Abstract
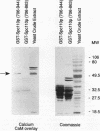
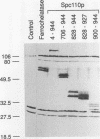
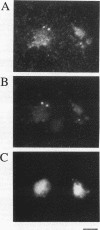
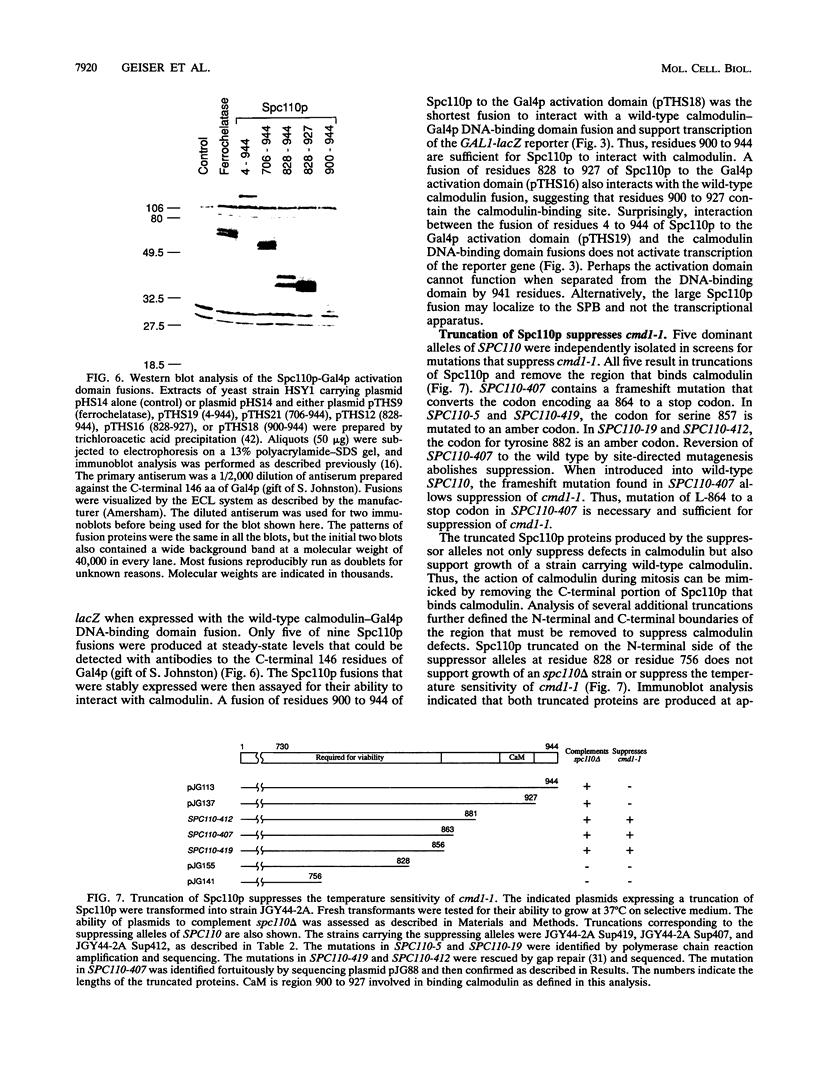
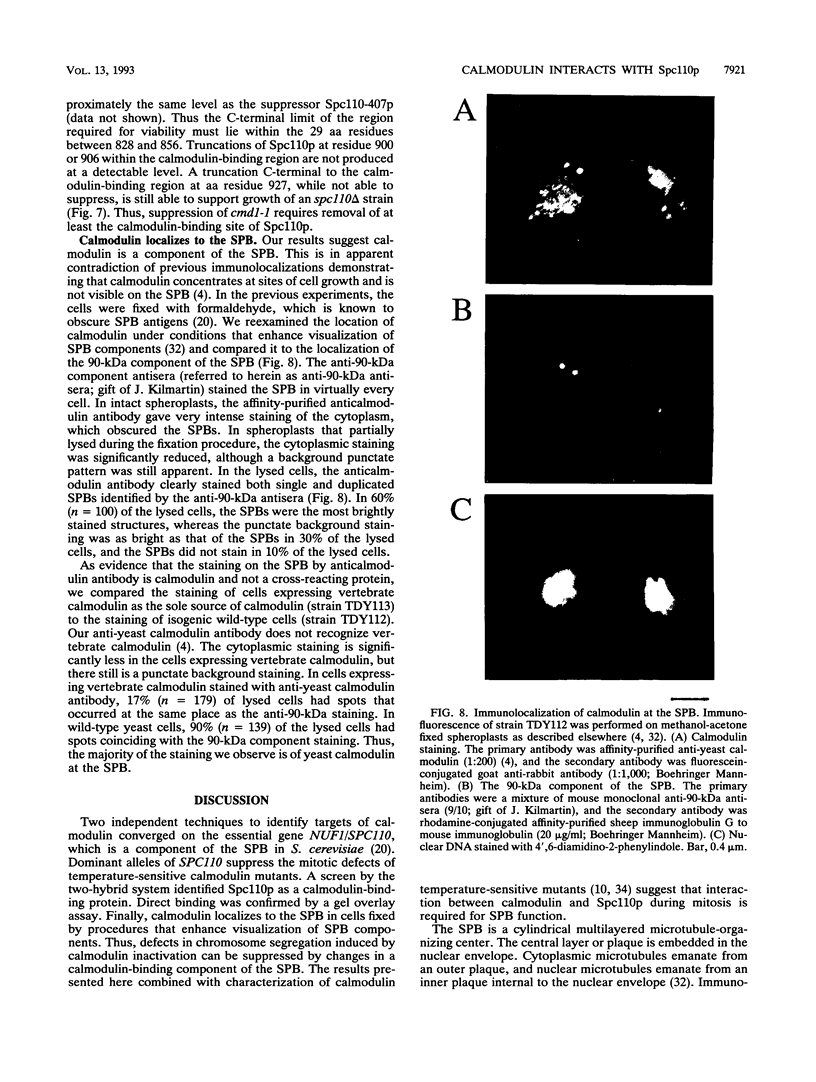
Two independent methods identified the spindle pole body component Nuf1p/Spc110p as the essential mitotic target of calmodulin. Extragenic suppressors of cmd1-1 were isolated and found to define three loci, XCM1, XCM2, and XCM3 (extragenic suppressor of cmd1-1). The gene encoding a dominant suppressor allele of XCM1 was cloned. On the basis of DNA sequence analysis, genetic cosegregation, and mutational analysis, XCM1 was identified as NUF1/SPC110. Independently, a C-terminal portion of Nuf1p/Spc110p, amino acid residues 828 to 944, was isolated as a calmodulin-binding protein by the two-hybrid system. As assayed by the two-hybrid system, Nuf1p/Spc110p interacts with wild-type calmodulin and triple-mutant calmodulins defective in binding Ca2+ but not with two mutant calmodulins that confer a temperature-sensitive phenotype. Deletion analysis by the two-hybrid system mapped the calmodulin-binding site of Nuf1p/Spc110p to amino acid residues 900 to 927. Direct binding between calmodulin and Nuf1p/Spc110p was demonstrated by a modified gel overlay assay. Furthermore, indirect immunofluorescence with fixation procedures known to aid visualization of spindle pole body components localized calmodulin to the spindle pole body. Sequence analysis of five suppressor alleles of NUF1/SPC110 indicated that suppression of cmd1-1 occurs by C-terminal truncation of Nuf1p/Spc110p at amino acid residues 856, 863, or 881, thereby removing the calmodulin-binding site.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes D. A., Thorner J. Genetic manipulation of Saccharomyces cerevisiae by use of the LYS2 gene. Mol Cell Biol. 1986 Aug;6(8):2828–2838. doi: 10.1128/mcb.6.8.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff S. E., Davis T. N. Calmodulin concentrates at regions of cell growth in Saccharomyces cerevisiae. J Cell Biol. 1992 Aug;118(3):619–629. doi: 10.1083/jcb.118.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff S. E., Edmonds C. G., Davis T. N. Structural analysis of wild-type and mutant yeast calmodulins by limited proteolysis and electrospray ionization mass spectrometry. Protein Sci. 1992 Apr;1(4):504–516. doi: 10.1002/pro.5560010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E. The Ca2+ pump of the plasma membrane. J Biol Chem. 1992 Feb 5;267(4):2115–2118. [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Davis T. N. A temperature-sensitive calmodulin mutant loses viability during mitosis. J Cell Biol. 1992 Aug;118(3):607–617. doi: 10.1083/jcb.118.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Erickson-Viitanen S., DeGrado W. F. Recognition and characterization of calmodulin-binding sequences in peptides and proteins. Methods Enzymol. 1987;139:455–478. doi: 10.1016/0076-6879(87)39106-2. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fishel B. R., Sperry A. O., Garrard W. T. Yeast calmodulin and a conserved nuclear protein participate in the in vivo binding of a matrix association region. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5623–5627. doi: 10.1073/pnas.90.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J. R., van Tuinen D., Brockerhoff S. E., Neff M. M., Davis T. N. Can calmodulin function without binding calcium? Cell. 1991 Jun 14;65(6):949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Schulman H. Neuronal Ca2+/calmodulin-dependent protein kinases. Annu Rev Biochem. 1992;61:559–601. doi: 10.1146/annurev.bi.61.070192.003015. [DOI] [PubMed] [Google Scholar]
- Hiraga K., Suzuki K., Tsuchiya E., Miyakawa T. Identification and characterization of nuclear calmodulin-binding proteins of Saccharomyces cerevisiae. Biochim Biophys Acta. 1993 May 8;1177(1):25–30. doi: 10.1016/0167-4889(93)90152-f. [DOI] [PubMed] [Google Scholar]
- Knighton D. R., Pearson R. B., Sowadski J. M., Means A. R., Ten Eyck L. F., Taylor S. S., Kemp B. E. Structural basis of the intrasteric regulation of myosin light chain kinases. Science. 1992 Oct 2;258(5079):130–135. doi: 10.1126/science.1439761. [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Labbe-Bois R. The ferrochelatase from Saccharomyces cerevisiae. Sequence, disruption, and expression of its structural gene HEM15. J Biol Chem. 1990 May 5;265(13):7278–7283. [PubMed] [Google Scholar]
- Mirzayan C., Copeland C. S., Snyder M. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992 Mar;116(6):1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. D., Means A. R. Calmodulin is required for cell-cycle progression during G1 and mitosis. EMBO J. 1989 Jan;8(1):73–82. doi: 10.1002/j.1460-2075.1989.tb03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. D., Means R. L., Lu K. P., May G. S., Means A. R. Characterization and expression of the unique calmodulin gene of Aspergillus nidulans. J Biol Chem. 1990 Aug 15;265(23):13767–13775. [PubMed] [Google Scholar]
- Rose M. D., Broach J. R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Kilmartin J. V. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990 Nov;111(5 Pt 1):1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. H., Hirata A., Ohya Y., Anraku Y. Mutations in yeast calmodulin cause defects in spindle pole body functions and nuclear integrity. J Cell Biol. 1992 Dec;119(6):1625–1639. doi: 10.1083/jcb.119.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet S. C., Rogers C. M., Welsh M. J. Calmodulin stabilization of kinetochore microtubule structure to the effect of nocodazole. J Cell Biol. 1988 Dec;107(6 Pt 1):2243–2251. doi: 10.1083/jcb.107.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet S. C., Welsh M. J. Calmodulin colocalization with cold-stable and nocodazole-stable microtubules in living PtK1 cells. Eur J Cell Biol. 1988 Oct;47(1):88–93. [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. J., Dedman J. R., Brinkley B. R., Means A. R. Calcium-dependent regulator protein: localization in mitotic apparatus of eukaryotic cells. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1867–1871. doi: 10.1073/pnas.75.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. J., Dedman J. R., Brinkley B. R., Means A. R. Tubulin and calmodulin. Effects of microtubule and microfilament inhibitors on localization in the mitotic apparatus. J Cell Biol. 1979 Jun;81(3):624–634. doi: 10.1083/jcb.81.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. P., Bruns M., Hartley B. S. Extraction and rapid inactivation of proteins from Saccharomyces cerevisiae by trichloroacetic acid precipitation. Yeast. 1989 Jan-Feb;5(1):51–53. doi: 10.1002/yea.320050107. [DOI] [PubMed] [Google Scholar]
- Zhu G., Muller E. G., Amacher S. L., Northrop J. L., Davis T. N. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol Cell Biol. 1993 Mar;13(3):1779–1787. doi: 10.1128/mcb.13.3.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]