Abstract
Eukaryotic cell function depends on the physical separation of nucleoplasmic and cytoplasmic components by the nuclear envelope (NE). Molecular communication between the two compartments involves active, signal-mediated trafficking, a role that is exclusively performed by nuclear pore complexes (NPCs). The individual NPC components and the mechanisms that are involved in nuclear trafficking are well documented and have become textbook knowledge. However, in addition to their roles as nuclear gatekeepers, NPC components, nucleoporins, have been shown to play critical roles in chromatin organization and gene regulation. These findings have sparked new enthusiasm to study the roles of this multi-protein complex in nuclear organization and explore novel functions that in some cases appear to go beyond a role in transport. Here, we discuss our current view of NPC biogenesis, which is tightly linked to proper cell cycle progression and cell differentiation. In addition we will summarize new data suggesting that NPCs represent dynamic hubs for the integration of gene regulation and nuclear transport processes.
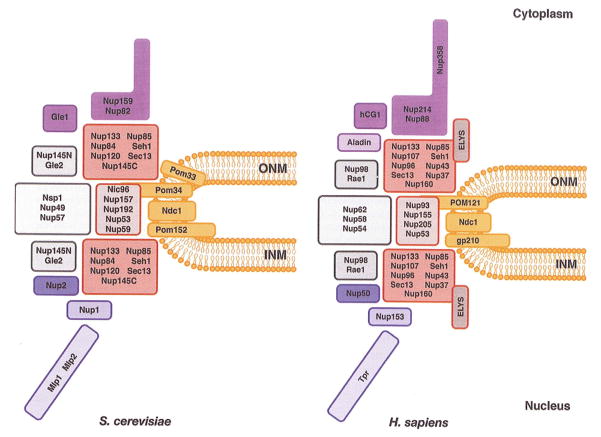
The nuclear envelope (NE) is perforated by nuclear pore complexes (NPCs), which are large multiprotein channels mediating macromolecular import and export by an active, signal-dependent process (Terry et al. 2007). Unlike other cellular transport channels, NPCs span a double lipid bilayer constituted by the outer (ONM) and inner nuclear membranes (INM) that are fused at sites of NPC insertion. The highly curved pore membrane is energetically unstable and thus nuclear pores play an important structural role in maintaining the integrity of the transport channels. Each nuclear pore has a diameter of ~ 50 nm, exhibits a total molecular mass of ~ 60 MDa and is thought to contain more than 500 polypeptides (Beck et al. 2004; Alber et al. 2007). This large number of NPC components is the result of the assembly of multiple copies of ~ 30 different nucleoporins (Nups), which give rise to an eight-fold symmetrical protein complex (Strambio-De-Castillia et al. 2010). The NPC is organized in a protein scaffold, which is anchored in the NE by at least three transmembrane Nups. Eight filaments extend from this core structure into the cytoplasm and the nucleoplasm, the latter being frequently referred to as the nuclear basket (Figure 1). In addition, the central core is coated by natively unfolded Nups containing up to 50 repeat motifs that are rich in phenylalanine and glycine residues (FG-repeats) (Alber et al. 2007; Terry and Wente 2009). These largely hydrophobic patches constitute the central channel of the pore and transiently bind to nuclear transport receptors during translocation of cargo molecules across the NPC. At the same time NPCs establish an efficient permeability barrier for macromolecules larger than 30–40 kDa and thus prevent the uncontrolled mixing of nucleoplasmic and cytoplasmic components. Since the role of Nups in nuclear trafficking and the various models of the physical properties of the permeability barrier have been discussed in many excellent reviews (Terry et al. 2007; Lim et al. 2008; Terry and Wente 2009; Strambio-De-Castillia et al. 2010; Walde and Kehlenbach 2010; Wozniak et al. 2010), we will focus on the biogenesis of the NPC and discuss its role in chromatin organization and gene regulation.
Figure 1. Composition of the NPC.
Schematic of the yeast (left) and mammalian (right) NPCs. The framed boxes represent individual Nups or subcomplexes. The transmembrane Nups are depicted in orange, the soluble scaffold components in red, the central core Nups in grey, the components of the cytoplasmic filaments in purple and the members of the nuclear basket in blue. Note that the relative position of the components is not meant to represent the structure of the NPC beyond this general organization.
THE BIOGENESIS OF THE NUCLEAR PORE
Considerable progress has been made in deciphering the molecular mechanisms involved in NPC biogenesis. Immunodepletion of Nups from Xenopus egg extracts, siRNA-mediated knockdown in cultured cells or knockout strategies in yeast and animals have led to the identification of individual Nups or subcomplexes involved in the NPC assembly process. The one NPC substructure that was consistently identified across different experimental strategies as being essential for NPC formation is the Nup107/160 subcomplex (Siniossoglou et al. 1996; Boehmer et al. 2003; Harel et al. 2003b; Walther et al. 2003a). The transmembrane Nups POM121 and Ndc1 in vertebrates have also been shown to decrease pore density, in a partially redundant manner (Mansfeld et al. 2006; Stavru et al. 2006a; Stavru et al. 2006b). Accordingly, the yeast transmembrane Nups Pom34, Pom152 and Ndc1 have been demonstrated to be involved in NPC assembly, Ndc1 being essential while Pom34 and Pom152 having partially redundant functions (Madrid et al. 2006; Miao et al. 2006). It thus appears that most core Nups are essential for NPC assembly or maintenance.
Studying the mechanisms of NPC formation at the molecular level is complicated by the fact that in metazoa, nuclear pores assemble at different cell cycle stages. At the end of mitosis, when the NE reforms around segregated chromosomes, NPCs reform from disassembled precursors (Antonin et al. 2008). In addition, new NPCs continuously assemble in interphase into the expanding NE (Maul et al. 1971). This ‘interphase’ NPC assembly, thought to provide a sufficient number of NPC components for daughter cells during the next cell division, occurs in an intact NE. Importantly, organisms with closed mitosis, such as budding and fission yeast, only assemble NPCs into an intact NE (Winey et al. 1997), suggesting the post-mitotic NPC assembly pathway evolved from the ‘interphase’ mechanism with the advent of open mitosis. It is important to note that the assembly of NPCs into an existing NE is not restricted to dividing cells but also occurs during cell differentiation. For instance, millions of NPCs are assembled during oogenesis, and cells undergoing endoreplication continue assembling NPCs during differentiation (Burke and Stewart 2002). Moreover, ‘interphase’ assembly occurs in response to changes in metabolic activities, as in lymphocytes where NPC density nearly doubles upon stimulation with phytohemaagglutinin in the absence of cell division (Maul et al. 1971). Very little is known about potential differences between post-mitotic and interphase NPC assembly, which can be explained at least in part by the wide use of an in vitro NPC assembly assay based on cell-free Xenopus eggs extracts. This experimental system recapitulates the post-mitotic assembly of the NE and NPCs (Dabauvalle and Scheer 1991; Forbes 1992). Moreover, as asynchronous cultured metazoan cells and whole organisms convey both post-mitotic and interphase NPC assembly, most studies carried out in these systems did not distinguish between the two pathways. In the past years, single cell live imaging provided cell-cycle specific information about NPC assembly (Bodoor et al. 1999; Hase and Cordes 2003; D’Angelo et al. 2006; Dultz et al. 2008; Doucet et al. 2010). Moreover, the Xenopus egg extracts system can be adapted to distinguish between post-mitotic NPC assembly (occurring during the early time points of the nuclear assembly reaction) and interphase assembly, which occurs later during the nuclear expansion phase (D’Angelo et al. 2006; Dawson et al. 2009; Doucet et al. 2010). Different experimental strategies using this system can also lead to the formation of pore-free NEs, in which the mechanisms of pore formation in an intact NE can be studied in vitro (Harel et al. 2003b).
Post-mitotic NPC assembly
In metazoa, post-mitotic NPC assembly is concomitant with NE reformation and involves the ordered recruitment of ER membranes and disassembled NPC components to chromatin (Anderson and Hetzer 2008b). Nucleoporins have open access to chromatin and transmembrane Nups can freely diffuse between both sides of the reforming nuclear membranes. Chromatin constitutes a matrix for pore formation and recruitment of Nups to chromatin has been shown to occur independently of membranes. For instance, the Nup107/160 complex is targeted to chromatin via ELYS/Mel-28, which directly binds to DNA through an AT-hook (Rasala et al. 2006; Franz et al. 2007; Rasala et al. 2008). This step is controlled by a crosstalk between the small GTPase Ran and the transport receptor Importin β (Harel et al. 2003a; Walther et al. 2003b; Rotem et al. 2009). Association of Importin β to the Nup107/160 complex prevents its targeting to chromatin, as well as further association with other NPC members. Interaction of RanGTP with Importin β releases the Nup107/160-ELYS complex and promotes its subsequent chromatin binding (Figure 2).
Figure 2. Model for mammalian NPC assembly mechanisms in mitosis and interphase.
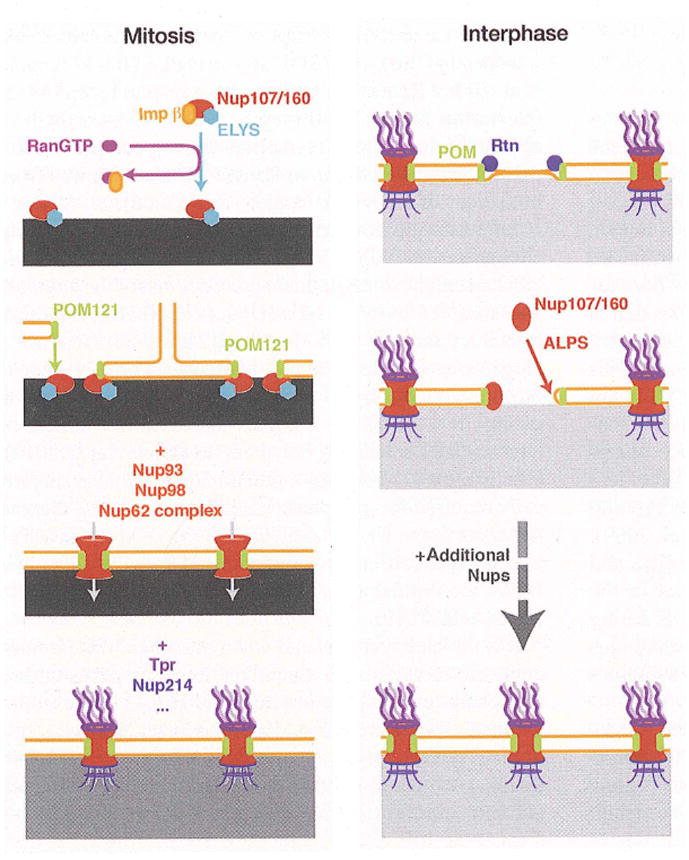
The sequences of events leading to post-mitotic (left) or interphase (right) NPC assembly are represented from top to bottom. The chromatin is depicted in grey, the intensity of the color reflecting its compaction state.
A second step in NPC formation involves the recruitment of transmembrane Nups to chromatin. In vitro, the recruitment of membrane vesicles containing Pom121 and Ndc1, but not gp210, was shown to depend on prior Nup107/160 recruitment (Rasala et al. 2008). Accordingly, an interaction between the cytoplasmic domain of Pom121 and the Nup107/160 complex was characterized (Rasala et al. 2008). It is now clear that membrane vesicles most likely result from fractionation of the ER occurring during preparation of cell-free extracts, and that post-mitotic NE formation actually results from reshaping of the ER around chromatin (Anderson and Hetzer 2007). INM proteins as well as the transmembrane Nups Pom121 and Ndc1 were shown to mediate this process, by driving the spreading of the mitotic ER on segregated chromatin through their capacity to bind DNA in a collaborative manner (Anderson et al. 2009). In the particular case of Pom121 this is likely mediated by its interaction with the chromatin-bound Nup107/160 complex. Indeed, a systematic analysis of the kinetics of Nups recruitment to the reforming NE revealed that Pom121 was targeted to chromatin after the Nup107/160 complex (Dultz et al. 2008). Moreover, the Pom121-Nup107/160 interaction may be important to initiate the connection between soluble members of the NPC and the reforming NE (Antonin et al. 2005). However, it was recently shown in vivo and in vitro that Pom121 is not rate-limiting for NPC assembly at the end of mitosis (Doucet et al. 2010); this observation may account for the previously suggested overlapping functions of Ndc1 and Pom121 in NPC assembly (Stavru et al. 2006a). Notably, the third transmembrane Nup gp210 is recruited later to the chromatin periphery (Bodoor et al. 1999), suggesting it is not involved in the early membrane association of the reforming pore. EM analysis of nuclear assembly reactions in Xenopus egg extracts revealed that membrane-free chromatin is coated with single copies of ELYS-bound Nup107/160 complexes, suggesting the lateral oligomerization of the complex is promoted by the interaction with membranes and/or Pom121 (Rotem et al. 2009).
A third step in NPC formation includes the recruitment of Nup93, Nup98 and the Nup62 complex (Bodoor et al. 1999; Dultz et al. 2008). Nup93 is in complex with other members including Nup53 and Nup155. Interestingly, Nup53 interacts with Ndc1 (Hawryluk-Gara et al. 2005; Mansfeld et al. 2006) and Nup155 was shown to be critical for NE formation in Xenopus extracts and in C. elegans embryos (Franz et al. 2005). This suggests that the recruitment of the Nup93 complex at this stage may be critical to stabilize the interaction between the NPC and the NE. As Nup93 and Nup98 briefly precede Nup58 (a member of the Nup62 complex) on chromatin, they might be involved in recruiting the Nup62 complex (Dultz et al. 2008), which is consistent with previous data showing that Nup98 depletion in mice leads to a defect in Nup62 assembly (Wu et al. 2001). Importantly, the association of these components was shown to coincide with the activation of nuclear import (Dultz et al. 2008). The Nup62 complex and Nup98 contain FG-repeats, which are known to interact with transport receptors (Terry and Wente 2009), and are sufficient to recapitulate selective nuclear transport (Frey and Gorlich 2007; Jovanovic-Talisman et al. 2009). Moreover, the Nup93 complex significantly contributes to the establishment of the size exclusion limit of the NPC (Shulga et al. 2000; Galy et al. 2003).
The last step of post-mitotic NPC assembly is the recruitment of peripheral Nups, including Nup214, Tpr, gp210 and the major pools of Nup50 and Nup153 (Bodoor et al. 1999; Haraguchi et al. 2000; Hase and Cordes 2003; Dultz et al. 2008; Katsani et al. 2008). This is consistent with studies showing that gp210 and Nup214 are dispensable for the assembly of the NPC core (Walther et al. 2001; Galy et al. 2008). While the scaffold core of the NPC has a two-fold symmetry about the NE plane, a number of peripheral Nups are asymmetrically localized (e.g. components of the nuclear basket and cytoplasmic filaments, see Figure 1). Since these components are incorporated after nuclear import has occurred, the observed asymmetry may be controlled
Interphase NPC assembly
In interphase, the topology of the NE, and thus the context of NPC assembly, differs from the situation at the end of mitosis, i.e. the NE is intact and therefore the nucleoplasm and cytoplasm are physically separated. Using Xenopus egg extracts it was shown that in intact nuclei, the Nup107/160 complex was required from the nuclear and cytoplasmic sides of the NE to support new pore assembly (D’Angelo et al. 2006). This study implies that at least a fraction of the Nup107/160 complex was assembled in new pores without interacting with chromatin. This idea is further supported by the finding that ELYS is not rate limiting for interphase NPC assembly. Notably, no homolog of Elys has been identified in yeast, which undergoes closed mitosis, but an ortholog of Elys was discovered in A. nidulans, a fungus undergoing partial NPC disassembly during mitosis (Liu et al. 2009). However, this protein is shorter than vertebrate Elys and lacks an AT-hook. Instead, An-Elys is involved in the maintenance of the An-Nup84 complex in the NE during mitosis, suggesting Elys is not restricted to vertebrates but may have been an early determinant in the evolution of organisms towards open mitosis.
Then how is the Nup107/160 complex targeted to new NPC assembly sites? At least part of the answer can be found in a membrane-curvature sensor domain that is present in Nup133, a member of the Nup107/160 complex (Drin et al. 2007). This so-called ALPS domain is an amphipathic alpha-helixes harbouring a hydrophobic patch and an uncharged polar face. As opposed to classical amphipathic helixes, interacting with lipid bilayers through hydrophobic and electrostatic interactions, interaction of these uncharged helix with membranes requires accessibility to the carbon tails of the lipids as they mainly bind through hydrophobic interactions. As a result, ALPS motifs preferentially associate with lipid bilayers exhibiting a loose lipid packing, which is typically achieved in curved membranes (Antonny 2006). The ALPS motif of Nup133 was shown to be specifically required for Nup107/160 recruitment to the NE and NPC assembly in interphase (Doucet et al. 2010). It is not clear yet if the ALPS targets completely fused sites, or fusion intermediates. In the latter case, it might participate in the completion of the fusion, by stabilizing curved patches in the membranes. However, in vitro data support that this motif does not have affinity towards flat membranes (Drin et al. 2007; Doucet et al. 2010), and is thus most likely not involved in promoting membrane fusion. The ability to directly sense the highly curved sites of ONM and INM fusion (or intermediates) ensures high specificity regarding the sequence of events and spatial restriction of pore assembly. It also provides an elegant mechanism for Nup107/160 targeting from both sides of the NE to new assembly sites. Interestingly, the ALPS domain of Nup133 is dispensable for post-mitotic assembly, suggesting the initial connection between soluble Nups and the nuclear membrane are mediated by different mechanisms.
Once targeted to new sites, the Nup107/160 complex is incorporated into the NE to establish the NPC scaffold structure. A critical step in this process is mediated by the RanGTPase and the transport receptors Importin β and transportin (Lusk et al. 2002; Ryan et al. 2003; D’Angelo et al. 2006). RanGTP interacts with transport receptors sequestering Nup107/160 members and releases them to allow protein-protein interactions between complexes. Importantly, the generation of RanGTP is also required from both sides of the NE (D’Angelo et al. 2006).
Interestingly, while Pom121 is recruited to new pore sites after the Nup107/160 complex during post-mitotic NPC assembly, it is recruited before Nup107 in intact NEs (Doucet et al. 2010). Indeed, pore intermediates harbouring Pom121 but not Nup107 or Nup133 were characterized. Moreover, Pom121 knock-down inhibited Nup107 recruitment to the NE, and mistargeting the Nup107/160 complex increased the number of intermediates containing only Pom121 (Doucet et al. 2010). Mirroring ELYS behaviour, Pom121 is specifically required for interphase NPC assembly, which is illustrated by the fact that knocking down simultaneously ELYS and Pom121 reduces further NPC formation, as compared to single knock-downs (Doucet et al. 2010). Finally, Pom121 contains a functional NLS which is specifically required for interphase assembly, suggesting the transmembrane Nup is delivered to the INM by an active transport process or that interactions of Pom121 with other Nups is regulated by Importin alpha (Doucet et al. 2010; Yavuz et al. 2010).
It is not clear yet how and when the other NPC components are recruited and assembled into new pores during interphase and how the permeability barrier is preserved while new pores are inserted in the intact NE? It is striking that interphase pore insertion is a long process (about 30–60 minutes) compared to post-mitotic assembly, which occurs in a few minutes (D’Angelo et al. 2006). One possibility is that narrow channels are initially formed and gradually expand as more components are assembled into the NPC. It is also possible that fusion intermediates are sufficient to allow the recruitment of the Nup107/160 complex as well as other Nups, and that completion of membrane fusion is ultimately coupled to assembly of FG-rich Nups. More work is needed to clarify these mechanisms.
Fusion of inner and outer nuclear membranes
NPC formation in an intact NE occurs by a de novo process (D’Angelo et al. 2006), which implies NPC formation involves fusion of the INM and ONM. Scanning EM studies have provided some potential intermediates including inward dimpling of the nuclear membranes and fusion of the bilayers (Goldberg et al. 1997). While further investigations are required to test if these NE structures represent INM-ONM fusion intermediates, they highlight the fact that INM and ONM fusion must involve intermediates harbouring a complex topology of positive and negative curvature to allow the close apposition of the membranes required for fusion. Curved lipid bilayers exhibit a differential packing of their two leaflets, which can be induced by a change in lipid composition (e.g. introduction of conical lipids) or by membrane bending proteins (Antonny 2006). Transmembrane Nups have long been suspected to play a critical role in NPC assembly and possibly INM and ONM fusion. In particular, the Nup gp210, which contains a large luminal domain that could in principle span the perinuclear space and bridge the ONM and INM, was suspected to be the key fusogenic component. However, the fact that gp210 is not expressed in many cell types argues against a general role of this Nup in membrane fusion. Instead, recent evidence in human cells suggests Pom121 might be involved in membrane fusion. As mentioned above, depletion of Pom121 by RNAi specifically inhibited interphase pore assembly in cultured mammalian cells, and a Pom121-specific inhibitory antibody prevented ONM and INM fusion in vitro in expanding nuclei (Doucet et al. 2010). Pom121 might thus induce or stabilize membrane fusion. Its luminal domain is only ~ 30 amino acids in length and therefore likely cannot span the perinuclear space of the NE. However, Pom121 might recruit luminal ER proteins to induce membrane curvature. Alternatively, Pom121 might cooperate with reticulons (Rtns), which have also been implicated in NPC assembly (Dawson et al. 2009).
The Rtns, structurally related to Yop1 and metazoan DP1, are thought to trigger or stabilize membrane curvature by inserting a wedge-like hydrophobic hairpin within one leaflet of a lipid bilayer (Sheetz et al. 1976; Oertle and Schwab 2003; De Craene et al. 2006; Voeltz et al. 2006; Shibata et al. 2008). Interestingly, Rtns have been implicated in NPC formation in an intact NE (Dawson et al. 2009), suggesting they might induce the NE bending required for INM and ONM fusion. Indeed, co-depletion of Rtn1 and Yop1 in yeast leads to NPC assembly phenotypes indicative of membrane fusion defects. Moreover, an inhibitory antibody directed against Rtn4 (Voeltz et al. 2006) prevents INM and ONM fusion in Xenopus egg extracts (Dawson et al. 2009). As flat membranes, and in particular the NE, are mostly devoid of Rtns which reside preferentially in the tubular ER (De Craene et al. 2006; Voeltz et al. 2006; Anderson and Hetzer 2008a), the mechanisms by which they may trigger nuclear membrane bending are not clear. An appealing model is that Rtns would adopt a transient ring-like organization in the NE and displace the lipids on the outer leaflet of the ONM, from the site of their insertion towards the centre of the ring. The lipid displacement would induce a combination of positive and negative curvature and result in the ONM getting closer to the INM. As NPCs exhibit an 8-fold radial symmetry, the ring-like organization of Rtns might result from their interaction with transmembrane Nups. Coincidently, a novel transmembrane Nup, Pom33, was recently identified in yeast and shown to interact with Rtn1 (Chadrin et al. 2010). Rtns are likely rapidly replaced by components able to stabilize the induced membrane curvature. Interestingly, multiple soluble Nups have putative membrane interaction domains (specifically vNup133, yNup170, and yNup53) (Marelli et al. 2001; Drin et al. 2007; Patel and Rexach 2008). In particular, the ALPS motif of Nup133 may replace Rtns in the outer leaflet of bent nuclear membranes through its membrane curvature sensing ability and stabilize the fusion sites/intermediates.
Once membrane fusion has occurred, additional interactions are likely required to stabilize the highly curved pore. Strikingly, several components of the yNup84/vNup107–160 complex exhibit structural similarities with proteins of the COPII complex, suggesting they may oligomerize in a similar structure to form a membrane coat and stabilize the pore membrane (Debler et al. 2008; Brohawn and Schwartz 2009; Whittle and Schwartz 2009). In addition in yeast, the transmembrane Nups physically interact with yNup170/yNup157 and yNup53/yNup59 proteins (Flemming et al. 2009; Makio et al. 2009; Onischenko et al. 2009). In addition, genetic interactions between this network of Nups and Rtn1/Yop1 further support the linkage between the transmembrane Nups, Rtns, yNup170/yNup157, and yNup53/yNup59 steps (Dawson et al. 2009). As previously mentioned, vNdc1 also functionally interacts with Nup53, ortholog of yNup53/yNup59, suggesting the functional interactions characterized in yeast may be conserved in vertebrates (Mansfeld et al. 2006). Although a potential role of Ndc1 in membrane fusion is not known, this transmembrane Nup may be crucial for the stabilization of the pore membrane through its interaction network with the soluble core of the NPC.
THE ROLE OF THE NPC IN REGULATION OF GENE EXPRESSION
Eukaryotic gene expression is regulated at multiple levels in the cell nucleus, from chromatin modification and compaction to the synthesis, processing and export of mRNA. The precise execution of transcriptional programs during development and in dividing differentiated cells relies on faithful reproduction of a specific gene expression state through mitosis, and is crucial for maintaining correct cellular identity. Nucleocytoplasmic transport, which occurs exclusively through the NPC, controls the access of chromatin modifying enzymes and transcription factors and the exit of RNA, and as such, is intimately tied to regulation of eukaryotic gene expression. Yet in addition to the more established roles of Nups in the transport process, it is becoming evident that the NPC components have additional functions in gene regulation and in nuclear organization of chromatin (Akhtar and Gasser 2007; Brown and Silver 2007). Recent evidence suggests that Nups are able to physically interact with specific genomic loci, and via such direct interaction, can regulate transcriptional status of its targets and participate in chromatin domain establishment. These uncovered additional functions of Nups suggest that they may play a critical role in setting up or maintaining transcriptional identity of developing and differentiated cells in a chromatin-associated manner. The major unresolved questions arising from these studies are the molecular nature and the cellular purpose of the Nup-gene contacts.
Interestingly, the proposed role of Nups in developmental gene regulation is supported by reports of several NPC components exhibiting tissue-specific expression and tissue-specific disease phenotypes (Capelson and Hetzer 2009). Given the indispensable role of the NPC in cellular function, tissue-specific requirements of Nups can appear unexpected. Nonetheless, both stable and dynamic Nups have been found to play a role in particular aspects of development and to underlie specific human pathologies. Some compelling examples include mouse Nup133, which was recently demonstrated to play a role in embryonic development of the neural lineage, such that Nup133 null neural progenitors failed to efficiently produce terminally differentiated neurons (Lupu et al. 2008), and a mutation in human Nup155, which leads to Atrial Fibrillation (AF), an inherited form of clinical arrhythmia that can lead to sudden cardiac death (Zhang et al. 2008). The Drosophila homologue of Nup155, Nup154, has also been shown to have a tissue-specific role, but here instead affecting gametogenesis in both sexes (Gigliotti et al. 1998; Grimaldi et al. 2007). Furthermore, a mutation in the zebrafish ELYS was described to affect normal development and proliferation of the retina and the intestine (Davuluri et al. 2008; de Jong-Curtain et al. 2008). Mutations in several plant Nups have been reported to cause diverse developmental defects, which often appear comparatively mild and relatively specific (Meier and Brkljacic 2009). One such example is a mutant allele of a nuclear basket Nup Tpr/NUA, which affects the flowering time, seed production and leaf morphology of A. thaliana (Xu et al. 2007).
Perhaps the best-studied examples of a Nup underlying a particular human disorder are the identified translocations of the Nup98 gene, which result in fusions of the N-terminal part of Nup98 to transcriptional regulators and other DNA-binding proteins and have been characterized as mutations leading to several types of leukemia (Nakamura et al. 1996; Saito et al. 2004; Slape and Aplan 2004). For instance, the oncogenic fusion of Nup98 to the transcription factor HOXA9 results in acute myelogenous leukemia and was shown to induce the aberrant expression of HOXA9 target genes, beyond the effects of over-expression of HOXA9 alone, and to block the differentiation of human hematopoietic cells (Ghannam et al. 2004; Takeda et al. 2006). Fusions of Nup98 to PHD finger proteins have been shown to produce very similar effects and to lock target genes into aberrantly active state, resulting in leukemic transformation (Wang et al, 2009). These findings agree with the notion that the normal function of Nup98 may include regulation of gene expression via direct interaction with target genes. Clearly, tissue-specific effects of the NPC can arise from its classic transport-associated or its recently identified chromatin-binding functions, or from the interplay of both. But in order to fully understand the mystery of how tissue-specific gene expression can be influenced by a particular NPC component, the chromatin-associated roles of Nups have to be considered.
Originally, a relationship between nuclear pores and nuclear organization of chromatin has been suggested by high-resolution images of mammalian nuclei that distinctly show the non-random association of de-condensed chromatin with nuclear pores. Such lighter-stained, de-condensed chromatin is thought to predominantly contain active genes or parts of the genome that are more open and permissive to transcription. The observed correlation between NPCs and open chromatin led to a notion, known as the ‘gene gating hypothesis’ (Blobel 1985), which proposed that nuclear pores are capable of specifically interacting with active genes to promote co-regulation of transcription with mRNA export. Several lines of evidence, subsequently obtained in yeast, supported this link of NPC to transcriptional activation. Genome-wide Chromatin Immunoprecipitation (ChIP) analysis in Saccharomyces cerevisiae demonstrated that some nucleoporins, such as Mlp1, Nup2 and Nup60 often occupy regions of highly transcribed genes (Casolari et al. 2004; Casolari et al. 2005). Furthermore, interaction between the NPC component Nup2 and promoters of active genes, termed the “Nup-PI” phenomenon, has been described (Schmid et al. 2006). Additionally, enhancing association of an inducible gene with the NPC boosted its transcript levels (Taddei et al. 2006), and genes reported to associate with Nups were observed to re-localize to the nuclear periphery upon transcriptional activation (Casolari et al. 2004; Taddei et al. 2006). These findings suggested that sites of active transcription are localized to the nuclear pores at the NE and appear to be positively regulated by the association with Nups.
Interestingly, functional involvement of Nups in gene activation has recently been discovered in multi-cellular organisms (Capelson et al. 2010; Kalverda et al. 2010; Vaquerizas et al. 2010), but with an unexpected twist. Analysis of chromatin binding behaviour of Drosophila Nups, achieved by different methods such as immunostaining of polytene chromosomes, ChIP and Dam-ID, revealed the presence of several NPC components at active genes and a functional requirement for their presence in transcription of their binding targets. Reducing levels of Nup98 or Sec13 by RNA interference (RNAi) resulted in decreased levels of transcriptional activity and mRNA levels of its target genes, which included the developmentally induced ecdysone-responsive genes (Capelson et al. 2010; Kalverda et al. 2010). Surprisingly, the Nup-chromatin contacts were commonly found to occur in the nucleoplasm, away from the NE-embedded NPCs. These observations, consistently reported by all 3 studies, suggest that Nups may retain the ability to regulate gene activity even when not associated with the actual nuclear pore. The Nups identified to participate in gene binding and regulation, namely Nup98, Sec13, Nup62 and other FG Nups, Nup50, Nup153 and Tpr, belong to the peripheral and commonly dynamic sub-group of Nups, with the exception of Sec13, which nevertheless has been reported to have an intra-nuclear population (Enninga et al. 2003). As discussed above, certain sub-complexes of the NPC, such as the Nup107–160 complex, form the main structural scaffold of the NPC. The components of such complexes have been found to be predictably stable at the NE-embedded formed NPCs, with low turn-over rates as judged from FRAP experiments with GFP-tagged Nups (Rabut et al. 2004). In contrast, peripheral Nups that are recruited later during pore assembly have been found to be highly dynamic, with high turn-over rates detected in similar FRAP experiments (Rabut et al. 2004). An intriguing possibility is that the dynamic behaviour of these Nups underlies their role in transcriptional activation. Dynamic Nups could come off the nuclear pore to interact with transcribing genes or with mRNA and associated machinery inside the nucleus — that is, not at NE contact sites — a notion supported by the reported transcription-dependent mobility of Nup153 and Nup98 (Griffis et al. 2004). Since the majority of active genes is located in the nuclear interior, this model could overcome a major limitation of the ‘gene gating hypothesis’, which argues that active genes have to be repositioned to the nuclear pores at the NE in order to be regulated by the NPC components.
Furthermore, one of the studies reported an additionally interesting type of Nup-chromatin interactions (Vaquerizas et al.). According to the genome-wide ChIP data, both Nup153 and Megator (Mtor), the Drosophila homologue of Tpr, appear to occupy unusually long stretches of DNA, sometimes hundreds of kilobases (Vaquerizas et al. 2010), instead of discrete binding peaks usually observed for transcriptional regulators and determined for Drosophila Nup98 (Capelson et al.). This distribution is intriguing as it implies a global structural function for Nup153 and Mtor, which may underlie a functional organization of active genomic areas. It is tempting to speculate that such long Mtor/Tpr and Nup153 chromatin-binding stretches, termed Nuclear pore Associated Regions (NARs) may be an extension of the previously described Tpr filaments, which extend from the nucleoplasmic face of the NPC into the nucleoplasm (Zimowska and Paddy 2002; Krull et al. 2004). In this case, these nuclear basket filaments may interact with the genome to assist genomic targeting of imported transcription factors or to fulfil the gene-gating role of coupling mRNA production and export, previously assigned to NE-embedded NPCs. In support of the latter idea, experiments in yeast have suggested a role for the homologues of Tpr, Mlp1/2 in down-regulating transcription of target genes in response to an mRNA export defect (Vinciguerra et al. 2005), implicating Mlp1/2 in co-ordinating the two processes. In flies however, although a potential correlation between Tpr/Nup153-defined NARs and gene activity was reported, it does not appear to be absolute (Vaquerizas et al.). Another recent study of Drosophila dosage compensation showed that Nup153 and Mtor/Tpr co-purify with the Dosage Compensation Complex and are necessary for the two-fold hyper-transcription of the male X (Mendjan et al. 2006), further implicating these two Nups in the establishment of active chromatin. Although the interaction itself may be indicative a physical link between transcriptional or epigenetic activators and Nups of the nuclear basket, which has been previously suggested in yeast ((Rodriguez-Navarro et al. 2004; Casolari et al. 2005; Menon et al. 2005), its functional role remains debated (Grimaud and Becker 2009). It is thus also possible that Tpr/Mtor and Nup153 NARs occupy regions of permissible, but not necessarily on-going transcription, regions of alternative processes, or that they primarily constitute a structural support to chromatin organization, reminiscent of the suggested function for the components of biochemically defined nuclear matrix (Vlcek et al. 2001; Marshall 2002).
These reports were the first to demonstrate a potential functional role for chromatin-associated Nups in animal cells, raising an exciting possibility that Nups execute a previously unexplored function in developmental gene activity. And although multiple studies on chromatin-associated roles of Nups in yeast and flies demonstrated their link to active genes, other reports have identified an association with genomic regions implicated in alternative chromatin states. In the high resolution images of nuclei mentioned above, where heterochromatin looks interspersed by nuclear pores, it seems similarly plausible that the NPC would have a role in setting up this organization by serving as a boundary between condensed and decondensed regions. Boundary elements are generally defined by their ability to prevent communication between active and silent chromatin, which they are thought to do by physically partitioning the chromatin fiber (Gerasimova et al. 1995; Burgess-Beusse et al. 2002). Interestingly, examples of this type of involvement have been reported for the NPC: yeast Nup2, Nup60 and Prp20/RanGEF were identified in a screen for chromatin boundary activities that can prevent the spreading of repressing factors into neighbouring genes (Ishii et al. 2002; Dilworth et al. 2005). Furthermore, the initial characterization of the genome-wide chromatin distribution of yeast Nups revealed that certain NPC components, such as Nup145C (homologue of mammalian Nup96) and Nup84 (homologue of Nup107), both considered stable Nups, did not preferentially localize to active genes, but instead were enriched on “neutral” chromatin (Casolari et al. 2004). Interestingly, the apparent exclusion of heterochromatin from the regions below the NPCs, mentioned above, is disrupted by a knock-down of Tpr in poliovirus-infected mammalian cells, allowing heterochromatin to form all along the NE (Krull et al.). These unexpected findings suggest a potentially important role for Tpr in delimiting heterochromatin boundaries and support the proposed function of Nups in setting up global chromatin organization.
Intriguingly, the only characterization of a genome-wide binding pattern of a stable Nup performed to date in metazoan cells demonstrated an enrichment for repressive heterochromatic histone marks (Brown et al. 2008). ChIP analysis of Nup93 in human HeLa cells showed that its binding sites correlate with regions of high tri-methylation of H3K9, H4K20 and H3K27. And although these results were obtained from a correlation between different cell types and thus should be interpreted with caution, they serve as further potential evidence for the association of nuclear pores with non-active chromatin regions. Moreover, genome-wide mapping of the Drosophila NPC components by DamID, which compared a full-length and an exclusively nucleoplasmic, NPC-uncoupled version of Nup98 (that had its NPC-interaction domain deleted), identified enrichment for gene activity only in the NPC-uncoupled data set (Kalverda et al.). Genes associated with the components of the NE-embedded NPCc appeared to be frequently non-active. These results correlate well with the observed genomic binding of the stable Nup93, which is predominantly present at the nuclear pores and not in the nucleoplasm (Brown et al. 2008), suggesting that at least in animal cells, the actual NPC may not preferentially bind active chromatin and that the association of Nups with active genes may be often carried out off-pore.
Multiple lines of evidence outlined above support the existence of physical contacts between NPC components and specific genomic loci, prompting the questions of what are of the molecular roles and mechanisms of these interactions? Do they exist to couple mRNA production to its eventual exit through the nuclear pores or to connect the entry of developmental transcription factors to their target genes? Can they alternatively involve interaction with non-active genomic regions for the possible purposes of chromatin organization and partitioning expression domains? Several studies, discussed below, have delved into the molecular mechanism of these interactions, shedding light on potential interacting partners and on possible cellular consequences arising from these contacts. The three main emerging links include a role for Nups in transcriptional initiation, in co-regulation of mRNA processing and export, and in establishment of chromatin domains to maintain transcriptional memory.
As what is perhaps the strongest evidence for the gene gating model, yeast Sus1, a member of the mRNA export complex that physically interacts with the NPC component Nup1, was shown to be part of the histone acetyltransferase complex SAGA that is directly linked to transcriptional activation (Rodriguez-Navarro et al. 2004; Luthra et al. 2007). Histone acetylation is thought to play a role in setting up a transcriptionally permissive chromatin state, is enriched at gene promoters and is associated with induction of transcription (MacDonald and Howe 2009). In addition, the recruitment of active GAL genes to the nuclear periphery and presumably to the NE-embedded nuclear pores was shown to dependent on Sus1 and other SAGA components (Cabal et al. 2006; Luthra et al. 2007). This interaction has recently been shown to be conserved, as E(y)2, the Drosophila homologue of a subunit of the SAGA complex, appears to anchor the heat shock protein 70 (hsp70) gene to the NPC and to be required for its efficient transcription (Kurshakova et al. 2007). Furthermore, mammalian Nup98 has been reported to physically interact with another histone acetyltransferase, CBP/p300 (Kasper et al. 1999). This interaction appears to be highly relevant to the cancer-associated roles of Nup98, as an oncogenic fusion of Nup98 to NSD1 histone methyltransferase was shown to produce aberrant activation, hyper-acetylation and recruitment of p300 to Hox-A genes in leukaemogenesis (Wang et al. 2007). Moreover, when analyzed in developing salivary glands, Drosophila Nup98 and Sec13 were found to be recruited early on to their target genes, often preceding or coinciding with the first appearance of phosphorylated RNA polymerase II (RNAP II) (Capelson et al.). This role in transcriptional induction agrees well with the report of interaction between Nups and gene promoters during the early steps of transcriptional initiation in yeast, which proposed that transient contact with NPC proteins may be a general feature of gene activation (Schmid et al. 2006).
Recently, two new interesting candidates have been implicated in the association between transcribing genes and NPCs: some components of the exosome appear to mediate post-transcriptional tethering of GAL genes to the NPC (Vodala et al. 2008), and the THO complex seems to link 3′ end RNA processing to nuclear pore association (Rougemaille et al. 2008). Interestingly, during development it appears that Drosophila FG Nups are recruited to their target genes subsequently to recruitment of Nup98 and Sec13 and after the appearance of active RNAP II. Consistently, unlike Nup98 and Sec13, binding of FG Nups to chromatin was found to be sensitive to treatment with Flavopiridol (Capelson et al.), an inhibitor of the kinase Positive Transcription Elongation Factor b (P-TEFb) that specifically phosphorylates Serine 2 of RNAP II CTD (Chao and Price 2001). Serine 2 phosophorylation has been proposed to be required for transition into efficient transcriptional elongation as well as be necessary for recruitment of mRNA processing factors (Cho et al. 2001; Shim et al. 2002). It appears plausible that FG Nups and likely other Nups as well may play a role in elongation and the mRNA processing that accompanies it.
Interestingly, sites of NPC residence on chromatin in yeast, identified by the ChIP approach (Casolari et al. 2004), and THO complex binding (Rougemaille et al. 2008) were preferentially located in the 3′ ends of active genes, unlike the promoter and 5′ region association observed in the Nup-PI phenomenon, which was discovered by mapping of the MNase-Nup2 fusion. One explanation for this apparent contradiction is the presence of both gene ends at the NPC through interactions with different Nups, which would result in DNA looping. Such chromatin looping at the NPC has been recently directly observed for a particular subset of genes and appears to play a role in “remembering” recent transcriptional activity and thus in rapid re-activation of inducible loci (Brickner et al. 2007; Tan-Wong et al. 2009). The idea of the NPC being involved in chromatin domain establishment (or gene looping) is further supported by the reports of certain yeast Nups exhibiting boundary activity, mentioned above (Ishii et al. 2002).
As described above, during mitosis in higher eukaryotes the NE and the nuclear pores break down and reform around newly segregated chromatin (Hetzer et al. 2005). Mitotic propagation of cell-specific chromatin architecture is intimately linked to this reassembly, yet how post-mitotic nuclear reformation around chromatin translates into interphase chromatin organization is almost completely unknown. As discussed previously, studies in Xenopus cell-free systems demonstrated that a sub-complex of the NPC, the Nup107–160 complex, as well as ELYS, Nup153 and Nup358 are recruited to chromatin early in the post-mitotic reassembly (Walther et al. 2003a; Walther et al. 2003b). Thus, binding of a subset of Nups to chromatin represents one of the initial events in nuclear reorganization, implicating metazoan nucleoporins as potential regulators of interphase chromatin architecture and thus, of epigenetic memory. This suggested role in epigenetic memory supports the studies in yeast, where the long-term association of an inducible gene with the nuclear pore was reported to occur for several cell generations after the initial activation in yeast (Brickner et al. 2007). And although yeast undergoes closed mitosis and do not need to reform their NE, a role of the NPC in positional or epigenetic memory remains an intriguing possibility.
A possible model to integrate the diverse findings presented above could involve several distinct types of mechanisms by which the NPC regulates gene expression in chromatin-associated manner (Figure 3). One is the physical association of the stable NE-embedded nuclear pore structure with chromatin, which could have a role in tethering a chromatin boundary or in the general establishment of the 3D organization of chromatin inside the nucleus. As the stable core of the NPC does not turn over during the entire life span of a cell (D’Angelo et al. 2009), this extreme stability makes the core components of the NPC ideal candidates for establishing long-term transcription programs. The highly stable NPC core may serve as a scaffold for nuclear architecture of chromatin and could be utilized as a basis for epigenetic memory in differentiated cells. It appears that although some genes in yeast and metazoa contact the NE-embedded nuclear pores for activation, at least the metazoan NPC does not preferentially or exclusively bind transcribing genes and may bind other genomic regions for purely organizational purposes.
Figure 3. A schematic representation of three alternative hypotheses for the functional roles of the genome-NPC contacts.
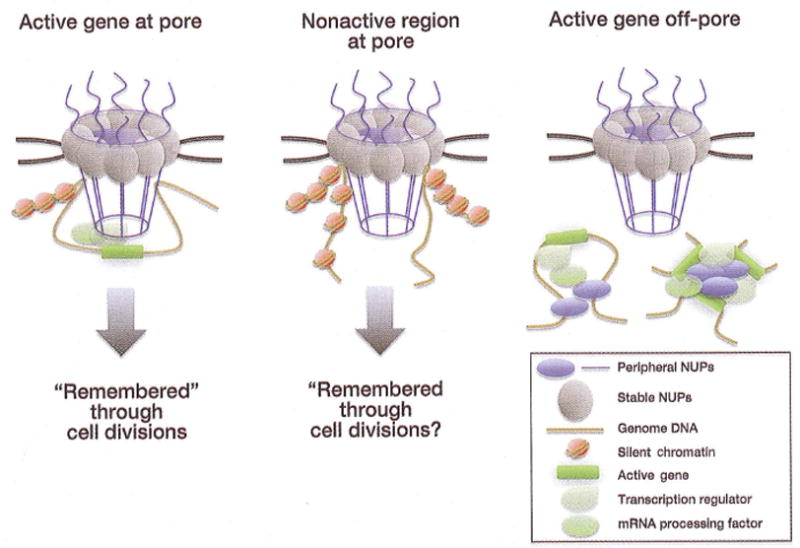
As demonstrated in yeast, the NE-embedded NPCs may bind active genes at opposing ends, forming gene loops, to establish assembled transcription and processing domains that are “remembered” through cell divisions. Alternatively, and as may be more prevalent in metazoa, the NE-associated NPCs may interact with boundary or non-transcribing genomic regions to similarly set up gene expression domains or general nuclear chromatin organization. An intriguing possibility would be the use of such contacts for “remembering” a particular chromatin organization in differentiating or differentiated cells, the latter of which do not turn over their NPCs and remain stable over long periods of time. Finally, NE-independent NPC components may bind active genes in the nucleoplasm for the similar purposes of setting up local transcription and processing organization, which may involve gene looping or even gene contacts between distant loci.
An alternative and additional mechanism would involve the binding of intranulcear Nups to active genes, irrespective of the nuclear position of the genes. It remains to be determined whether intranuclear chromatin-binding Nups shuttle between genomic sites and the nuclear periphery, which would be consistent with the dynamic behaviour of some Nups (Rabut et al. 2004). It is tempting to speculate that the mobility of NPC components may establish a mechanism of communication between sites of production of mRNA and sites of its final exit, similarly to what has been originally proposed for the NPC-chromatin relationship (Blobel 1985). In yeast, the intranuclear Nup-gene contacts have not been reported, and active genes have been observed to re-position to the membrane-associated nuclear pores in several instances. It is thus unclear whether yeast only utilizes one mode for chromatin-associated function of Nups, which involves transcriptionally active genes being recruited to the NE-embedded NPCs. It is possible that intranuclear chromatin binding of Nups has evolved preferentially in higher eukaryotes to compensate for larger nuclear size or for greater complexity of gene regulation. In Drosophila and possibly mammals, intranuclear Nups may have retained the ability to interact with transcription initiation apparatus and the mRNA processing machinery, and perhaps with both gene ends. In this manner, their role at genes undergoing induction may be to create an expression domain and set up a co-regulation of the start and finish events of the transcription process. The long unstructured GLFG domain of Nup98 and FG repeats of FG Nups may be perfect for this role as a scaffolding platform for mRNA production. In support of this hypothesis, leukemic fusions of Nup98 has been recently shown to self-interact with endogenous Nup98 in the intranuclear compartment (Xu and Powers). It is possible to envision that the self-interactions of Nup98 may even drive the clustering of distant gene loci onto a transcription platform, similarly to the model of transcription factories (Osborne et al. 2004; Mitchell and Fraser 2008). These diverse functional links lend to an intriguing model of the NPC as a potential nexus of key nuclear processes, integrating chromatin organization, transcription and transport (Kohler and Hurt 2010). Assembly of transcription machinery and distant gene loci, mRNA processing and export process, and finally, the memory of these activating or chromatin tethering events through cell divisions may all be in some ways connected to the NPC.
Acknowledgments
We thank members of the Hetzer lab for excellent assistance and helpful comments. M.C. was supported by a Damon Runyon fellowship (DRG 1952-07). C.D. and M.H are supported by grants from the National Institute of Health (RO1GM057438) and the American Cancer Society RSG 09-178-01-DDC.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8 (7):507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450(7170):695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nature cell biology. 2007;9(10):1160–1166. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. The Journal of cell biology. 2008a;182(5):911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. The life cycle of the metazoan nuclear envelope. Curr Opin Cell Biol. 2008b;20(4):386–392. doi: 10.1016/j.ceb.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. The Journal of cell biology. 2009;186(2):183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS letters. 2008;582(14):2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The Integral Membrane Nucleoporin pom121 Functionally Links Nuclear Pore Complex Assembly and Nuclear Envelope Formation. Mol Cell. 2005;17(1):83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Antonny B. Membrane deformation by protein coats. Current opinion in cell biology. 2006;18(4):386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306(5700):1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A. 1985;82(24):8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. Journal of cell science. 1999;112 ( Pt 13):2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(3):981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5(4):e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU. A lattice model of the nuclear pore complex. Communicative & integrative biology. 2009;2(3):205–207. doi: 10.4161/cib.2.3.7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22(5):627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17(2):100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3(8):575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441(7094):770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10(7):697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140(3):372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140(3):372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19(10):1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117(4):427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. The Journal of cell biology. 2010;189(5):795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276(34):31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15(24):3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312(5772):440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136(2):284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle MC, Scheer U. Assembly of nuclear pore complexes in Xenopus egg extract. Biology of the cell / under the auspices of the European Cell Biology Organization. 1991;72(1–2):25–29. doi: 10.1016/0248-4900(91)90074-w. [DOI] [PubMed] [Google Scholar]
- Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, Dolan AC, Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4(10):e1000240. doi: 10.1371/journal.pgen.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. The Journal of cell biology. 2009;184(5):659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, Yates JR, 3rd, Ferro-Novick S, Novick P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17(7):3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong-Curtain TA, Parslow AC, Trotter AJ, Hall NE, Verkade H, Tabone T, Christie EL, Crowhurst MO, Layton JE, Shepherd IT, et al. Abnormal Nuclear Pore Formation Triggers Apoptosis in the Intestinal Epithelium of elys-Deficient Zebra Fish. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Molecular cell. 2008;32(6):815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171(6):955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141(6):1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nature structural & molecular biology. 2007;14(2):138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. The Journal of cell biology. 2008;180(5):857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga J, Levay A, Fontoura BM. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol. 2003;23(20):7271–7284. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D, Sarges P, Stelter P, Hellwig A, Bottcher B, Hurt E. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. The Journal of cell biology. 2009;185(3):387–395. doi: 10.1083/jcb.200810016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DJ. Structure and function of the nuclear pore complex. Annual review of cell biology. 1992;8:495–527. doi: 10.1146/annurev.cb.08.110192.002431. [DOI] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. The EMBO journal. 2005;24(20):3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO reports. 2007;8 (2):165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130(3):512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I. A role for gp210 in mitotic nuclear-envelope breakdown. Journal of cell science. 2008;121(Pt 3):317–328. doi: 10.1242/jcs.022525. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14(12):5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82(4):587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Ghannam G, Takeda A, Camarata T, Moore MA, Viale A, Yaseen NR. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J Biol Chem. 2004;279(2):866–875. doi: 10.1074/jbc.M307280200. [DOI] [PubMed] [Google Scholar]
- Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, Graziani F, Malva C. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J Cell Biol. 1998;142(5):1195–1207. doi: 10.1083/jcb.142.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Wiese C, Allen TD, Wilson KL. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. Journal of cell science. 1997;110 ( Pt 4):409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15(4):1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi MR, Cozzolino L, Malva C, Graziani F, Gigliotti S. nup154 genetically interacts with cup and plays a cell-type-specific function during Drosophila melanogaster egg-chamber development. Genetics. 2007;175(4):1751–1759. doi: 10.1534/genetics.106.062844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev. 2009;23(21):2490–2495. doi: 10.1101/gad.539509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. Journal of cell science. 2000;113 ( Pt 5):779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003a;14(11):4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003b;11(4):853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14(5):1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16 (5):2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M, Walther TC, Mattaj IW. Pushing the Envelope: Structure, Function, and Dynamics of the Nuclear Periphery. Annu Rev Cell Dev Biol. 2005 doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109(5):551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457(7232):1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 140(3):360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140(3):360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19(1):764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani KR, Karess RE, Dostatni N, Doye V. In vivo dynamics of Drosophila nuclear envelope components. Mol Biol Cell. 2008;19(9):3652–3666. doi: 10.1091/mbc.E07-11-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell. 2010;38(1):6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29(10):1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and tpr as the architectural element of the nuclear basket. Mol Biol Cell. 2004;15(9):4261–4277. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26 (24):4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Ullman KS, Fahrenkrog B. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol. 2008;267:299–342. doi: 10.1016/S1937-6448(08)00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, De Souza CP, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84–120 complex. Mol Biol Cell. 2009;20(2):616–630. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14(6):831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. The Journal of cell biology. 2002;159(2):267–278. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282(5):3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- MacDonald VE, Howe LJ. Histone acetylation: where to go and how to get there. Epigenetics. 2009;4(3):139–143. doi: 10.4161/epi.4.3.8484. [DOI] [PubMed] [Google Scholar]
- Madrid AS, Mancuso J, Cande WZ, Weis K. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. The Journal of cell biology. 2006;173(3):361–371. doi: 10.1083/jcb.200506199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K, Wozniak RW. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. The Journal of cell biology. 2009;185(3):459–473. doi: 10.1083/jcb.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Molecular cell. 2006;22(1):93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. The Journal of cell biology. 2001;153(4):709–724. doi: 10.1083/jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. Order and disorder in the nucleus. Curr Biol. 2002;12(5):R185–192. doi: 10.1016/s0960-9822(02)00724-8. [DOI] [PubMed] [Google Scholar]
- Maul GG, Price JW, Lieberman MW. Formation and distribution of nuclear pore complexes in interphase. The Journal of cell biology. 1971;51(21):405–418. doi: 10.1083/jcb.51.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Brkljacic J. The nuclear pore and plant development. Curr Opin Plant Biol. 2009;12(1):87–95. doi: 10.1016/j.pbi.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21(6):811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102(16):5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Ryan KJ, Wente SR. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 2006;172(3):1441–1457. doi: 10.1534/genetics.105.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22(1):20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, Chen SJ, Willman CL, Chen IM, Feinberg AP, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- Oertle T, Schwab ME. Nogo and its paRTNers. Trends in cell biology. 2003;13(4):187–194. doi: 10.1016/s0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. The Journal of cell biology. 2009;185(3):475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36(10):1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Patel SS, Rexach MF. Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol Cell Proteomics. 2008;7(1):121–131. doi: 10.1074/mcp.M700407-MCP200. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6(11):1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(47):17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19(9):3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116(1):75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin Beta Regulates the Seeding of Chromatin with Initiation Sites for Nuclear Pore Assembly. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135(2):308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Ryan KJ, McCaffery JM, Wente SR. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. The Journal of cell biology. 2003;160(7):1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Miyaji-Yamaguchi M, Nagata K. Aberrant intracellular localization of SET-CAN fusion protein, associated with a leukemia, disorganizes nuclear export. Int J Cancer. 2004;111(4):501–507. doi: 10.1002/ijc.20296. [DOI] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21(3):379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Painter RG, Singer SJ. Biological membranes as bilayer couples. III. Compensatory shape changes induced in membranes. The Journal of cell biology. 1976;70(1):193–203. doi: 10.1083/jcb.70.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. The Journal of biological chemistry. 2008;283(27):18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16(16):2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS. Yeast nucleoporins involved in passive nuclear envelope permeability. The Journal of cell biology. 2000;149(5):1027–1038. doi: 10.1083/jcb.149.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84(2):265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Slape C, Aplan PD. The role of NUP98 gene fusions in hematologic malignancy. Leuk Lymphoma. 2004;45(7):1341–1350. doi: 10.1080/10428190310001659325. [DOI] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. The Journal of cell biology. 2006a;173(4):509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Nautrup-Pedersen G, Cordes VC, Gorlich D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. The Journal of cell biology. 2006b;173(4):477–483. doi: 10.1083/jcb.200601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11(7):490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441(7094):774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Takeda A, Goolsby C, Yaseen NR. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006;66(13):6628–6637. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23(22):2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8(12):1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6(2):e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6(2):e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 2005;24 (4):813–823. doi: 10.1038/sj.emboj.7600527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlcek S, Dechat T, Foisner R. Nuclear envelope and nuclear matrix: interactions and dynamics. Cell Mol Life Sci. 2001;58(12–13):1758–1765. doi: 10.1007/PL00000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodala S, Abruzzi KC, Rosbash M. The nuclear exosome and adenylation regulate posttranscriptional tethering of yeast GAL genes to the nuclear periphery. Mol Cell. 2008;31(1):104–113. doi: 10.1016/j.molcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Walde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20(8):461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003a;113(2):195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003b;424(6949):689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. The EMBO journal. 2001;20(20):5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9(7):804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, Jr, Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8(11):2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67(13):2215–2230. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Powers MA. Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis. Mol Biol Cell. 21(9):1585–1596. doi: 10.1091/mbc.E09-07-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19(5):1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz S, Santarella-Mellwig R, Koch B, Jaedicke A, Mattaj IW, Antonin W. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010;584(15):3292–3298. doi: 10.1016/j.febslet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135(6):1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Paddy MR. Structures a nd dynamics of Drosophila Tpr inconsistent with a static, filamentous structure. Exp Cell Res. 2002;276(2):223–232. doi: 10.1006/excr.2002.5525. [DOI] [PubMed] [Google Scholar]