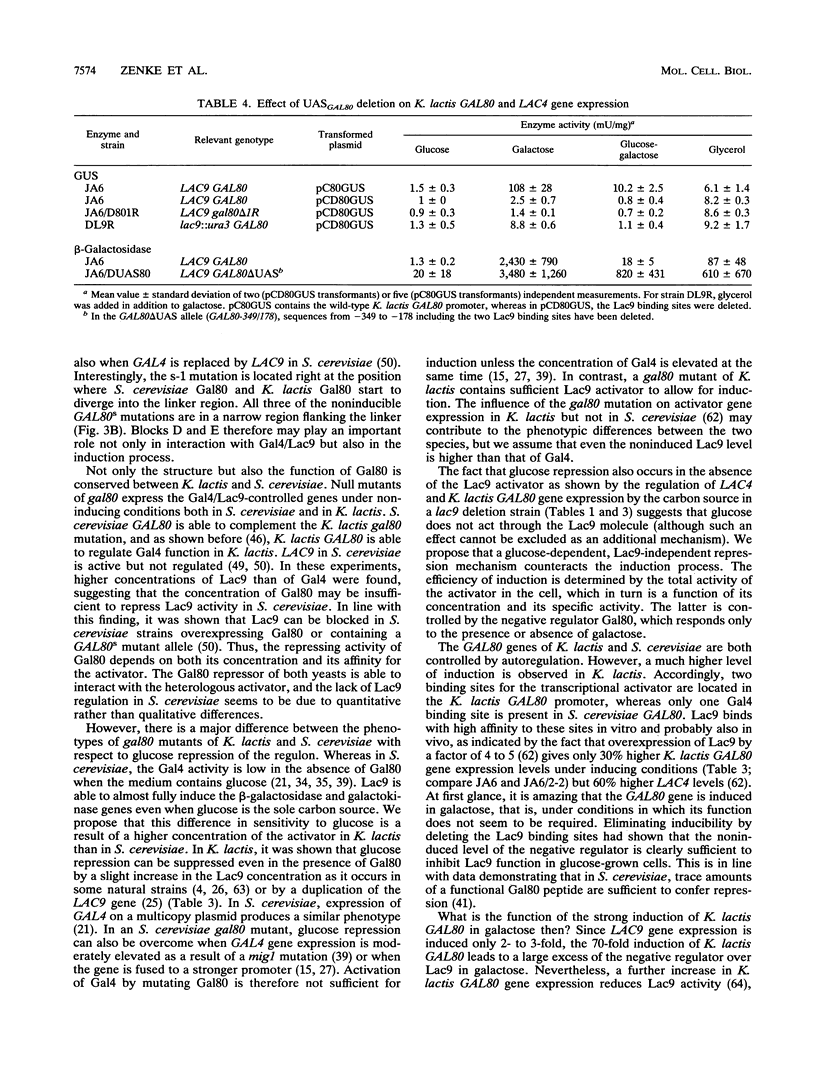
We cloned the GAL80 gene encoding the negative regulator of the transcriptional activator Gal4 (Lac9) from the yeast Kluyveromyces lactis. The deduced amino acid sequence of K. lactis GAL80 revealed a strong structural conservation between K. lactis Gal80 and the homologous Saccharomyces cerevisiae protein, with an overall identity of 60% and two conserved blocks with over 80% identical residues. K. lactis gal80 disruption mutants show constitutive expression of the lactose/galactose metabolic genes, confirming that K. lactis Gal80 functions in essentially in the same way as does S. cerevisiae Gal80, blocking activation by the transcriptional activator Lac9 (K. lactis Gal4) in the absence of an inducing sugar. However, in contrast to S. cerevisiae, in which Gal4-dependent activation is strongly inhibited by glucose even in a gal80 mutant, glucose repressibility is almost completely lost in gal80 mutants of K. lactis. Indirect evidence suggests that this difference in phenotype is due to a higher activator concentration in K. lactis which is able to overcome glucose repression. Expression of the K. lactis GAL80 gene is controlled by Lac9. Two high-affinity binding sites in the GAL80 promoter mediate a 70-fold induction by galactose and hence negative autoregulation by Gal80. Gal80 in turn not only controls Lac9 activity but also has a moderate influence on its rate of synthesis. Thus, a feedback control mechanism exists between the positive and negative regulators. By mutating the Lac9 binding sites of the GAL80 promoter, we could show that induction of GAL80 is required to prevent activation of the lactose/galactose regulon in glycerol or glucose plus galactose, whereas the noninduced level of Gal80 is sufficient to completely block Lac9 function in glucose.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet. 1989 Apr;216(2-3):422–427. doi: 10.1007/BF00334386. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987 Dec;7(12):4400–4406. doi: 10.1128/mcb.7.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Kornberg R. D. GAL4 protein: purification, association with GAL80 protein, and conserved domain structure. Mol Cell Biol. 1990 Jun;10(6):2916–2923. doi: 10.1128/mcb.10.6.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. J., Saliola M., Falcone C., Bianchi M. M., Fukuhara H. Sequence organization of the circular plasmid pKD1 from the yeast Kluyveromyces drosophilarum. Nucleic Acids Res. 1986 Jun 11;14(11):4471–4481. doi: 10.1093/nar/14.11.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Gerardot C. J., Martin A. K. Genetic evidence for similar negative regulatory domains in the yeast transcription activators GAL4 and LAC9. Nucleic Acids Res. 1990 Sep 11;18(17):5213–5217. doi: 10.1093/nar/18.17.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Sheetz R. M., Lacy L. R. Genetic regulation: yeast mutants constitutive for beta-galactosidase activity have an increased level of beta-galactosidase messenger ribonucleic acid. Mol Cell Biol. 1981 Nov;1(11):1048–1056. doi: 10.1128/mcb.1.11.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R. J., Strasser A. W., Höner C. B., Hollenberg C. P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991 Oct;7(7):691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne D. C. Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics. 1966 Sep;54(3):911–916. doi: 10.1093/genetics/54.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves P. M., Maurer K., Mager W. H., Planta R. J. Kluyveromyces contains a functional ABF1-homologue. Nucleic Acids Res. 1992 May 11;20(9):2211–2215. doi: 10.1093/nar/20.9.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gödecke A., Zachariae W., Arvanitidis A., Breunig K. D. Coregulation of the Kluyveromyces lactis lactose permease and beta-galactosidase genes is achieved by interaction of multiple LAC9 binding sites in a 2.6 kbp divergent promoter. Nucleic Acids Res. 1991 Oct 11;19(19):5351–5358. doi: 10.1093/nar/19.19.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M., Segawa T., Nogi Y., Suzuki Y., Fukasawa T. Autogenous regulation of the Saccharomyces cerevisiae regulatory gene GAL80. Mol Gen Genet. 1987 May;207(2-3):273–279. doi: 10.1007/BF00331589. [DOI] [PubMed] [Google Scholar]
- Jacobson A. Purification and fractionation of poly(A)+ RNA. Methods Enzymol. 1987;152:254–261. doi: 10.1016/0076-6879(87)52028-6. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987 Jul 3;50(1):143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Johnston S. A., Zavortink M. J., Debouck C., Hopper J. E. Functional domains of the yeast regulatory protein GAL4. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6553–6557. doi: 10.1073/pnas.83.17.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe R. J., Harriss J. V., Sharp Z. D., Douglas M. G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983 Nov;25(2-3):333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- Kuger P., Gödecke A., Breunig K. D. A mutation in the Zn-finger of the GAL4 homolog LAC9 results in glucose repression of its target genes. Nucleic Acids Res. 1990 Feb 25;18(4):745–751. doi: 10.1093/nar/18.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhandaivelu N., Jones W. K., Martin A. K., Dickson R. C. The signal for glucose repression of the lactose-galactose regulon is amplified through subtle modulation of transcription of the Kluyveromyces lactis Kl-GAL4 activator gene. Mol Cell Biol. 1992 May;12(5):1924–1931. doi: 10.1128/mcb.12.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lamphier M. S., Ptashne M. Multiple mechanisms mediate glucose repression of the yeast GAL1 gene. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5922–5926. doi: 10.1073/pnas.89.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuther K. K., Johnston S. A. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992 May 29;256(5061):1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Pelham H. R. The sequence of the Kluyveromyces lactis BiP gene. Nucleic Acids Res. 1990 Nov 11;18(21):6438–6438. doi: 10.1093/nar/18.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Chasman D. I., Buchman A. R., Kornberg R. D. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987 Oct;7(10):3446–3451. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Yoshimatsu T., Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylin L. M., Bhat J. P., Hopper J. E. Regulated phosphorylation and dephosphorylation of GAL4, a transcriptional activator. Genes Dev. 1989 Aug;3(8):1157–1165. doi: 10.1101/gad.3.8.1157. [DOI] [PubMed] [Google Scholar]
- Mylin L. M., Gerardot C. J., Hopper J. E., Dickson R. C. Sequence conservation in the Saccharomyces and Kluveromyces GAL11 transcription activators suggests functional domains. Nucleic Acids Res. 1991 Oct 11;19(19):5345–5350. doi: 10.1093/nar/19.19.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylin L. M., Johnston M., Hopper J. E. Phosphorylated forms of GAL4 are correlated with ability to activate transcription. Mol Cell Biol. 1990 Sep;10(9):4623–4629. doi: 10.1128/mcb.10.9.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Fukasawa T. Functional domains of a negative regulatory protein, GAL80, of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jul;9(7):3009–3017. doi: 10.1128/mcb.9.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Fukasawa T. Nucleotide sequence of the yeast regulatory gene GAL80. Nucleic Acids Res. 1984 Dec 21;12(24):9287–9298. doi: 10.1093/nar/12.24.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Shimada H., Matsuzaki Y., Hashimoto H., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. II. The isolation and dosage effect of the regulatory gene GAL80. Mol Gen Genet. 1984;195(1-2):29–34. doi: 10.1007/BF00332719. [DOI] [PubMed] [Google Scholar]
- Parthun M. R., Jaehning J. A. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol Cell Biol. 1992 Nov;12(11):4981–4987. doi: 10.1128/mcb.12.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun M. R., Jaehning J. A. Purification and characterization of the yeast transcriptional activator GAL4. J Biol Chem. 1990 Jan 5;265(1):209–213. [PubMed] [Google Scholar]
- Perlman D., Hopper J. E. Constitutive synthesis of the GAL4 protein, a galactose pathway regulator in Saccharomyces cerevisiae. Cell. 1979 Jan;16(1):89–95. doi: 10.1016/0092-8674(79)90190-9. [DOI] [PubMed] [Google Scholar]
- Riley M. I., Hopper J. E., Johnston S. A., Dickson R. C. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol. 1987 Feb;7(2):780–786. doi: 10.1128/mcb.7.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saliola M., Shuster J. R., Falcone C. The alcohol dehydrogenase system in the yeast, Kluyveromyces lactis. Yeast. 1990 May-Jun;6(3):193–204. doi: 10.1002/yea.320060304. [DOI] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Langdon S. D., Johnston S. A. Interaction between transcriptional activator protein LAC9 and negative regulatory protein GAL80. Mol Cell Biol. 1989 Jul;9(7):2950–2956. doi: 10.1128/mcb.9.7.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Leuther K. K., Johnston S. A. GAL4 mutations that separate the transcriptional activation and GAL80-interactive functions of the yeast GAL4 protein. Genetics. 1990 May;125(1):21–27. doi: 10.1093/genetics/125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 May 25;18(10):3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Mutations affecting synthesis of beta-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980 Aug;95(4):877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Milner J. S. Cloning and analysis of the Kluyveromyces lactis TRP1 gene: a chromosomal locus flanked by genes encoding inorganic pyrophosphatase and histone H3. Yeast. 1989 Jan-Feb;5(1):35–50. doi: 10.1002/yea.320050106. [DOI] [PubMed] [Google Scholar]
- Tajima M., Nogi Y., Fukasawa T. Duplicate upstream activating sequences in the promoter region of the Saccharomyces cerevisiae GAL7 gene. Mol Cell Biol. 1986 Jan;6(1):246–256. doi: 10.1128/mcb.6.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia T. E., Hamilton R. W., Cano C. L., Hopper J. E. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol Cell Biol. 1984 Aug;4(8):1521–1527. doi: 10.1128/mcb.4.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T. D., Dickson R. C. The organization and transcription of the galactose gene cluster of Kluyveromyces lactis. Nucleic Acids Res. 1988 Aug 25;16(16):8011–8028. doi: 10.1093/nar/16.16.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowski-Louvel M., Tanguy-Rougeau C., Fukuhara H. A nuclear gene required for the expression of the linear DNA-associated killer system in the yeast Kluyveromyces lactis. Yeast. 1988 Mar;4(1):71–81. doi: 10.1002/yea.320040108. [DOI] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W., Breunig K. D. Expression of the transcriptional activator LAC9 (KlGAL4) in Kluyveromyces lactis is controlled by autoregulation. Mol Cell Biol. 1993 May;13(5):3058–3066. doi: 10.1128/mcb.13.5.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W., Kuger P., Breunig K. D. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9 (K1GAL4) [corrected]. Nucleic Acids Res. 1993 Jan 11;21(1):69–77. doi: 10.1093/nar/21.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]