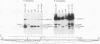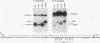Abstract
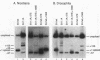
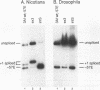
cis-spliced nuclear pre-mRNA introns found in a variety of organisms, including Tetrahymena thermophila, Drosophila melanogaster, Caenorhabditis elegans, and plants, are significantly richer in adenosine and uridine residues than their flanking exons are. The functional significance of this intronic AU richness, however, has been demonstrated only in plant nuclei. In these nuclei, 5' and 3' splice sites are selected in part by their positions relative to AU-rich elements spread throughout the length of an intron. Because of this position-dependent selection scheme, a 5' splice site at the normal (+1) exon-intron boundary having only three contiguous consensus nucleotides can compete effectively with an enhanced exonic site (-57E) having nine consensus nucleotides and outcompete an enhanced site (+106E) embedded within the AU-rich intron. To determine whether transitions from AU-poor exonic sequences to AU-rich intronic sequences influence 5' splice site selection in other organisms, alleles of the pea rbcS3A1 intron were expressed in Drosophila Schneider 2 cells, and their splicing patterns were compared with those in tobacco nuclei. We demonstrate that this heterologous transcript can be accurately spliced in transfected Drosophila nuclei and that a +1 G-to-A knockout mutation at the normal splice site activates the same three cryptic 5' splice sites as in tobacco. Enhancement of the exonic (-57) and intronic (+106) sites to consensus splice sites indicates that potential splice sites located in the upstream exon or at the 5' exon-intron boundary are preferred in Drosophila cells over those embedded within AU-rich intronic sequences. In contrast to tobacco, in which the activities of two competing 5' splice sites upstream of the AU-rich intron are modulated by their proximity to the AU transition point, D. melanogaster utilizes the upstream site which has a higher proportion of consensus nucleotides. The enhanced version of the cryptic intronic site is efficiently selected in D. melanogaster when the normal +1 site is weakened or discrete AU-rich elements upstream of the +106E site are disrupted. Selection of this internal site in tobacco requires more drastic disruption of these motifs. We conclude that 5' splice site selection in Drosophila nuclei is influenced by the intrinsic strengths of competing sites and by the presence of AU-rich intronic elements but to a different extent than in tobacco.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aroian R. V., Levy A. D., Koga M., Ohshima Y., Kramer J. M., Sternberg P. W. Splicing in Caenorhabditis elegans does not require an AG at the 3' splice acceptor site. Mol Cell Biol. 1993 Jan;13(1):626–637. doi: 10.1128/mcb.13.1.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Cherbas L., Lee K., Cherbas P. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev. 1991 Jan;5(1):120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- Conrad R., Liou R. F., Blumenthal T. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J. 1993 Mar;12(3):1249–1255. doi: 10.1002/j.1460-2075.1993.tb05766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R., Liou R. F., Blumenthal T. Functional analysis of a C. elegans trans-splice acceptor. Nucleic Acids Res. 1993 Feb 25;21(4):913–919. doi: 10.1093/nar/21.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C., Taylor F. M., Martindale D. W. Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res. 1990 Sep 11;18(17):5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr Robert, Moses Phyllis, Morelli Giorgio, Coruzzi Gloria, Chua Nam-Hai. Expression dynamics of the pea rbcS multigene family and organ distribution of the transcripts. EMBO J. 1986 Sep;5(9):2063–2071. doi: 10.1002/j.1460-2075.1986.tb04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991 Sep;10(9):2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol. 1990 May;14(5):727–733. doi: 10.1007/BF00016505. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guo M., Lo P. C., Mount S. M. Species-specific signals for the splicing of a short Drosophila intron in vitro. Mol Cell Biol. 1993 Feb;13(2):1104–1118. doi: 10.1128/mcb.13.2.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Elmer J. S., Rogers S. G. Transient expression of heterologous RNAs using tomato golden mosaic virus. Nucleic Acids Res. 1988 Nov 25;16(22):10511–10528. doi: 10.1093/nar/16.22.10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shaw J. E. A variant Tc4 transposable element in the nematode C. elegans could encode a novel protein. Nucleic Acids Res. 1993 Jan 11;21(1):59–67. doi: 10.1093/nar/21.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., McCullough A. J., Schuler M. A. 3' splice site selection in dicot plant nuclei is position dependent. Mol Cell Biol. 1993 Aug;13(8):4485–4493. doi: 10.1128/mcb.13.8.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., McCullough A. J., Schuler M. A. Expression of maize Adh1 intron mutants in tobacco nuclei. Plant J. 1993 Mar;3(3):393–403. doi: 10.1046/j.1365-313x.1993.t01-22-00999.x. [DOI] [PubMed] [Google Scholar]
- Lührmann R., Kastner B., Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990 Nov 30;1087(3):265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- McCullough A. J., Lou H., Schuler M. A. Factors affecting authentic 5' splice site selection in plant nuclei. Mol Cell Biol. 1993 Mar;13(3):1323–1331. doi: 10.1128/mcb.13.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough A. J., Lou H., Schuler M. A. In vivo analysis of plant pre-mRNA splicing using an autonomously replicating vector. Nucleic Acids Res. 1991 Jun 11;19(11):3001–3009. doi: 10.1093/nar/19.11.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Burks C., Hertz G., Stormo G. D., White O., Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992 Aug 25;20(16):4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallette F., Mege E., Reiss A., Adesnik M. Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res. 1989 Jan 25;17(2):723–733. doi: 10.1093/nar/17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr Nuclear pre-mRNA splicing in yeast. Yeast. 1989 Nov-Dec;5(6):439–457. doi: 10.1002/yea.320050604. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]