Abstract
Here we present results that investigate the origins of signals observed in tip-enhanced Raman (TERS) measurements of functionalized nanoparticles. Surface enhanced Raman scattering (SERS) is known to give the largest enhancements in gap junctions. Similarly, gap-mode TERS also produces significant enhancements. The methodology developed here provides gap-mode like enhancements in TERS measurements without the need for a metal surface. Using a combination of aggregated nanoparticle SERS and TERS detection of functionalized nanoparticles, we assess the chemical origins of the observed peaks and show that molecules outside of gap junctions are also enhanced using our methodology. Our experiments use biotin and streptavidin as a model system for protein- ligand binding. Different size functionalized nanoparticles (20, 50, 80 nm) show changes in intensity in both SERS and TERS measurements. SERS measurements indicate that streptavidin has a larger Raman cross-section than biotin and is preferentially observed. The specific streptavidin peaks observed by TERS vary depending on whether streptavidin is attached to the nanoparticle and located in the gap or bound to the substrate surface. This methodology suggests a route to enhancing TERS signals associated with protein receptors in biological systems that cannot be isolated to a metallic surface.
INTRODUCTION
Increased understanding of surface enhanced Raman spectroscopy (SERS) over the last 35 years has transformed Raman into an ultrasensitive detection method. Excitation of a localized surface plasmon resonance (LSPR) results in a substantial local electromagnetic field around metallic nanostructures that gives rise to enhanced Raman scattering1, 2 SERS has shown single-molecule sensitivity3–5 and in some applications is more sensitive than fluorescence detection.6 The ultimate sensitivity depends critically upon both the generation of a strong and locally confined enhanced electromagnetic field and the analyte molecule residing in this enhanced field. Results show that these maximum enhancements originate from `hotspots', which are formed from junctions and crevices between nanoparticles.7–10 An optimum distance of a few nanometers between nanoparticles has been associated with the maximum enhancements.11 These hotspots are attributable to the strong coupling between the discrete particles within a dimer or aggregate structures, resulting in Raman signal enhancements of 6 to 8 orders of magnitude.1, 12
The high sensitivity of SERS combined with the non-destructive nature and chemical specificity of Raman spectroscopy has inspired other strategies, such as tip-enhanced Raman spectroscopy (TERS)13–16 and shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS)17–19, to provide spatial specificity and expand the utility. SHINERS uses scattered core-shell nanoparticles to generate Raman scattering from molecules at the optimum distance between adjacent particles. The novel innovation is the protective coating prevents nonspecific interactions and allows enhancements from any surface. TERS employs a scanning metallic or metallized tip as a nano-antenna to enhance the incident and the emitted fields, thus providing high spatial resolution and enhanced Raman signals simultaneously. The limitation of classical TERS is the lower enhancement associated with a single nano-probe. TERS is commonly used to investigate carbon nanotubes,20, 21 dye molecules,22, 23 and semiconductors24–26 due to intrinsically stronger Raman responses. The enhancements attainable with TERS are also sufficient for detection of molecules in biological samples.27–29
As noted above, the highest Raman enhancements are obtained in hotspots. In TERS, gap-modes that arise from image dipoles as the tip approaches a metal surface have also been shown to produce substantial enhancements.30–32 Independent work by groups of Pettinger and Deckert have demonstrated gap-mode TERS for the detection of single molecules.33, 34 One limitation of gap-mode TERS is the need for a metallic surface. To address this limitation it has been shown that nanoparticles on dielectric substrates can interact with the TERS tip to generate enhancements comparable to gap-modes.35, 36 The combination of nanoparticle probes and TERS detection using radial polarization provides spatial resolution on the dimensions of the nanoparticle probe that can be used to investigate intact cellular membranes and other biological samples.37
The signals observed from nanoparticles detected by TERS raise questions about where the molecules are located. The hotspot model suggests that the molecules in the gap will give rise to the largest signals; however, in our previous report signals associated with surface molecules dominated the TERS spectrum.35 It has been both theoretically calculated and experimentally demonstrated that substantial Raman signals can be generated outside of the hotspots.38–40 This suggests that nanoparticles bound to proteins can provide additional TERS enhancement without the target residing in the gap junction.
In the present work we utilize biotin and streptavidin as a model protein-ligand system.41 Streptavidin contains a bacterial recognition motif sequence RYD, similar to the mammalian RGD motif. Biotin-streptavidin binding has become ubiquitous throughout science and provides a well-studied system to validate this methodology.
The goal of this article is to elucidate what is enhanced and provide evidence regarding the location of the enhanced molecule with respect to the gap. In the present report we investigate the effect of nanoparticle probe size on the detected TERS signal, the signal observed from different functionalized gold nanoparticles (GNPs), and compare the observed TERS signals to spectra obtained from aggregated nanoparticle SERS. Our results suggest that functionalized GNPs with TERS detection can distinguish between a protein in the gap junction and one bound to the nanoparticle outside the gap. These results support the use of targeted nanoparticles and TERS for investigating bio-molecular interactions, such as ligand-receptor binding that are of paramount importance in understanding cellular activities.
EXPERIMENTAL
TERS sample preparation
Samples used for TERS measurements were prepared using a previously reported protocol.35 In summary, GNPs functionalized with either biotin or streptavidin (Nanocs Inc.) were deposited onto glass slides functionalized with the streptavidin or biotin, respectively. Thus two different geometries were achieved, one with the biotinylated-GNPs on top and streptavidin on the glass slide, the other with streptavidin-GNPs adsorbed to a biotin-functionalized slide. Typically, 20~50 μL of biotinylated-GNPs solution (0.05%) were spin-coated or drop-coated onto a streptavidin functionalized glass slide and then allowed to air dry for 30 minutes. Similarly, the inverse geometry was realized by coating a biotin derivatized microscope slide with 20–50 μL of streptavidin-GNPs (50nm) solution (0.05%), followed by rinsing with nano-pure water with a resistivity of 18.2 MΩ cm, and drying at room temperature. These sample slides were used as prepared and stored at 4 °C after the TERS measurement.
TERS microscopy
TERS measurements utilized a system comprised of a commercial atomic force microscope (AFM, Nanonics MV4000) and a home-built Raman microscope, as previously described.35, 37 The TERS tip is a chemical mechanical polished (CMP) Au-nanoparticle affixed onto the apex of a transparent optical-fiber AFM cantilever (Nanonics Supertips, Ltd.). The manufacturer specified the diameter of the Au-nanoparticle to be 150–300nm. A 633nm HeNe laser beam is used to irradiate the TERS tip at normal incidence to the sample with radial polarization achieved through a liquid crystal mode converter (ArcOptix). The advantage of a focused, radial polarized laser beam has been previously reported.35, 37, 42, 43 A maximum laser power of 2 mW was used in TERS and SERS measurements in order to avoid sample damage. The upright microscope was modified to accommodate dark-field microscopy (Olympus LMPlan FLN 50× 0.5 BD objective, 150W tungsten halogen lamp), enabling identification of the region of interest (ROI) where single GNPs are distinguished from GNP aggregates through differences in the observed scattering.37 The back-scattered signals were detected by the same microscope objective used for excitation, passed through an edge filter, sent to a spectrograph (Horiba Jobin Yvon iHR320), and recorded by a CCD camera (Jobin Yvon, Synapse). Each TERS map was carried out by scanning the sample area of interest under the TERS tip fixed in the laser focus. After acquiring a TERS map, the tip position was checked to see if it remained in the focused spot and then retracted from the surface to decrease the possibility of contaminating or breaking it.
SERS of nanoparticle clusters
50–100 μL of functionalized GNPs were centrifuged at 15000 rcf for 15 minutes to prepare SERS-active nanoparticle clusters. Biotinylated-GNPs with varying sizes (20, 50, 80 nm) and 50 nm streptavidin-GNPs were used as received. After sufficient centrifuging, we removed the bulk of the supernatant, leaving the clustered nanoparticle pellets at the bottom of the centrifuge tube. These pellets were re-dispersed into a very small volume of nano-pure water and deposited onto a clean microscope slide and subsequently covered with a glass cover slip to prevent possible contamination during SERS measurements. SERS measurements of biotin and streptavidin functionalized GNPs were carried out on these nanoparticle clusters, as observed through an Olympus UPlanFLN 40×/0.75 NA objective on our home-built Raman microscope.44
UV-Vis and SERS of biotin-streptavidin complex
UV-Vis extinction spectra were obtained using a Hitachi U-2910 spectrophotometer. The binding of streptavidin to 80nm biotinylated-GNPs was monitored spectroscopically. Following the binding between biotinylated-GNPs and streptavidin, clusters of the streptavidin/biotinylated-GNPs complex were prepared with the same protocol as assembling biotinylated-GNPs and streptavidin-GNPs nano-clusters. These clusters were then deposited onto a clean microscope slide and sealed with a cover slip for SERS measurement.
Data analysis
All TERS data acquired were plotted or mapped without further manipulation like smoothing, filtering, or background subtraction. The reported SERS spectra are the average of 6–10 measurements to represent the features observed.
RESULTS AND DISCUSSION
Functionalized GNP probes were located on a streptavidin substrate using reflective dark-field microscopy. Based on the color and brightness of the reflected scattering from those probes, an ROI was identified for TERS microscopy. Once the ROI was identified, a routine AFM scan on a relatively larger area was performed first to make compensations to the slope and height of the sample surface and also pinpoint the location of the single biotinylated-GNP probes for TERS mapping. Figure 1A shows a 3D AFM image of a single biotinylated-GNP probe captured within a 2×2 μm TERS map.
Figure 1.
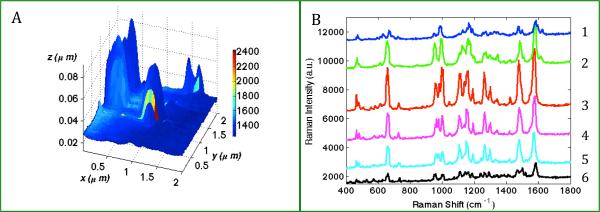
(A) TERS map with simultaneously obtained topographic features and Raman characteristics from 80nm biotinylated-GNP probe adsorbed onto a streptavidin functionalized slide. Scan size: 2×2 μm, TERS step-size 55nm. Color represents the Raman intensity of a streptavidin marker band (965cm−1). (B) Plot of the representative TERS spectra extracted from 6 pixels along the colored line y= 1.08 μm in (A), where (1) x=1.31 μm, (2) x=1.25 μm, (3) x=1.14 μm, (4) x=1.08 μm, (5) x=0.97 μm, (6) x=0.86 μm.
In Figure 1A, the TERS map was constructed by superimposing the Raman map at 965 cm−1 onto the simultaneously acquired AFM image. From the topography, a single object with the height of ~80 nm and width of ~250 nm was clearly observed in the center of the image. The height matches that of the 80 nm biotinylated-GNP used. The width, a convolution of the nanoparticle and the TERS tip, is consistent with the tip and nanoparticle size used. The Raman color map in Figure 1A provides chemical characteristics of the same ROI. At each point (pixel) of the ROI, a full TERS spectrum was recorded therefore allowing for the differentiation of chemical composition pixel by pixel. The TERS map shows a high-degree of correlation between the location of a biotinylated-GNP probe and Raman signal enhancement. It confirms the result of our previous study35 that only the area with the biotinylated-GNP probe experiences a significant enhancement of electromagnetic field and generates an appreciable TERS signal. The observed asymmetry in intensity, as evident in Figure 1A, is consistent in nearly all TERS scans on different samples, and has been modeled and explained previously.37 The TERS spectrum observed is highly reproducible and the observed peaks only vary in intensity depending on the distance from the centroid of the probe as shown in Figure 1B. Most of the peaks observed can be assigned to aromatic amino acid residues in streptavidin such as tryptophan, tyrosine, and phenylalanine. Biotin peaks were also observed, such as at 1051 cm−1, but these peaks are far less prominent in comparison to the peaks from the amino acid residues in streptavidin. Detailed peak assignments are presented in Table 1.
Table 1.
Peak Assignments for normal Raman, SERS and TERS spectra
| Raman | SERS | TERS | Details | |||||
|---|---|---|---|---|---|---|---|---|
| SAa | SAa | SA-Bb | Biotin | 50nm b-GNPc | 80nm b-GNPc | 50nm sa-GNPd | Assignment | Refs |
| 473 | 476 | 469 | Phe | 52 | ||||
| 490 | 503 | 503 | 506 | 489 | Tyr | 52 | ||
| 538 | 539 | 540 | Trp, Val | 57 | ||||
| 566 | 567 | 567 | 580 | 565 | Trp | 57 | ||
| 619 | 624 | 624 | 624 | 620 | Phe | 57 | ||
| 661 | 667 | 663 | ν-C-S | 49 | ||||
| 704 | 704 | 706 | Trp(W18) | 49 | ||||
| 715 | Biotin | |||||||
| 754 | 742 | 740 | Trp(W18) | 57 | ||||
| 813 | 812 | 813 | 817 | 813 | Biotin/Streptavidin | 58 | ||
| 827 | Tyr(Y1) | 57 | ||||||
| 849 | 854 | 860 | Tyr(Y1) | 57 | ||||
| 876 | 883 | 887 | 885 | Trp(W17) | 57 | |||
| 925 | 941 | 942 | 934 | 932 | Trp(W25),Glu,Asp | 57 | ||
| 959 | 971 | 967 | 965 | 973 | Trp, Val | 57 | ||
| 996 | 999 | 992 | 993 | 993 | Biotin/Phe | |||
| 1007 | 1014 | 1010 | 1007 | 1010 | Trp(W16),Phe,Ala | 57 | ||
| 1048 | 1051 | 1050 | 1052 | Biotin | 59 | |||
| 1075 | 1077 | 1077 | 1072 | Glu,Thr | 57 | |||
| 1082 | ν-C-C | 46 | ||||||
| 1095 | 1103 | Ala,Lys | 57 | |||||
| 1123 | 1125 | 1126 | 1130 | 1133 | 1125 | 1124 | ν-C-N,Trp(W13),Val,Glu | 57 |
| 1154 | 1149 | ν-C-N, Val, Lys | 57 | |||||
| 1175 | 1178 | 1176 | 1176 | 1171 | 1173 | Phe,Tyr,Val | 57 | |
| 1204 | 1203 | Streptavidin | ||||||
| 1221 | 1219 | 1218 | Tyr(Y7a) | 46 | ||||
| 1235 | 1237 | 1238 | AmideIII | 57 | ||||
| 1242 | 1247 | Ureido ring | 46 | |||||
| 1257 | 1274 | 1273 | 1274 | 1272 | 1272 | Biotin, AmideIII | 57,60 | |
| 1293 | 1304 | 1298 | ω-CH2 | 46 | ||||
| 1322 | 1310 | 1316 | 1317 | 1312 | Ser(γ-CH2) | 57 | ||
| 1332 | 1335 | Trp(W7) | 57 | |||||
| 1356 | 1351 | 1350 | Trp(W7) | 57 | ||||
| 1372 | 1376 | ω-CH2 | 46 | |||||
| 1384 | 1380 | 1392 | ν-COO-, ν-Cα-H | 49 | ||||
| 1411 | 1411 | 1411 | 1408 | Biotin/Streptavidin | ||||
| 1448 | 1453 | 1453 | 1448 | σ-CH2, CH3 | 57 | |||
| 1461 | ν-CH2 ring | 46 | ||||||
| 1482 | 1476 | 1473 | 1486 | 1485 | Streptavidin | |||
| 1492 | 1496 | 1493 | Biotin | 60 | ||||
| 1504 | 1509 | 1504 | 1502 | NA | ||||
| 1547 | 1560 | 1560 | 1561 | 1562 | Trp(W3) | 57 | ||
| 1586 | 1580 | 1582 | ν-C-N | 46 | ||||
| 1591 | 1591 | 1592 | Trp(W2) | 57 | ||||
| 1639 | 1639 | ν−C=O | 46 | |||||
SA - streptavidin;
SA-B - streptavidin-biotin complex;
b-GNP - biotinylated GNP;
sa-GNP - streptavidin-GNP
Investigations using different size biotinylated-GNPs yield interesting results. TERS image of 50 nm biotinylated-GNPs on streptavidin functionalized microscope slide and the corresponding TERS spectra are shown in Figure S1. TERS spectra of the 50 nm biotinylated-GNP probe on streptavidin slide show similar spectral patterns as the 80 nm probe, especially at 473, 661, 1050, 1133, 1272, 1580 cm−1, (See Table 1 and Figure S2) bands indicating the presence of biotin and various amino acid residues in streptavidin. Though the absolute intensities of these Raman bands vary, their relative intensity patterns resemble each other. Another spectral characteristic associated with the 50 nm biotinylated-GNP probe is its diminished Raman intensity compared with the 80nm biotinylated-GNPs. Attempts were made to obtain TERS spectra from 20 nm biotinylated-GNPs; however, we have not successfully detected enhanced Raman scattering from individual 20nm biotinylated-GNPs. The challenges are two-fold. First, for 20nm biotinylated-GNPs absorption, instead of scattering, strongly contributes to its optical response,45 making it very difficult to identify single biotinylated-GNP probes under the dark field microscope. Second, SERS is known to be a short-range effect, enhancing modes in a short distance related to the nanoparticle diameter,1 therefore the field enhanced region of 20 nm probes might be too small to reach the bound protein.
The prominence of streptavidin bands in Figure 1 is not expected. The experimental configuration consists of biotin attached to the probe and a bare gold particle TERS tip. Thus biotin should be detected in the gap. One possibility for the observation of streptavidin bands in the above experiments is streptavidin migration into the gap between the biotinylated-GNP and TERS tip. To further understand the molecular origins of the TERS spectra, the experiment was performed with the inverse probe-substrate configuration; a biotinylated surface and streptavidin-GNP probe. Shown in Figure 2A, the TERS map obtained from the inverse geometry again correlates the presence of the nanoparticle with the enhanced Raman signals. Figure 2B shows the change of the spectra as the TERS tip scanned across the streptavidin-GNP probe on the substrate. The observed Raman scattering from streptavidin-GNPs on biotin surface is different from the biotinylated-GNPs on streptavidin functionalized slide (Figure 1). Specifically, many Raman peaks observed with the biotinylated-GNP/streptavidin-surface geometry are not visible in Figure 2. However, nearly all of the peaks observed here can be correlated to the peaks observed in SERS measurements of clustered streptavidin-GNPs, as discussed below.
Figure 2.
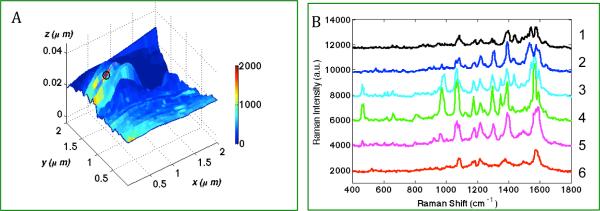
(A) TERS map with simultaneously obtained topographic features and Raman characteristics from 50nm streptavidin-GNP probe coated on the biotin functionalized microscope slide. Scan size: 2×2 um, TERS step-size = 55nm. Color-bar represents the intensity difference at a streptavidin marker band (965cm−1); (B). Plot of the representative TERS spectra extracted from 6 consecutive pixels along the line y=1.25 μm, (1) x=0.53 μm, (2) x=0.47 μm, (3) x=0.42 μm, (4) x=0.36 μm, (5) x=0.31 μm, (6) x=0.25 μm.
To identify distinctive Raman bands for biotin and streptavidin, SERS measurements were obtained from: 1) aggregated biotinylated-GNPs, 2) aggregated streptavidin-GNPs, and 3) streptavidin/biotinylated-GNP complex formed by titrating biotinylated-GNPs with streptavidin.
In Figure 3, biotin SERS spectra were obtained from clusters of biotinylated nanoparticles of different, specific sizes. As illustrated, the size of the nanoparticle for the most part influences only the intensity of the biotin Raman peaks. Peaks observed from different modes of biotin are highly reproducible. Peaks are observed at 1082, 1242, 1376, 1448, 1586, 1639 cm−1 that correspond to ν-C-C, ureido ring stretching, ω-CH2, ν-CH2 ring, ν-C-N, ν-C=O, respectively, and agree with previous reports.46 In agreement with our nanoparticle TERS experiments, 50 and 80 nm biotinylated-GNPs seem to generate significant SERS enhancement, while the 20 nm biotinylated-GNPs evince much lower to negligible SERS signal. Indeed, it is difficult to distinguish prominent SERS peaks when clusters of 20 nm biotinylated-GNPs are used.
Figure 3.
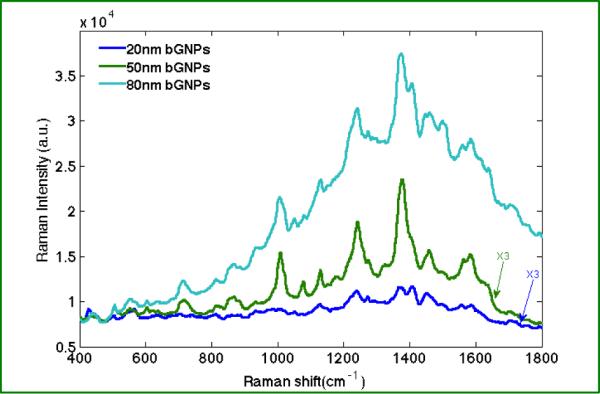
SERS spectra obtained from colloidal clusters comprised of 20, 50, and 80 nm biotinylated-GNPs. Each spectrum is the averaged of 6–10 spectra retrieved from different nanoparticle clusters. (5s for each acquisition).
Figure 4 shows the spectrometric titration of biotinylated-GNPs with free streptavidin. 10 μL aliquots of 5g/L streptavidin solution was added sequentially into 250 μL of 0.05% biotinylated-GNPs (d=80nm) solution and the extinction spectrum was measured after each addition and sufficient mixing. The biotin-streptavidin complex results from high affinity binding of streptavidin to the biotinylated-GNPs. The intensity of the extinction spectra shows a monotonic decrease as the concentration of the conjugated nanoparticles was diluted by the addition of streptavidin solution. A small shift in wavelength from 540nm to 546nm was observed upon the initial addition of streptavidin, indicating the adsorption of streptavidin onto the surface of individual biotinylated-GNPs.47 Since streptavidin has four binding sites, multiple biotinylated-GNPs can be bound, as evidenced by the emergence of a shoulder peak at 583nm right after adding streptavidin to the biotinylated-GNPs solution. Coupling between nanoparticles in close proximity results in a shift in the plasmon resonance observed in the extinction spectrum. The 40 nm redshift of the wavelength results from this plasmonic coupling and agrees with previous reports.48
Figure 4.
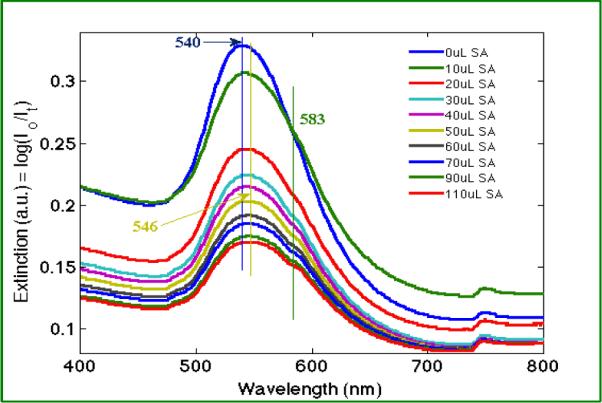
The UV-visible extinction spectra measured from serial addition of streptavidin (SA) solution to the biotinylated-GNP solution show a peak shift associated with nanoparticle aggregation. The vertical lines indicate the peak positions in the extinction spectra.
To distinguish the enhanced Raman bands of biotin and streptavidin, SERS measurements were obtained from streptavidin and streptavidin-biotin complex. Figure 5 presents Raman bands of streptavidin obtained from SERS on 50nm streptavidin-GNPs clusters, streptavidin bound to biotinylated-GNPs as well as normal Raman of streptavidin as a powder. Detailed peak assignments are tabulated in the Table 1. Differences are observed between Raman spectrum of streptavidin powder and SERS spectrum of streptavidin-GNPs, including frequency shifts of certain peaks, disappearance of some and emergence of other peaks. These differences could be attributed to the change of symmetry of streptavidin upon interacting with GNPs. SERS is known to preferentially enhance the Raman modes closest to the nanostructure.49 Additionally, SERS is reported to relax selection rules and make infrared active vibrations observable in Raman measurements.50 Of note, the amide I vibration was not observed in the SERS or TERS data in agreement with a recent report.51
Figure 5.
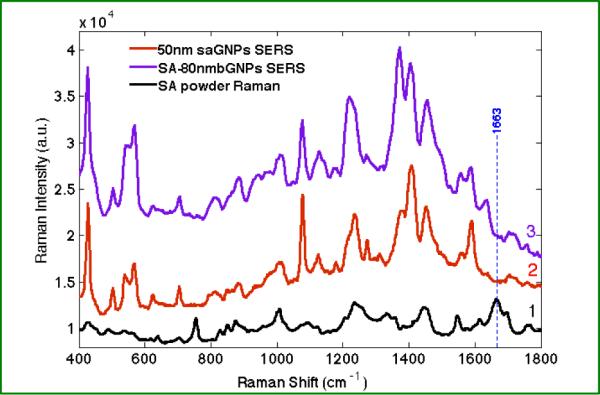
(1) Normal Raman spectrum of streptavidin (SA) powder, averaged from 6 spectra (30s for each acquisition, 10 accumulations); (2) and (3) show the averaged spectrum of 5–10 SERS spectra of streptavidin-GNPs (saGNP, 50 nm) and streptavidin bound biotinylated-GNPs (80 nm, SA-80nmbGNPs) complex, respectively (5s acquisition). The dotted blue line denotes the amide I peak of streptavidin powder.
In Figure 5, the normal Raman spectrum of streptavidin is dominated by residues of aromatic amino acids such as tryptophan, tyrosine and phenylalanine due to inherently larger Raman cross sections. In the SERS spectrum of streptavidin, while aromatic modes still dominate, other modes are observed, such as 1077, 1411, 1591cm−1, attributed to Glu/Thr, ν-COO-, Trp (W2) respectively. These residues are likely enhanced because they are closer to the nanoparticle surface. The SERS spectra obtained from streptavidin-GNPs and from the titration of biotinylated-GNPs with streptavidin are nearly identical. Differences are observed, such as the prominent peaks at 1048, 1372, 1639 cm−1, which are attributed to biotin moiety in the titration product and are evident in the biotinylated-GNPs SERS spectrum. The dominance of streptavidin peaks in these SERS spectra indicates a much larger Raman cross section for streptavidin compared with biotin.
The enhancement mechanism of TERS, in most respects, is the same as that of SERS. More specifically in these experiments, the TERS tip and the single GNP probe on the slide approach during scanning and can be effectively considered as an interacting nanoparticle dimer pair. This leads to the possibility of interpreting the complex TERS spectra of biomolecules by comparing with their SERS counterparts.52 To elucidate the observed TERS spectra, we compared these TERS spectra with those observed in the SERS spectra of biotinylated-GNPs, streptavidin-GNPs and the streptavidin/biotinylated-GNP complex in Figure 6.
Figure 6.
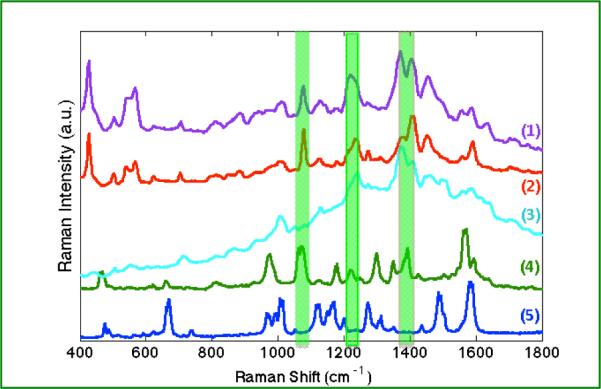
A comparison of SERS and TERS spectra is shown. SERS spectra obtained from (1) streptavidin/biotinylated-GNPs complex (purple), (2) streptavidin-GNPs (red), (3) biotinylated-GNPs (lt. blue), are compared with TERS spectra from (4) a single streptavidin-GNP on biotinylated surface (green), (5) and a single biotinylated-GNP on streptavidin derivatized surface (blue). Peaks observed at 1072, 1218, 1392cm−1 (green highlights) are indicative of streptavidin in the gap region.
In Figure 6, differences are clearly observed when comparing the TERS and SERS results. One rationale for these differences is a more controlled geometry is achieved in the TERS experiment. In the TERS measurement a single nanoparticle probe interacts with the TERS tip to form an enhancing nanoparticle dimer. The polarization dependence of the induced coupled-plasmon is strongly directional and thus, plasmonic interactions correspondingly leads to a reduced dataset.37 Fewer peaks and less background are observed in the TERS measurements. While SERS has molecules in multiple configurations around the nanoparticle clusters, TERS on isolated particles enhances a limited subset of vibrational modes.
It is worth noting here that not all SERS modes are observed in single particle TERS. For instance, as listed in Table 1, SERS peaks at 503, 540, 624, 706, 887, 1407, 1639 cm−1 are not observed in our TERS spectra. Despite reports of long-range enhancements in nanocylinders and OWL nanodisks,53 SERS is considered a short-range effect. The enhancement has been experimentally shown to vanish rapidly within 2–3 nm for various 2-D nanostructures according to the relation:
| Eq. 1 |
where a is the mean radius of the nanoparticle and r the distance from the molecule to surface.54 Moskovits and coworkers corroborated this dependence with the observation that only the amino acid residues in the very immediate vicinity of the nanoparticle surface are preferentially enhanced.49 Therefore it is expected that only amino acid residues near the nanoparticle probe experience the enhanced electric field and subsequently generate the observed TERS signals.
In the case of streptavidin-biotin binding, TERS signals, as attributed in Table 1, arises from residues of Trp, Tyr, Phe, etc., which comprise the aromatic girdle at the binding pocket.55 The prominence of these modes in the observed spectrum argues for the binding pocket being in the immediate vicinity of the probe.
Differences in the TERS results with biotinylated-GNPs and streptavidin-GNPs suggests the TERS measurement is sensitive to these changes, and can distinguish biotin bound to a streptavidin surface from streptavidin migrating to the gap region. Specifically, peaks are observed in the spectra between 1000–1600 cm−1 of the streptavidin-GNP TERS. These peaks are associated with carbon/hetero-atom bonds and amino acid residues in streptavidin.40 An examination of the streptavidin structure suggests that these additional peaks arise from amino acids on the periphery of streptavidin. Using biotinylated-GNPs appears to selectively enhance the aromatic residues associated with the binding pocket, while the streptavidin-GNPs TERS spectrum includes peak contributions from aromatic as well as non-aromatic amino acids. This indicates that the protein moieties interacting with the enhanced electric field are different.
In both the SERS and TERS experiments, biotin peaks are rarely seen in the presence of streptavidin, likely due to biotin's small cross section in comparison with the aromatic amino acid residues of streptavidin. Biotin peaks at 1051, 1304, 1493 cm−1 are present in the TERS spectra, particularly the biotinylated-GNP/streptavidin experiments where biotin is in the geometric junction of the probe-tip dimer. This suggests that only when biotin is in the gap that it experiences sufficient enhancement to be observed.
Our results indicate that the size of the nanoparticle probe influences the extent of the enhanced electric field, consistent with equation 1, and not the chemical origins of the TERS signals. In comparison, a change of molecular binding geometry excites different modes, leading to the different spectra. The TERS responses observed from 50nm biotinylated-GNP/streptavidin-surface are less intense than the 80nm one, but most spectral characteristics are retained. The difference in intensity may arise from either a difference in the number of molecules probed or a change in the effective enhancement. In the case of the streptavidin-GNP/biotin-surface configuration, different peaks are observed in the TERS spectrum. Further, the TERS spectrum of streptavidin-GNP/ biotin-surface configuration shows a higher degree of correspondence with the streptavidin SERS results.
The differences in the TERS spectrum, shown in Figure 6, with streptavidin known to be in the gap (viz. streptavidin-GNPs/biotin functionalized surface configuration) and the case where it is unclear (viz. biotinylated-GNPs/streptavidin functionalized surface) suggest enhancements outside of gap junctions. Discrete dipole calculations have shown high electric fields outside the gap along the direction of polarization.40 Recent experiments also suggest that significant enhancements are achieved outside of gap junctions.39, 56 These experiments and calculations suggest that streptavidin peaks present with the biotinylated-GNPs arise from the streptavidin on the glass surface. Thus, the use of targeting nanoparticles to receptors may provide a route to gap-mode like enhancements without the restriction of a metallic surface.
CONCLUSIONS
Our results show that comparisons of SERS from clustered GNPs with single nanoparticle TERS provide evidence for detection of streptavidin peaks. Differences observed when the streptavidin was functionalized to the nanoparticle surface, rather than the glass substrate, suggest that the peaks detected with biotinylated-GNPs arise from streptavidin distal to the gap junction. The size of the nanoparticle probe altered the absolute signal observed, which may indicate either larger enhancements with larger particles or an increased number of molecules sampled. The use of functionalized nanoparticles to controllably enhance Raman scattering from molecules outside of gap junctions provides a way to increase sensitivity in TERS imaging experiments from complex samples such as protein receptors in cellular membranes.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by the National Institutes of Health Award RR024367 to ZDS, and by the University of Notre Dame.
Footnotes
Electronic Supplementary Information (ESI) available: TERS image and representative TERS spectra of 50 nm biotinylated-GNP probe on streptavidin derivatized slide are shown in Figure S1–2. This material is available free of charge via the Internet at http://pubs.rsc.org
REFERENCES
- 1.Stiles PL, Dieringer JA, Shah NC, Van Duyne RR. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 2.Camden JP, Dieringer JA, Zhao J, Van Duyne RP. Accounts Chem. Res. 2008;41:1653–1661. doi: 10.1021/ar800041s. [DOI] [PubMed] [Google Scholar]
- 3.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari R, Feld MS. Phys Rev Lett. 1997;78:1667–1670. [Google Scholar]
- 4.Nie SM, Emery SR. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 5.Xu HX, Bjerneld EJ, Kall M, Borjesson L. Phys Rev Lett. 1999;83:4357–4360. [Google Scholar]
- 6.McGuinness CD, Macmillan AM, Karolin J, Smith WE, Graham D, Pickup JC, Birch DJS. Analyst. 2007;132:633–634. doi: 10.1039/b706409h. [DOI] [PubMed] [Google Scholar]
- 7.Doering WE, Nie SM. Journal of Physical Chemistry B. 2002;106:311–317. [Google Scholar]
- 8.Dieringer JA, Lettan RB, Scheidt KA, Van Duyne RP. Journal of the American Chemical Society. 2007;129:16249–16256. doi: 10.1021/ja077243c. [DOI] [PubMed] [Google Scholar]
- 9.Fang Y, Seong NH, Dlott DD. Science. 2008;321:388–392. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 10.Michaels AM, Jiang J, Brus L. J. Phys. Chem. B. 2000;104:11965–11971. [Google Scholar]
- 11.Alexander KD, Skinner K, Zhang SP, Wei H, Lopez R. Nano Letters. 2010;10:4488–4493. doi: 10.1021/nl1023172. [DOI] [PubMed] [Google Scholar]
- 12.Willets KA, Van Duyne RP. Annual Review of Physical Chemistry. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 13.Anderson MS. Appl Phys Lett. 2000;76:3130–3132. [Google Scholar]
- 14.Hayazawa N, Inouye Y, Sekkat Z, Kawata S. Opt Commun. 2000;183:333–336. [Google Scholar]
- 15.Pettinger B, Picardi G, Schuster R, Ertl G. Electrochemistry. 2000;68:942–949. [Google Scholar]
- 16.Stockle RM, Suh YD, Deckert V, Zenobi R. Chem Phys Lett. 2000;318:131–136. [Google Scholar]
- 17.Anema JR, Li JF, Yang ZL, Ren B, Tian ZQ. Annu Rev Anal Chem (Palo Alto Calif) 2011;4:129–150. doi: 10.1146/annurev.anchem.111808.073632. [DOI] [PubMed] [Google Scholar]
- 18.Li JF, Ding SY, Yang ZL, Bai ML, Anema JR, Wang X, Wang A, Wu DY, Ren B, Hou SM, Wandlowski T, Tian ZQ. Journal of the American Chemical Society. 2011;133:15922–15925. doi: 10.1021/ja2074533. [DOI] [PubMed] [Google Scholar]
- 19.Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS, Fan FR, Zhang W, Zhou ZY, Wu de Y, Ren B, Wang ZL, Tian ZQ. Nature. 2010;464:392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 20.Verma P, Inouye Y, Kawata S. Top Appl Phys. 2006;103:241–260. [Google Scholar]
- 21.Hartschuh A, Sanchez EJ, Xie XS, Novotny L. Phys Rev Lett. 2003;90:095503 1–4. doi: 10.1103/PhysRevLett.90.095503. [DOI] [PubMed] [Google Scholar]
- 22.Domke KF, Zhang D, Pettinger B. Journal of the American Chemical Society. 2006;128:14721–14727. doi: 10.1021/ja065820b. [DOI] [PubMed] [Google Scholar]
- 23.Bharadwaj P, Beams R, Novotny L. Chemical Science. 2011:136–140. [Google Scholar]
- 24.Lee N, Hartschuh RD, Mehtani D, Kisliuk A, Maguire JF, Green M, Foster MD, Sokolov AP. J. Raman Spectrosc. 2007;38:789–796. [Google Scholar]
- 25.Hayazawa N, Motohashi M, Saito Y, Ishitobi H, Ono A, Ichimura T, Verma P, Kawata S. J. Raman Spectrosc. 2007;38:684–696. [Google Scholar]
- 26.Mehtani D, Lee N, Hartschuh RD, Kisliuk A, Foster MD, Sokolov AP, Maguire JF. J. Raman Spectrosc. 2005;36:1068–1075. [Google Scholar]
- 27.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev IK. Journal of the American Chemical Society. 2012;134:13323–13329. doi: 10.1021/ja303263y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood BR, Bailo E, Khiavi MA, Tilley L, Deed S, Deckert-Gaudig T, McNaughton D, Deckert V. Nano Letters. 2011;11:1868–1873. doi: 10.1021/nl103004n. [DOI] [PubMed] [Google Scholar]
- 29.Cialla D, Deckert-Gaudig T, Budich C, Laue M, Moller R, Naumann D, Deckert V, Popp J. J. Raman Spectrosc. 2009;40:240–243. [Google Scholar]
- 30.Pettinger B, Domke KF, Zhang D, Picardi G, Schuster R. Surf. Sci. 2009;603:1335–1341. [Google Scholar]
- 31.Pettinger B, Domke KF, Zhang D, Schuster R, Ertl G. Phys Rev B. 2007;76:113409 1–4. [Google Scholar]
- 32.Deckert-Gaudig T, Bailo E, Deckert V. Phys. Chem. Chem. Phys. 2009;11:7360–7362. doi: 10.1039/b904735b. [DOI] [PubMed] [Google Scholar]
- 33.Steidtner J, Pettinger B. Phys Rev Lett. 2008;100:236101 1–4. doi: 10.1103/PhysRevLett.100.236101. [DOI] [PubMed] [Google Scholar]
- 34.Bailo E, Deckert V. Angewandte Chemie International Edition. 2008;47:1658–1661. doi: 10.1002/anie.200704054. [DOI] [PubMed] [Google Scholar]
- 35.Carrier SL, Kownacki CM, Schultz ZD. Chemical Communications. 2011;47:2065–2067. doi: 10.1039/c0cc05059h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olk P, Renger J, Wenzel MT, Eng LM. Nano Letters. 2008;8:1174–1178. doi: 10.1021/nl080044m. [DOI] [PubMed] [Google Scholar]
- 37.Alexander KD, Schultz ZD. Analytical Chemistry. 2012;84:7408–7414. doi: 10.1021/ac301739k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titus EJ, Weber ML, Stranahan SM, Willets KA. Nano Lett. 2012;12:5103–5110. doi: 10.1021/nl3017779. [DOI] [PubMed] [Google Scholar]
- 39.Willets KA, Stranahan SM, Weber ML. J. Phys. Chem. Lett. 2012;3:1286–1294. doi: 10.1021/jz300110x. [DOI] [PubMed] [Google Scholar]
- 40.Hao E, Schatz GC. Journal of Chemical Physics. 2004;120:357–366. doi: 10.1063/1.1629280. [DOI] [PubMed] [Google Scholar]
- 41.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 42.Schultz ZD, Stranick SJ, Levin IW. Analytical Chemistry. 2009;81:9657–9663. doi: 10.1021/ac901789w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz ZD, Stranick SJ, Levin IW. Appl. Spectrosc. 2008;62:1173–1179. doi: 10.1366/000370208786401635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asiala SM, Schultz ZD. Analyst. 2011;136:4472–4479. doi: 10.1039/c1an15432j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohren CF, Huffman DR. Absorption and Scattering of Light by Small Particles. John Wiley and Sons; 1983. [Google Scholar]
- 46.Galarreta BC, Norton PR, Lagugne-Labarthet F. Langmuir. 2011;27:1494–1498. doi: 10.1021/la1047497. [DOI] [PubMed] [Google Scholar]
- 47.Barbillon G, Bijeon JL, Bouillard JS, Plain J, Lamy De la Chapelle M, Adam PM, Royer P. Journal of microscopy. 2008;229:270–274. doi: 10.1111/j.1365-2818.2008.01898.x. [DOI] [PubMed] [Google Scholar]
- 48.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 49.Pavel I, McCarney E, Elkhaled A, Morrill A, Plaxco K, Moskovits M. The Journal of Physical Chemistry C. 2008;112:4880–4883. doi: 10.1021/jp710261y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campion A, Kambhampati P. Chem. Soc. Rev. 1998;27:241–250. [Google Scholar]
- 51.Blum C, Schmid T, Opilik L, Metanis N, Weidmann S, Zenobi R. The Journal of Physical Chemistry C. 2012;116:23061–23066. [Google Scholar]
- 52.Blum C, Schmid T, Opilik L, Weidmann S, Fagerer SR, Zenobi R. J. Raman Spectrosc. 2012:1895–1904. [Google Scholar]
- 53.Wei W, Li SZ, Millstone JE, Banholzer MJ, Chen XD, Xu XY, Schatz GC, Mirkin CA. Angew Chem Int Edit. 2009;48:4210–4212. doi: 10.1002/anie.200806116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dieringer JA, McFarland AD, Shah NC, Stuart DA, Whitney AV, Yonzon CR, Young MA, Zhang XY, Van Duyne RP. Faraday Discuss. 2006;132:9–26. doi: 10.1039/b513431p. [DOI] [PubMed] [Google Scholar]
- 55.Freitag S, LeTrong I, Klumb L, Stayton PS, Stenkamp RE. Protein Sci. 1997;6:1157–1166. doi: 10.1002/pro.5560060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wustholz KL, Henry A-I, McMahon JM, Freeman RG, Valley N, Piotti ME, Natan MJ, Schatz GC, Duyne RPV. Journal of the American Chemical Society. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- 57.Fagnano C, Torreggiani A, Fini G. Biospectroscopy. 1996;2:225–232. [Google Scholar]
- 58.Li PW, Zhang J, Zhang L, Mo YJ. Vib Spectrosc. 2009;49:2–6. [Google Scholar]
- 59.Liu FF, Gu HM, Yuan XJ, Lin Y, Dong X. Journal of Physics: Conference Series. 2011;277:012–025. [Google Scholar]
- 60.Emami M, Teimouri A, Chermahini AN. Spectrochim Acta. 2008;71:1516–1524. doi: 10.1016/j.saa.2008.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


