Abstract
The DNA mismatch repair (MMR) pathway corrects specific types of DNA replication errors that affect microsatellites and thus is critical for maintaining genomic integrity. The genes of the MMR pathway are highly conserved across different organisms. Likewise, defective MMR function universally results in microsatellite instability (MSI) which is a hallmark of certain types of cancer associated with the Mendelian disorder hereditary nonpolyposis colorectal cancer. (Lynch syndrome). To identify previously unrecognized deleted genes or loci that can lead to MSI, we developed a functional genomics screen utilizing a plasmid containing a microsatellite sequence that is a host spot for MSI mutations and the comprehensive homozygous diploid deletion mutant resource for Saccharomyces cerevisiae. This pool represents a collection of non-essential homozygous yeast diploid (2N) mutants in which there are deletions for over four thousand yeast open reading frames (ORFs). From our screen, we identified a deletion mutant strain of the PAU24 gene that leads to MSI. In a series of validation experiments, we determined that this PAU24 mutant strain had an increased MSI-specific mutation rate in comparison to the original background wildtype strain, other deletion mutants and comparable to a MMR mutant involving the MLH1 gene. Likewise, in yeast strains with a deletion of PAU24, we identified specific de novo indel mutations that occurred within the targeted microsatellite used for this screen.
Introduction
The DNA mismatch repair (MMR) pathway corrects specific types of replication errors caused by DNA polymerase slippage and is critical for maintaining genomic integrity. Given its importance, the canonical genes of the MMR pathway are highly conserved among different species including Escherichia coli, Saccharomyces cerevisiae and Homo sapiens. Microsatellite (MS) sequences are composed of homopolymers and tracts of di- or trinucleotide repeats among others. Defective MMR function increases the rate of insertion and deletion (indels) mutations in microsatellites and this molecular phenotype is commonly referred as microsatellite instability (MSI). In H. sapiens, the consequences of defective DNA MMR is dramatically apparent in the Mendelian cancer syndrome, hereditary nonpolyposis colorectal cancer (HNPCC), otherwise known as Lynch Syndrome [1]. Affected individuals have germline mutations in the human MMR genes MSH2, MLH1, PMS2 and MSH6, and are at substantially increased risk for developing MSI-positive colorectal carcinoma as well as other malignancies including endometrial, gastrointestinal and genitourinary cancers [2]. While the role of these MMR genes is understood in great molecular detail, there are very few studies that have explored the possible influence on MSI of genes other than the known components of the canonical MMR pathway. Furthermore, not all clinical cases of Lynch syndrome have been explained by germline mutations in one of these four genes, suggesting that other genes may contribute.
In S. cerevisiae, the MMR genes encode for the proteins Msh2p, Mlh1p, Msh3p, Pms1p and Msh6p, all of which make up two basic protein complexes that mediate MMR. Also, there is substantial experimental evidence that EXO1, a 5′→3′ exonuclease, is involved in MMR given evidence of physical interactions with Msh2p and Mlh1p [3]. To identify other genes that increase MSI outside of these canonical MMR genes, we developed a functional genomics screen using the diploid homozygous deletion mutant resource for S. cerevisiae. This mutant pool represents a collection of non-essential homozygous yeast diploid mutants in which there are over four thousand yeast open reading frames (ORFs) [4]. Using homologous recombination, each ORF has been systematically deleted with a kanamycin resistance gene cassette flanked by two unique DNA barcode sequences [4]. For high-throughput identification and quantitative analysis of individual deletion mutants, one can PCR amplify these barcode tags from yeast genomic DNA and hybridize the amplicon products to an oligonucleotide barcode microarray (e.g. TAG3 or TAG4, Affymetrix). One can identity and determine the relative quantitative level of any given yeast mutant based on the intensity of the complementary array probes.
As previously described, we constructed a series of plasmids (the pHJ series) in which a segment of a tumor suppressor cDNA sequence with a coding microsatellite is placed upstream and in-frame of the selectable marker gene URA3 [5]. These human microsatellite sequences were chosen to model the human MSI process in yeast as they were found to be targets of MSI mutations in primary MMR-deficient colorectal cancers. If one transforms a MMR defective haploid S. cerevisiae strains with these plasmids, de novo indels occur in the introduced human microsatellite sequences at a 100-fold higher rate than background [5]. De novo microsatellite indels generally lead to frameshifts in the downstream URA3 marker and thus provide a selectable phenotype. In our previous application of this MSI assay, we measured the mutation rates (mutations per cell division) of specific human tumor suppressor coding sequence microsatellites in yeast msh2 and mlh1 deletion strains, both of which have defective MMR [5]. From our current screen, we identified a deletion mutant straing of the PAU24 gene loci (formerly referred to as DAN3) that has a MSI phenotype. In a series of validation experiments with independently created deletion mutants from the original screen, we determined that this pau24 mutant has increased MSI-specific mutation rates in comparison to the original background wildtype strain and comparable to an mlh1 deletion mutant. Likewise, we identified specific de novo indel mutations consistent with MSI that occurred within the targeted microsatellite region of the experimental plasmid for this deletion mutant.
Results
Functional genomics screen
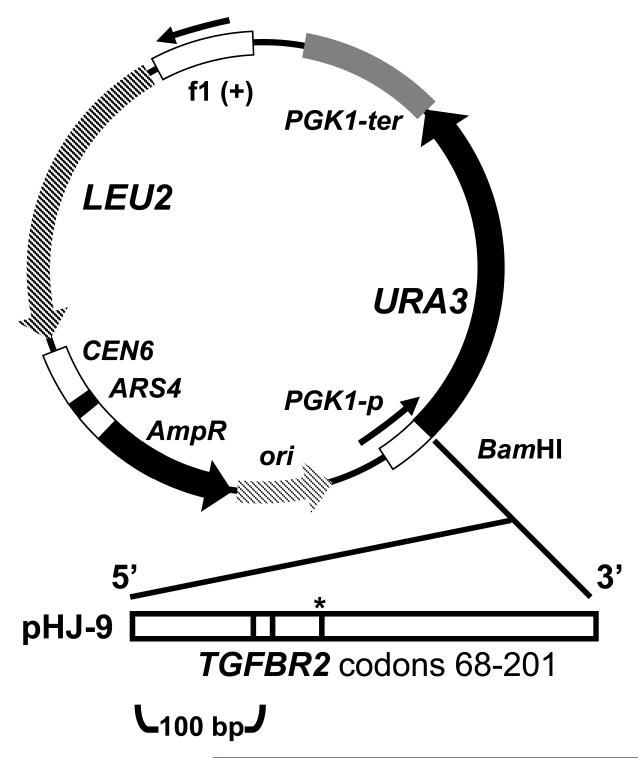
For the MSI functional genomics screen, we used the plasmids pHJ-9 (MS-positive experimental vector) and pCI-HA (MS-negative control vector). The pHJ9 plasmid is derived from pCI-HA which is a low copy number centromeric vector, has a unique BamHI site located upstream of URA3 and contains an additional LEU2 marker for plasmid retention [6]. Using the BamHI site for subcloning, pHJ-9 contains 400 bp of the coding region of the human TGFRBR2 gene upstream and in-frame of the URA3 marker (Fig. 1). Within this segment of TGFBR2 coding sequence lies a homopolymer (A)10 tract which is commonly mutated in MSI-positive colorectal cancers [1]. We previously demonstrated that this chimeric Ura3p protein permits growth on media lacking uracil (Ura-) and failure to grow in 5-fluorotic acid (FOA) containing media [5]. With the exception of three bp indels, de novo indel mutations in the (A)10 tract cause frameshifts that disrupt the URA3 coding sequence, enable selectable FOAR and allow cell growth on FOA containing media [7]. A variety of genetic assays can use this resistance phenotype for scoring. As a control and baseline for screening the homozygous deletion pool, we used the original vector, pCI-HA, to determine the background MSI mutations that directly affected the URA3 gene as opposed to the homopolymer (A)10 tract in pHJ-9. As we previously demonstrated, the spontaneous mutation rate of the URA3 gene in pCI-HA is exceptionally low, ranging on the order of 2 × 10−7 mutations per cell division [5].
Figure 1.
Description of the pHJ-9 microsatellite plasmid. The pHJ9 plasmid incorporates a region of the TGFBR2 between codons 68-201 with one microsatellite. Microsatellites are designated as bars within the gene insert and the (A)10 homopolymer is also marked with a star. Codons refer to the location within the human TGFBR2 tumor suppressor gene ORF. The control vector pCI-HA is essentially the same without the upstream gene sequence. These are centromeric plasmids with a LEU2 and URA3 marker.
We used the diploid yeast pool representing homozygous deletion mutant strains for 4,728 non-essential genes [4]. Each deletion strain has two unique barcodes, referred to as the DOWNTAG and UPTAG, flanking the deleted gene. On a special oligonucleotide barcode array (e.g. TAG3, Affymetrix), each DOWNTAG and UPTAG sequence has both a complementary sense and antisense probe [4]. This allows for identification of a given deletion mutant strain through at least 4 different barcode probes on the array; 1) DOWNTAG -sense, 2) DOWNTAG -antisense, 3) UPTAG -sense, 4) UPTAG -antisense. Ideally, for any given strain, all four barcode probes should demonstrate increased intensity in the experimental condition (pHJ9) compared to the control condition (pCI-HA).
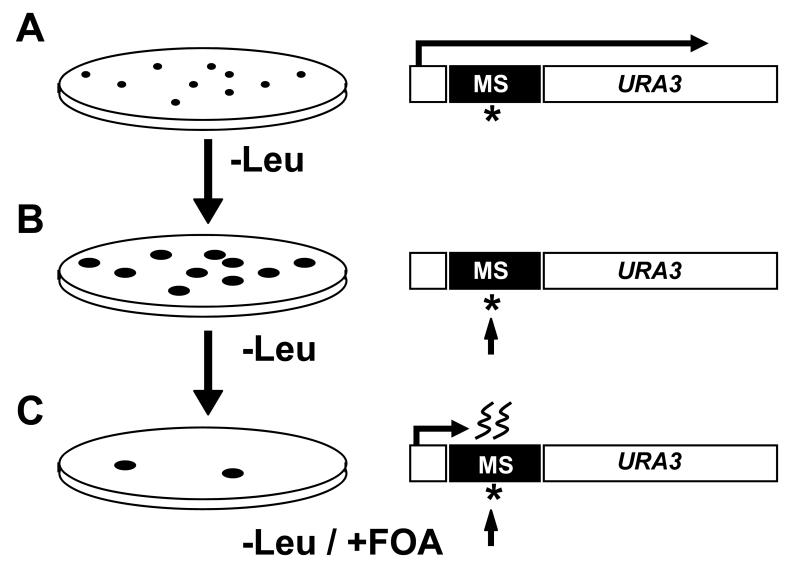
We performed five independent screens with the pHJ-9 and pCI-HA plasmids. The screen has several steps as shown in Figure 2. For each replicate experiment, we transformed either pHJ-9 or pCI-HA into the combined pool of all of the homozygous deletion mutant pool and subsequently plated the cells on synthetic dropout (SD) plates lacking leucine (Leu-). During the initial growth period on Leu-media, the plasmids undergo replication without selective pressure to maintain the wildtype URA3 and have the opportunity to accumulate mutations in the individual plasmid. After the initial transformation of the deletion mutant pool, we observed over 7,000 colonies per a replicate experiment. After three days of growth on SD-leu plates, transformed colonies are distinctly visible and we used replica plating to transfer the colonies to a dual selection media plates (Leu-, FOA). After selection on FOA media, we harvested all of the FOAR colonies, extracted genomic DNA from the collected cells, amplified the deletion mutation barcodes with PCR, hybridized the PCR amplicons to the TAG3 barcode microarrays and scanned the arrays post-hybridization [4].
Figure 2.
The MSI genomics screen. (A) After transformation with the experimental (pHJ-9) or control (pCI-HA) plasmids, cells were spread on single auxotrophic selection plates (-Leu). This growth permits multiple rounds of DNA replication and accumulation of new MSI-related mutations in the vulnerable cDNA microsatellite sequence where the location is shown as a star. (B) To select for mutations, the colonies were replica plated on FOA plates (FOA, -Leu). A de novo mutation in the microsatellite sequence is shown as an arrow. (C) Mutations in the pHJ-9 microsatellite (MS) sequence cause frameshifts, prevent these cells from producing functional Ura3p and thus enable growth on FOA-containing media.
Analysis of TAG3 barcode microarray data
A normalization procedure on the microarray barcode intensities was applied for all further data analysis [8, 9]. From the array barcode data, we identified the FOAR deletion mutants that showed significant fold change increase compared to pCI-HA control. To assess the false discovery rate (FDR) when analyzing for fold change (FC) differences on the five replicate pairs of experiments the program PaGE 5.1 was used [10]. Initially, 32 strains were identified that had a significant fold change increase in one or more of the barcode tags when comparing the intensity of the MS-positive condition (pHJ-9) to the control (pCI-HA) FOAR mutants. Among the 32, only four mutants (pau24, hxt3, gyp8 and skn1) had three or more significant barcode tags compared to the control condition and these four mutants were prioritized for further analysis. As generated by PaGE 5.1, the results in Table 1 include the confidence level that is one minus the FDR. Table 1 lists the most significant results and the average mean intensity for the tag barcodes of the experimental versus the control condition.
Table 1.
| Gene | Strain Open Reading Frame and Barcode |
Mean Intensity pCI-HA (control) |
Mean Intensity pHJ-9 (MS-positive) |
Confidence (1- FDR) |
|---|---|---|---|---|
| PAU24 | YBR301W downtag antisense | 504.70 | 4381.10 | 0.52 |
| YBR301W downtag sense | 1233.50 | 10866.08 | 0.47 | |
| YBR301W uptag antisense | 607.88 | 7201.59 | 0.52 | |
| YBR301W uptag sense | 811.12 | 9768.15 | 0.52 | |
| DNF2 | YDR345C downtag antisense | 222.58 | 1070.08 | 0.52 |
| YDR345C downtag sense | 286.95 | 2700.63 | 0.52 | |
| YDR345C uptag antisense | 243.56 | 2034.80 | 0.43 | |
| GYP8 | YFL027C downtag sense | 175.96 | 1300.91 | 0.52 |
| YFL027C uptag antisense | 780.84 | 5998.65 | 0.52 | |
| YFL027C uptag sense | 691.09 | 4981.96 | 0.52 | |
| SKN1 | YGR143W downtag antisense | 1498.32 | 9947.86 | 0.44 |
| YGR143W downtag sense | 2855.64 | 17787.49 | 0.38 | |
| YGR143W uptag antisense | 891.47 | 3442.59 | 0.43 | |
| YGR143W uptag sense | 976.28 | 4711.49 | 0.47 |
The FDR analysis did not yield the msh2 and mlh1 deletion mutants among the top ranked tag barcodes. Among the replicate experiments and array data, we examined at the average fold change for the barcode intensities for the msh2 and mlh1 mutants. No statistically significant FC increase was noted for msh2 but mlh1 did show a FC increase of 2.7 (p < 0.1) and 3.27 (p < 0.05) for the UPTAG sense and antisense barcodes respectively. Therefore, the combination of our FDR analysis and eliminating strains showing statistical significance in less than three barcodes was too stringent to identify the mlh1 mutation. The fact that we could not identify msh2 may be attributable to variation in the transformational efficiency of msh2 related to its growth and variance in the array hybridization conditions.
Fluctuation analysis to validate MSI-related mutants and determine mutation rates
Individual validation experiments on the four candidate deletion mutants were carried out using strains that were independently isolated during the original creation of the mutant. Each deletion strain was obtained from separate archived glycerol stocks and the cells were colony purified. These replicate strains had not been part of the original homozygous diploid pool was used. Prior to transformation, pHJ-9 was sequenced and confirmed that no MSI mutations existed in the homopolymer tract. pHJ-9 or pCI-HA deletion mutant strains, were individually transformed and selected for transformants on dual selection SD media plates for Leu- and Ura-conditions. The transformed strains were colony purified prior to determining MSI mutation rates. The original diploid wildtype background strain BY4743 and an mlh1 deletion mutant strain were used as a negative control and a positive control respectively.
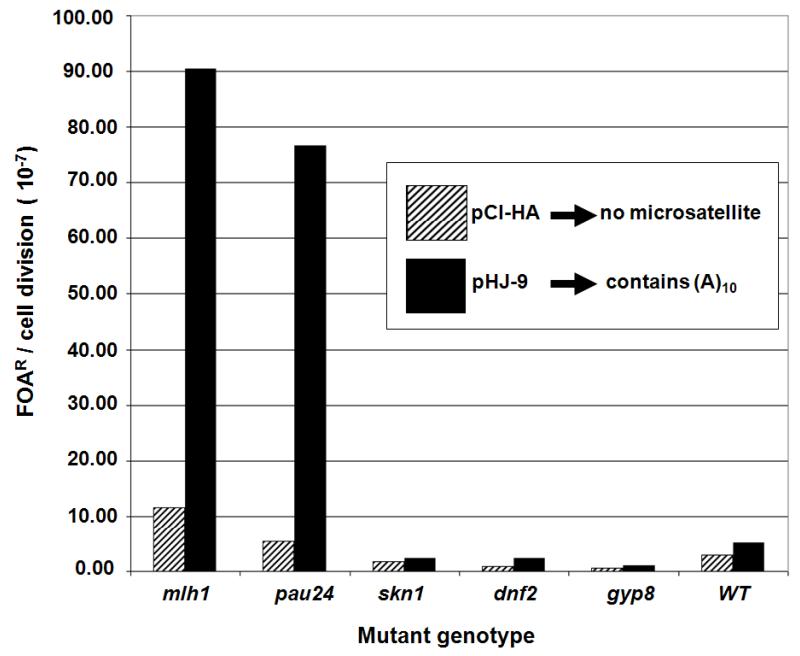
The mutation rates for each individual strain were measured using the method-of-the-median for fluctuation analysis. This required both pHJ9 (MS-positive experimental) and pCI-HA (control) [11]. Mutation rates were calculated as the number of FOAR events per cell division and the results are described in Figure 3. To validate these results, we conducted multiple independent fluctuation analyses on the wildtype strain as a negative control, the mlh1 mutant as a positive control and any deletion mutants from our screen. The results for the elevated mutation rate represent an average of several experiments. Among the four strains identified by the initial screen, only the pau24 strain had an elevated mutation rate. The other three deletion mutants did not have elevated MSI rates and were similar to the wildtype. Therefore, all three acted as a negative control for MSI.
Figure 3.
Tumor suppressor mutation rates for individual homozygous diploid deletion mutants and the wildtype strain BY4743. Mutation rates represent FOAR/cell division. The solid black columns represent mutation rates determined from the experimental microsatellite containing plasmid pHJ-9 and the hatched columns represent the control plasmid pCI-HA.
The fold increase of the mutation rate for a given deletion mutant was calculated by dividing the mutation rate for pHJ-9 in the mutant versus the wildtype background. The mlh1 mutant had a 37.8 fold elevation of mutation rate compared to the wildtype strain. The pau24 deletion mutant had a 32.1 fold elevation compared to the wildtype strain. The other deletion mutants demonstrated a two-fold or less increase of the mutation rates.
To verify that the pau24 mutant strain used for these MSI mutation rate experiments had the appropriate deletion of the PAU24 gene, we sequenced the junctions of the deletion cassettes with the adjacent genomic sequence from the pau24 homozygous deletion mutant. The sequences from the diploid homozygous mutant as well as the used haploid parental strains were amplified. Specific PCR primers were used to amplify out the deletion cassette and the adjacent yeast genomic sequence. These amplicons were subsequently Sanger sequenced. It was confirmed that the specific PAU24 deletion had occurred based on comparing the known genomic sequence flanking the PAU24 ORF and the correctly assigned deletion tag barcode.
DNA sequencing confirmation of de novo MSI mutations in the pau24 mutant
In the pau24 background, we determined if FOAR strains had MSI-related indel mutations in the homopolymer (A)10 tract of the pHJ-9 plasmid (MS-positive). First, the plasmids from independent experiments and different FOAR colonies were recovered. Based on these criteria, these plasmids had completely independent mutation events. From these recovered plasmids, the target MS homopolymer tract were sequenced and the mutations in the target MS region were identified. The same recovery procedure and DNA sequencing analysis was also carried out for the mlh1 FOAR strains. The results of the DNA sequencing analysis are summarized in Table 2. 81% of the 58 plasmids, recovered from the mlh1 deletion mutants, had indel mutations in the homopolymer (A)10 tract of pHJ9. 60% of the 40 plasmids recovered from the PAU24 mutant, had indel mutations. Overall, the predominant mutations were 2 bp deletions followed by 1 bp deletions or 1 bp insertions. From transformation of the wildtype strain grown on Leu-media, we also recovered and sequenced ten pHJ-9 plasmids, none of which demonstrated MSI mutations in the homopolymer tract. This eliminated the possibility that any of the pHJ-9 clones had spontaneously developed a MSI mutation. We also sequenced multiple colony-purified clones of pHJ-9 from our original plasmid preparation and did not identify any mutations.
Table 2.
| pau24 deletion mutant | mlh1 deletion mutant | |||
|---|---|---|---|---|
| Type of indel for each rescued plasmid |
number of mutations |
percentage | number of mutations |
percentage |
| 2 bp deletions | 12 | 30.0% | 24 | 41.4% |
| 1 bp deletions | 6 | 15.0% | 12 | 20.7% |
| 1 bp insertions | 6 | 15.0% | 11 | 19.0% |
| Total plasmids sequenced | 40 | - | 58 | - |
| Plasmids with mutations | 24 | 60.0% | 47 | 81.0% |
Discussion
As noted previously, Lynch syndrome or HNPCC is caused by germline mutations in one of several DNA mismatch repair (MMR) genes, namely MLH1, MSH2, MSH6 and PMS2 [1]. Overall, if one relies on the standard clinical guidelines such as the Amsterdam criteria, mutation analysis of MSH2 and MLH1 has a sensitivity of 61% and a specificity of 67% [12]. This leaves a significant fraction of patients who do not have identifiable mutations in the MMR genes. Mutations in the genes MSH3, EXO1, and TGFBR2 have been reported in some families with HNPCC but generally, these genes have not been demonstrated to contribute significantly to HNPCC [12]. Population studies of familial colorectal cancer have confirmed an unusually high number of pedigrees with a Mendelian pattern of inheritance but not attributable to the known syndromes [13]. Therefore, this data is highly suggestive of other genes which contribute to MSI.
To address the question about the contribution of other genes to MSI we used S. cerevisiae as a model organism system. Utilizing the homozygous diploid deletion mutant pool resource that has frequently been used for functional genomics studies in yeast, a screen identifying homozygous deletion mutants that demonstrate a MSI phenotype was developed (4). An experimental plasmid with a homopolymer (A)10 tract that is found in the human TGFBR2 gene and frequently mutated in human MSI-positive colorectal cancers was used. This plasmid models the sequence context that is frequently seen in MSI-positive cancers. We identified a deletion mutant strain of the PAU24 gene that has a MSI phenotype. This finding was confirmed in separate validation experiments using independently derived deletion mutants; the pau24 deletion mutant shows elevated MSI mutation rate compared to wildtype strains and other deletion mutant strains. Through sequencing rescued plasmids, we also demonstrated microsatellite specific indel mutations in the introduced homopolymer tract from strains showing the appropriate MSI phenotype. In addition, it was confirmed that the PAU24 gene had been appropriately deleted in our experimental mutants.
Relatively little is known about the function of PAU24 in S. cerevisiae. Given its sequence homology, it has been generally classified as a cell wall mannoprotein and a member of the seripauperin multigene family [14]. However, at the time this manuscript is prepared, there are no published reports in regards to determining cellular location experimentally or detailed functional characterization of this gene. In addition, there are no homologues in other organisms which have been reported. The majority of information about PAU24 has been derived based on its classification within the PAU gene family and genomics studies. For example, gene expression analysis has demonstrated that the PAU gene family members are induced under anaerobic conditions and low temperatures [15]. Related genes such as DAN1 have been characterized more thoroughly and shown to be a cell wall protein. There is also a paucity of information available even from extensive resources such as the Saccharomyces Genome Database [16]. Additional experiments in regards to cellular localization and protein function will need to be carried out to elucidate the exact role that PAU24 may have in microsatellite instability or whether another feature specific to the loci are causing the phenotype. To determine if homologues exist, we are pursuing efforts to determine if specific protein domains can better delineate related genes in other organisms.
One aspect of this genomics screen which may have affected the selection of mutants for MSI is the characteristics of the microsatellite [17]. In our case, a (A)10 homopolymer was used which may have influenced the selection for deletion mutants. In S. cerevisiae and E. coli, MMR function has been demonstrated to have relationships to the length and the general composition of the sequence context [18]. For example, it has been reported that variability in MMR efficiency can lead to mutation rate variations in GT/CA loci within the yeast genome [19]. These mutation frequency biases within microsatellites may have affected the type of genes that were identified in our functional genomics screen.
There is a previous report of a bystander mutation in the mismatch repair gene MSH3 that caused MSI among a small subset of yeast deletion haploid mutants from the original consortium [20]. This represented a spontaneous MMR gene mutation. Our screen was less susceptible to this artifact because the original screen and subsequent mutation rate assays used only diploid strains with homozygous deletions in both alleles. Therefore, a new, spontaneous mutation in a mismatch repair gene would have to have occurred in both copies of a DNA mismatch repair gene or have a strong autosomal dominant effect for a loss of DNA repair function. Either way, our use of a diploid strain reduced the probability of a complete loss of DNA mismatch repair function from random mutations.
In addition, the bystander MSH3 mutation arose from a single laboratory and a single wildtype haploid strain specific to that laboratory (Angela Chu, Personal Communication 2012). Most notably, these mutants were created by transformation into a haploid (1N) strain, then mating two haploids from the initial transformation to make the homozygous diploid. This approach is substantially more prone to producing bystander mutations with a specific phenotype given the initial use of a haploid (1N) strain. Generally, most of the deletion consortium members started with a diploid strain (2N) transformation followed by dissection for haploids and then mating to creating the diploid mutants. This is the exact reverse of what was done in the case of the bystander MSH3 mutant. The pau24 mutant strain was not generated by the group that produced the MSH3 background mutant. Furthermore, the YBR301W strain with the PAU24 deletion were made by transformation into the diploid, and dissected to get haploids, which were subsequently mated to make the homozygous diploid. For the MSI analysis, we used deletion mutant replicates derived from separate transformations and matings, thus lessening the chances of a random bystander mutation in a mismatch repair gene. We are continuing to investigate the relations between the pau24 deletion strain and the MSI phenotype.
Materials and Methods
General genetic methods, plasmids and strains
SD media were obtained from Bio101 Systems (Santa Ana, CA). 5-Fluoroorotic acid (FOA) was obtained from Zymo Research (Orange, CA). The SD-leu-ura plates contained FOA at a concentration of 1 gm per liter and uracil at 50 mg per liter. This concentration was empirically determined to be optimal for selecting FOA resistant (FOAR) colonies. Homozygous diploid deletion strains including pau24 (strain 37177), skn1 (strain 23773), dnf2 (34182), gyp8 (35646) and the wildtype strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0) were obtained from the Stanford Genome Technology Center. The diploid BY4743 was the original strain used in the construction of all deletion mutants (4). The construction of pHJ-9 and features of pCI-HA are as previously described (5). Plasmids were grown in LB-carbenicillin and plasmids were extracted using a Qiagen Maxiprep protocol (Valencia CA).
Functional genomics MSI screen and TAG3 microarray analysis
For each replicate experiment, we transformed either pHJ-9 (MSI-experimental) or pCI-HA (control) into the combined pool of the homozygous deletion mutant pool and subsequently spread the cells on SD-leu plates at 30 °C. The characteristics of the pool are as previously described (4). Transformation procedures are as previously described (21). The transformants were spread at a density of greater than 300 colonies per plate on 24 150-mm Petri plates. After three days of growth, transformed colonies are replica plated to SD-leu+FOA. After four days of growth on FOA media at 30 °C, we harvested all of the FOAR colonies and extracted genomic DNA from collected yeast cells using protocol I of the Zymo Research YeaStar kit (Zymo Research, Organe CA). We conducted the microarray experiments as has been previously described using TAG3 arrays (Affymetrix, Santa Clara CA) (22). After extracting genomic DNA from the harvested cells, two separate PCR reactions were used to amplify the UPTAG and DOWNTAG barcode for a given experiment with primer sequences as previously reported (23). PCR reaction conditions used 33 μl of dH2O, 6 μl of 10 × PCR buffer without MgCl2, 3 μl of 50 mM MgCl2, 1.2 μl of 10 mM dNTPs, 1.2 μl of 50 mM UPTAG or DOWNTAG primer mix, 0.6 μl of 5 U per μl Taq polymerase, 15 μl of genomic DNA. The thermocycler conditions for PCR were 94 °C for 3 min; followed by 30 repeat cycles of the following: 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s; 72 °C for 3 min; hold at 4 °C. As previously described, array hybridization was carried out using the TAG3 array (Affymetrix, Santa Clara CA) with the UPTAG and DOWNTAG PCR products (23). After scanning each array, we visually inspected the images and repeated the hybridization with a new array if gross defects (e.g. bubbles with reduced signal areas) were apparent.
For the array preprocessing two Perl scripts were used as previously described (9). The first step involved using the script “raw_file_data.pl” to map the raw intensity data in each input CEL file to the UPTAG barcode, the DOWNTAG barcodes and ultimately the strain. The second step involved normalizing the array data for each replicate experiment with the script “normalize_data.pl” using the output from the first step. The barcode tags were separated into four categories based on UPTAG versus DOWNTAG and sense versus antisense. Normalization was based on the appropriate category rather than the mean of the entire array. Afterwards, we used the program PaGE 5.1 (www.cbil.upenn.edu/PaGE/) to conduct our analysis on the five replicate pairs of microarray experiments with pHJ9 (MSI-experimental) and pCI-HA (control) (10). For PaGE5.1, the following settings were used: i) T-test, ii) log paired settings and iii) a confidence level of 0.3 that translates into a FDR of 0.7. FDR was set at a high level to improve sensitivity. We also calculated the average fold change (FC) between the experimental and control condition for increases in any given deletion barcode in the data set and applied a Student’s T-test to evaluate statistical significance.
Mutation rate analysis
Each deletion strain was obtained from separately archived glycerol stocks and colony purified the cells, making sure to use a replicate deletion mutant strain that had not been part of the original homozygous diploid pool. Using either pHJ-9 or pCI-HA, we individually transformed these fresh deletion mutant strains and selected for transformants on dual selection SD media plates for Leu- and Ura-conditions at 30 °C. We colony purified transformants prior to determining MSI mutation rates. The MSI rate for individual yeast deletion mutant strains was determined by fluctuation analysis using the method of the median from samples of 15 independent cultures [11, 24]. Yeast was transformed on SD-leu-ura plates, colony purified and patched. Transformed yeast strains were subsequently grown on SD-leu plates for 3 days. Individual colonies were isolated, suspended in water and dilutions were spread on SD-leu+FOA plates to measure FOAR and SD-leu to monitor viable cells. For strains showing elevated mutations rates, fluctuation analysis was repeated and mutation rates were determined by averaging the results.
Plasmid rescue, PCR and DNA sequencing
FOAR colonies were patched on SD-leu+FOA plates. Plasmids were recovered using methods as previously described [25]. Primers specific for pHJ-9 encompassing the tumor suppressor insert region were designed: 5′-GTTCCTGACTATGCGGGCTA-3′ and 5′-AATGTCTGCCCATTCTG-3′. The tumor suppressor MS sequence was amplified by PCR using the following protocol: 35 cycles of 94 °C for 30s, 55 °C for 30s and 72 °C for 30s. The PCR product was purified with the Qiagen QuickPCR kit (Qiagen, Valencia CA). DNA sequencing of pCI-HA and pHJ9 PCR products was performed using the first primer listed and Sanger terminator sequencing chemistry as per the manufacturer’s protocol (Life Sciences, Foster City CA). For yeast genomic sequencing to confirm the pau24 deletion mutant, PCR primers specific to PAU24 deletion knockout were used to PCR amplify the adjacent genomic region of the gene from the diploid and haploid deletion strains. These primers are as previous reported (4) and are available on the yeast deletion website (http://www-sequence.stanford.edu/group/yeast_deletion_project/Deletion_primers_PCR_sizes.txt). Sequencing reactions were run on an Applied Biosystems 9700 DNA sequencer (Life Sciences, Foster City CA). Traces were analyzed with the Bioedit program (www.mbio.ncsu.edu/bioedit) or DNAstar program (DNASTAR, Madison WI).
Acknowledgements
We would like to thank Angela Chu for providing yeast deletion strains, assistance in confirming the pau24 deletion mutant and general guidance about using the homozygous diploid deletion pools. This work was supported by the following grants from the NIH: 5K08CA96879-6 to H.P.J., DK56339 to H.P.J, 2P01HG000205 to K.W. and H.P.J.
Footnotes
Conflicts of Interest
The authors have no competing financial interests in relation to the work described.
References
- 1.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010 Jun;138(6):2073–87 e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–58. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jun SH, Kim TG, Ban C. DNA mismatch repair system. Classical and fresh roles. FEBS J. 2006;273(8):1609–19. doi: 10.1111/j.1742-4658.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 4.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 5.Ji HP, King MC. A functional assay for mutations in tumor suppressor genes caused by mismatch repair deficiency. Human Molecular Genetics. 2001;10(24):2737–43. doi: 10.1093/hmg/10.24.2737. [DOI] [PubMed] [Google Scholar]
- 6.Ishioka C, Suzuki T, FitzGerald M, Krainer M, Shimodaira H, Shimada A, et al. Detection of heterozygous truncating mutations in the BRCA1 and APC genes by using a rapid screening assay in yeast. PNAS. 1997;94(6):2449–53. doi: 10.1073/pnas.94.6.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Molecular & General Genetics. 1984;197(2):345–6. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 8.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 9.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320(5874):362–5. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant GR, Liu J, Stoeckert CJ., Jr. A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21(11):2684–90. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- 11.Lea DA, Coulson CA. The distribution of the numbers of mutants in bacterial populations. Journal of Genetics. 1949;49:264–85. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 12.Jeter JM, Kohlmann W, Gruber SB. Genetics of colorectal cancer. Oncology (Williston Park) 2006;20(3):269–76. discussion 85-6, 88-9. [PubMed] [Google Scholar]
- 13.Woods MO, Younghusband HB, Parfrey PS, Gallinger S, McLaughlin J, Dicks E, et al. The genetic basis of colorectal cancer in a population-based incident cohort with a high rate of familial disease. Gut. 2010;59(10):1369–77. doi: 10.1136/gut.2010.208462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Z, van Vuuren HJ. Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology. 2009;155(Pt 12):4036–49. doi: 10.1099/mic.0.030726-0. [DOI] [PubMed] [Google Scholar]
- 15.Rintala E, Toivari M, Pitkanen JP, Wiebe MG, Ruohonen L, Penttila M. Low oxygen levels as a trigger for enhancement of respiratory metabolism in Saccharomyces cerevisiae. BMC Genomics. 2009;10:461. doi: 10.1186/1471-2164-10-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38(Database issue):D433–6. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah SN, Hile SE, Eckert KA. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 2010;70(2):431–5. doi: 10.1158/0008-5472.CAN-09-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog. 2009;48(4):379–88. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci U S A. 2005;102(24):8639–43. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehner KR, Stone MM, Farber RA, Petes TD. Ninety-six haploid yeast strains with individual disruptions of open reading frames between YOR097C and YOR192C, constructed for the Saccharomyces genome deletion project, have an additional mutation in the mismatch repair gene MSH3. Genetics. 2007;177(3):1951–3. doi: 10.1534/genetics.107.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 22.Chu AM, Davis RW. High-throughput creation of a whole-genome collection of yeast knockout strains. Methods Mol Biol. 2008;416:205–20. doi: 10.1007/978-1-59745-321-9_14. [DOI] [PubMed] [Google Scholar]
- 23.Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protoc. 2007;2(11):2958–74. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- 24.Wierdl M, Dominska M, Petes TD. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics. 1997;146(3):769–79. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robzyk K, Kassir Y. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Research. 1992;20(14):3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]