Abstract
The conserved family of Cdc14 phosphatases targets cyclin-dependent kinase substrates in yeast, mediating late mitotic signaling events. To discover substrates and regulators of the Schizosaccharomyces pombe Cdc14 phosphatase Clp1, TAP-tagged Clp1, and a substrate trapping mutant (Clp1-C286S) were purified from asynchronous and mitotic (prometaphase and anaphase) cells and binding partners were identified by 2D-LC-MS/MS. Over 100 Clp1-interacting proteins were consistently identified, over 70 of these were enriched in Clp1-C286S-TAP (potential substrates) and we and others detected Cdk1 phosphorylation sites in over half (44/73) of these potential substrates. According to GO annotations, Clp1-interacting proteins are involved in many essential cellular processes including mitosis, cytokinesis, ribosome biogenesis, transcription, and trafficking among others. We confirmed association and dephosphorylation of multiple candidate substrates, including a key scaffolding component of the septation initiation network called Cdc11, an essential kinase of the conserved morphogenesis-related NDR kinase network named Shk1, and multiple Mlu1-binding factor transcriptional regulators. In addition, we identified Sal3, a nuclear β-importin, as the sole karyopherin required for Clp1 nucleoplasmic shuttling, a key mode of Cdc14 phosphatase regulation. Finally, a handful of proteins were more abundant in wild type Clp1-TAP versus Clp1-C286S-TAP, suggesting that they may directly regulate Clp1 signaling or serve as scaffolding platforms to localize Clp1 activity.
Cdc14 family phosphatases have been identified in eukaryotes ranging from yeast to mammals (reviewed in (1, 2)). They are proline-directed phosphatases (3) and in yeast, they target mitotic cyclin-dependent kinase 1 (Cdk1) substrates (reviewed in (1, 2), making them key regulators of late mitotic events. Consistent with their cell cycle role, Cdc14 family phosphatases localize dynamically to a variety of cellular structures in a cell cycle dependent manner. During interphase they are located at yeast spindle pole bodies (SPB)1 or centrosomes of higher organisms, and/or are sequestered in the nucleolus. During mitosis Cdc14 proteins are present on the SPBs/centrosomes, at kinetochores, on the mitotic spindle, at the site of cell division, and the midbody, depending on the different species (reviewed in (4)). At these different sites, Cdc14 phosphatases presumably dephosphorylate a wide variety of substrates that are hyperphosphorylated by Cdk1 during mitosis.
Many Cdc14 phosphatase substrates have now been identified in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mammalian cells. Thus far, yeast Clp1 substrates have also been Cdk1 substrates, but it is unclear whether Cdc14 phosphatases exclusively dephosphorylate Cdk1 substrates. Furthermore, it is not known whether all Cdk1 phosphorylation events can be reversed by Cdc14 family phosphatases because Cdc14 exhibits target site preference both in vitro and in vivo (5, 6). Tethering molecules involved in Cdc14 family sequestration and inhibition have also not been comprehensively identified. Outside of S. cerevisiae, for example, the nucleolar binding partners for this family are unknown (reviewed in (1, 2).
The sole Cdc14 phosphatase family member in S. pombe, termed Clp1/Flp1 (Clp1 for simplicity), regulates several aspects of mitotic exit but is not important for rDNA segregation as in S. cerevisiae (7). Clp1 promotes mitotic exit by antagonizing the auto-amplification loop of Cdk1 by dephosphorylating Cdc25 (8, 9). It also functions together with Aurora kinase to regulate chromosome biorientation (10), and stabilizes the cytokinetic ring through an interaction with anillin-like Mid1 (11). Like S. cerevisiae Cdc14, Clp1 is important for proper mitotic spindle function (12–14), and coordination of the nuclear division cycle with cell division through a cytokinesis checkpoint (15). Importantly, S. pombe Clp1 is not essential for cell viability, unlike S. cerevisiae Cdc14 (16, 17), making it amenable to clp1 gene manipulation.
In this study, we took advantage of the nonessential nature of clp1 and used a proteomics approach to comprehensively define Clp1-interacting proteins in S. pombe. In addition to identifying known Clp1 substrates and interacting proteins, we found new substrates, mapped their phosphorylation sites, and investigated the effect of dephosphorylation on a specific target, Cdc11. We also identified the karyopherin Sal3 as the importin responsible for Clp1 nuclear import and its ability to shuttle between important sites of cellular action.
EXPERIMENTAL PROCEDURES
Purification of TAP Complexes
Tandem affinity purifications (TAPs) were conducted as described (18) from the following strains: clp1-TAP, clp1-TAP nda3-KM311, clp-C286S-TAP, and clp-C286S-TAP nda3-KM311 (supplemental Table S1). The nda3-KM311 strains were arrested in prometaphase at 19 °C for 6 h before harvest. TAPs were also done from clp-C286S-TAP nda3-KM311 cells that were first blocked at 19 °C for 6 h and then released to 32 °C for 30 min (anaphase).
Analysis of TAP Complexes by Mass Spectrometry
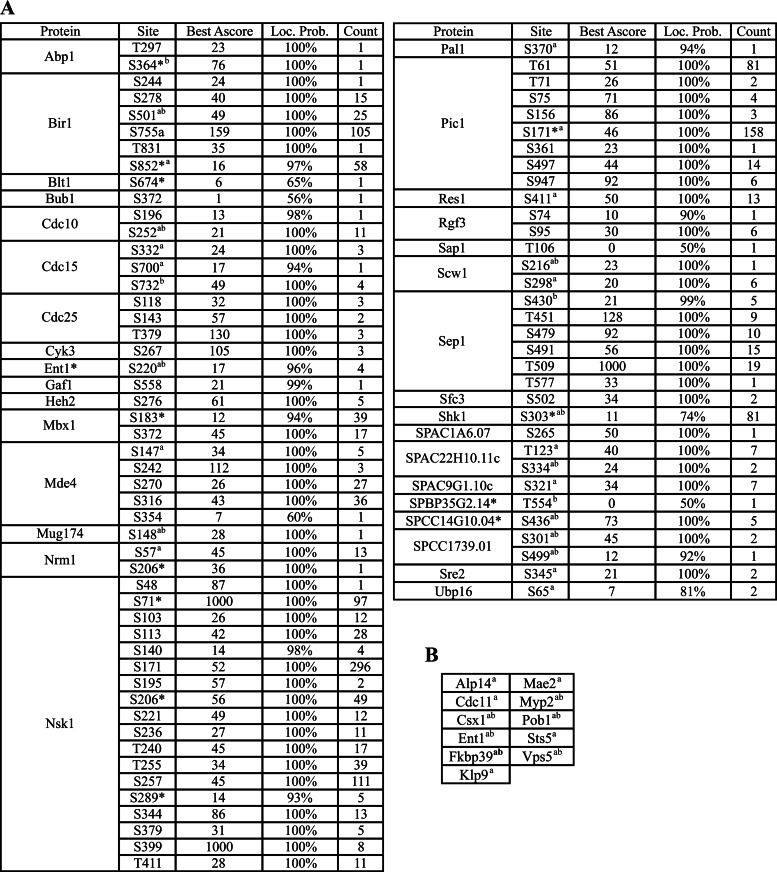
Proteins were digested and analyzed by two-dimensional liquid chromatography tandem MS (2D-LC-MS/MS) as previously described (19, 20). All RAW files can be accessed in Peptide Atlas (PASS00121). MS2 and MS3 spectra were extracted separately from RAW files and converted to DTA files using Scansifter software (21) (v2.1.25). Spectra with less than 20 peaks were excluded and the remaining spectra were searched using the SEQUEST algorithm (TurboSequest v.27 rev12) against the S. pombe protein database (created in May 2011 from pombase.org). Common contaminants were added and all sequences were reversed to estimate the false discovery rate (FDR), yielding 10352 total entries. Variable modifications (C+57, M+16, [STY]+80 for all spectra and [ST]-18 for MS3), strict trypsin cleavage, <10 missed cleavages, fragment mass tolerance: 0.00 Da (because of rounding in SEQUEST, this results in 0.5 Da tolerance), and parent mass tolerance: 2.5 Da were allowed. Peptide identifications were assembled and filtered in Scaffold (v3.6.4, Proteome Software, Portland, OR) using the following criteria: minimum of 99.9% protein identification probability; minimum of three unique peptides; minimum of 95% peptide identification probability; minimum peptide length of five amino acids; minimum number of one tryptic terminus; DeltaCN ≥ 0.1; and Xcorr minimum threshold values of 1.8, 2, and 2.5 for charge states +1, +2, and +3, respectively. FDRs were estimated in Scaffold 3 based on the percentage of decoy sequences identified after using the above filtering criteria; for the combined MudPITs (10 in total), the protein level FDR was 0.9% and the peptide level FDR was 0.0%. Proteins containing the same or similar peptides that could not differentiated based on MS/MS alone were grouped to satisfy the principles of parsimony; this is relatively rare in S. pombe. All proteins meeting the above criteria were exported to Excel for further filtering. (1) contaminant proteins (e.g. keratin) were removed (2) nonspecific proteins that were also identified in TAP preparations from a strain lacking the TAP tag or in multiple unrelated TAPs were demarcated by shading in Supplemental Tables and (3) proteins identified in only 1 experiment were removed (i.e., proteins must be identified in at least two experiments with at least three unique peptides per experiment). supplemental Table S2 is the final protein interactor list from all 10 Clp1-TAP runs. Phosphorylation sites were filtered to 50% Scaffold localization using Scaffold PTM (v2.0, Proteome Software, Portland, OR) and Ascores (22) are reported without further filtering (Fig. 3 and supplemental Tables S5 and S6).
Fig. 3.
Potential Clp1 substrates contain Cdk1 phosphorylation sites. A, Clp1 substrates containing Cdk1 consensus sites identified in our study. Localization probability = Scaffold score for phosphorylation sites localization, Ascore = describes how well a phosphorylation site is localized (22). Asterisks (*) indicate sites only identified by additional loss of water in MS3, indicating phosphorylation. Superscript letters (a and b) denote Cdk1 consensus sites identified in references (81) and/or (82) respectively. B, Clp1 substrates containing Cdk1 consensus sites identified in other studies (see A).
GO Annotation and Network Analysis of Clp1-interacting Proteins
GO annotations of the resulting proteins were collected from the S. pombe gene database (www.pombase.org) and categorized into the most general process in which the protein participates (Fig. 1C). Previously reported S. pombe protein–protein interactions were downloaded from BioGRID (v3.1.94, www.thebiogrid.org/). Physical interactions (excluding colocalization) of Clp1-interacting proteins were used to create an interaction network (Fig. 2) using Cytoscape, an open source platform for complex network analysis and visualization (version 2.8.3, (23).
Fig. 1.
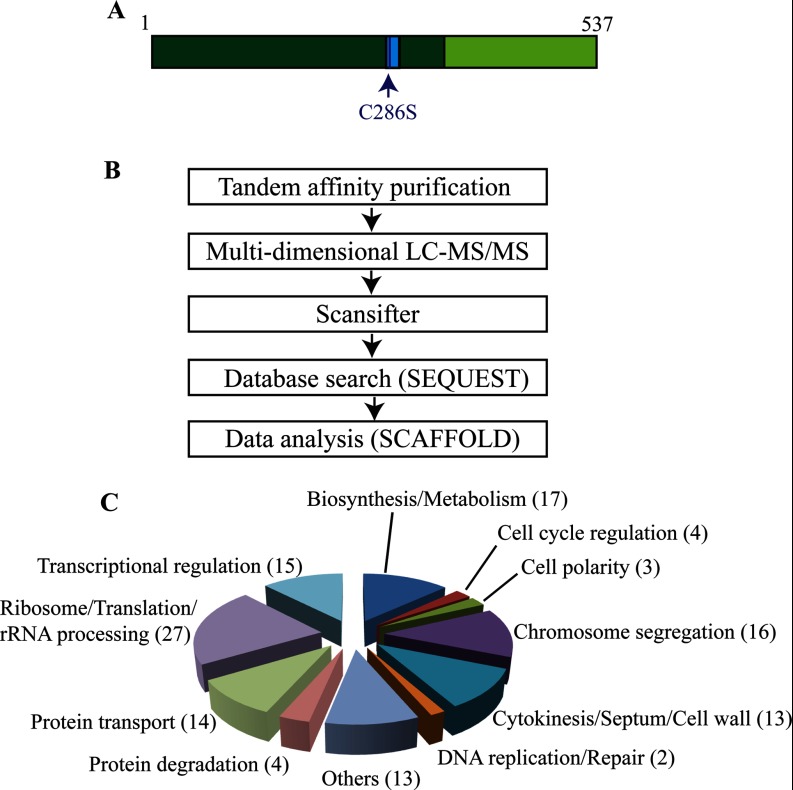
Identification of Clp1 interacting proteins. A, Schematic of Clp1's domain structure. The catalytic domain is shown in dark green. The PTP motif (residues 281–294), a signature of phosphatases, contains the catalytic site and is highlighted in blue. In the Clp1 substrate trapping mutant, the catalytic site cysteine 286 is replaced by serine. The regulatory domain is shown in lime green. B, Workflow used to identify Clp1-interacting proteins. C, Pie chart showing the different GO categories of all Clp1-interacting proteins (supplemental Table S2). The number of Clp1 interactors with each GO annotation are in indicated in parentheses.
Fig. 2.
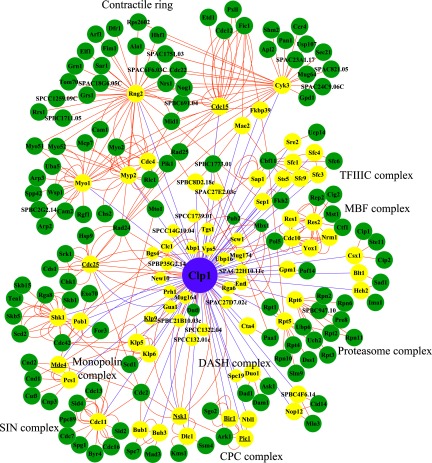
Network diagram of physical interactions of proteins enriched in Clp1-C286S-TAP (candidate substrates). The network and BioGRID physical interactions were organized using Cytoscape (see Experimental Procedures for details). The Clp1 node is purple, candidate Clp1 substrate nodes are yellow (those with known interaction partners depicted as larger nodes), and all other nodes are green (i.e., protein interactions from BioGRID). Node names that are underlined indicate previously known Clp1 substrates. Each edge (line between nodes) indicates an interaction. Purple edges indicate interactions found in this study and orange edges are BioGRID interactions.
Yeast Cell Culture and Strains
All S. pombe strains (supplemental Table S1) were grown in yeast extract (YE) or minimal media with required supplements (24). Transformations were performed using the lithium acetate method or electroporation (25, 26). Fusion epitope-tagged proteins were generated by tagging the 3′ end of the endogenous gene loci with epitope-tag-KanR cassette as previously described (27). Strain construction and tetrad analysis were accomplished through standard methods. For induction from the nmt81 promoter (28), cells were grown overnight in minimal media containing 5 μg/ml thiamine, washed three times with media lacking thiamine and then allowed to grow for 18 h in thiamine-free media (induction). For spot assays, cells were grown to mid-log phase at 25 °C, 8 million cells were resuspended in 1 ml of water and then 1:10 serial dilutions were plated on YE agar and incubated at 25 °C, 27 °C, 29 °C, 32 °C, or 36 °C for 4 days. The cdc11–8D mutant was made by site-directed mutagenesis (Multisite Quickchange kit, Agilent) and integrated in place of cdc11::ura4.
Immunoprecipitation and Immunoblotting
Cell pellets were collected from cells in log phase growth or arrested in mitosis and snap frozen in a dry ice/ethanol bath before lysis. Whole-cell S. pombe lysates were prepared in Nonidet P-40 buffer or HEPES buffer (50 mm HEPES-NaOH, pH 8.0, 150 mm NaCl, 5 mm EDTA, 1 mm EGTA, 50 mm β-glycerol phosphate, 0.1 mm Na3VO4, 1 mm dithiotreitol, 1 mm phenylmethylsulfonyl fluoride, 1% Nonidet P-40), and immunoprecipitations were performed as previously described (29), except 2 μg of antibody was used in each immunoprecipitation. In some cases cell lysates were made using the FastPrep-24 instrument (MP Biomedical, Santa Ana, CA). Protein samples were resolved by SDS-PAGE and transferred to PVDF membrane (Immobilon P, Millipore, Bedford, MA). Blocked membranes were incubated with primary antibodies (1:1,000 dilution) for 1 h at room temperature or overnight at 4 °C. Goat anti-mouse or goat anti-rabbit Alexa Fluor 680 (Invitrogen, Carlsbad, CA) secondary antibodies were used at a 1:5,000 dilution. Immunoblots were scanned using the infrared imaging system and protocol (Odyssey, LI-COR Biosciences) and images were exported from Odyssey software in TIFF format. Antibodies used included anti-myc (9E10), anti-HA (12CA5), anti-GFP (Roche), anti-Flag (Invitrogen), and anti-Cdc11 (30).
In Vitro Phosphatase Assays
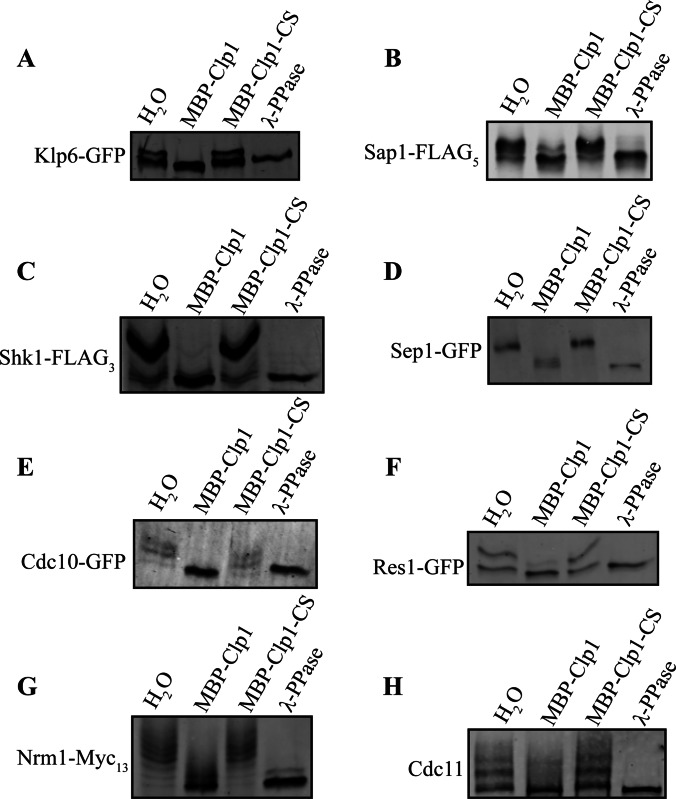
Phosphatase assays of immunoprecipitants were conducted as previously described (31) with the following modifications. After immunoprecipitation from native cell lysates, Sepharose-bound proteins were washed three times with 1 ml HEPES buffer (see above for buffer composition) and twice with 1 ml of Clp1 phosphatase buffer (50 mm imidazole, pH 6.9, 1 mm EDTA, and 1 mm dithiothreitol), divided equally into four parts. Three parts were treated with H2O, 100 ng MBP-Clp1, or 100 ng MBP-Clp1-C286S, respectively, in 20-μl reactions composed of 1× Clp1 phosphatase buffer. The fourth part was washed with λ-protein phosphatase buffer (25 mm Hepes-NaOH, pH 7.4, 150 mm NaCl, and 0.1 mg/ml BSA), and treated with 1 μl of λ-protein phosphatase (New England Biolabs, Inc., Ipswich, MA) in a 20-μl reaction composed of 1× λ-protein phosphatase buffer and 2 mm MnCl2. All reactions were incubated at 30 °C for 30 min with gentle mixing and terminated by adding 5 μl of 5× SDS sample buffer. Protein samples were resolved by SDS-PAGE in the absence or presence of 20, 50, or 100 μm Phos-tag acrylamide per the manufacturer's protocol (Wako Chemicals USA, Inc. Richmond, VA).
Microscopy
All microscopy images were acquired using a personal DeltaVision system (Applied Precision, Issaquah, WA). This system includes an IX71 microscope (Olympus, Center Valley, PA), 60 × NA 1.42 PlanApo objective, a CoolSnap HQ2 camera (Photometrics, Tucson, AZ), and softWoRx imaging software. Z series optical sections were taken at 0.5 μm steps. After image acquisition, Z stacks were combined (Quick Projection), deconvoluted, saved in softWoRx, and imported into Adobe® Creative Suite (CS4). Images of clp1Δ, csc1Δ and clp1Δcsc1Δ cells were acquired after fixation in 70% ethanol and staining with DAPI and methyl blue (septa/cell wall).
RESULTS
Identification of Clp1 Interacting Proteins
To identify Clp1 substrates and/or interacting partners, tandem affinity purifications (TAPs) (18, 32) of Clp1 were performed and the resultant complexes were analyzed by 2D-LC MS/MS (19) (Fig. 1B). Because interaction between Clp1 and its substrates might be transient, a catalytically inactive mutant of Clp1, Clp1-C286S, (Fig. 1A) (9, 33), and wild-type Clp1, were used as bait in the purifications. The Clp1-C286S mutant should act as a substrate trap, able to bind its substrates but unable to dephosphorylate them (34). To enhance the chances of identifying the largest number of interacting proteins, Clp1 was purified from asynchronously growing cells in which Clp1 is primarily at the SPB and nucleolus, and mitotically arrested cells in which Clp1 has spread throughout the cell, decorating the mitotic spindle, the kinetochores, and the contractile ring. To enrich for cells in mitosis, we used the β-tubulin mutant nda3-KM311 to arrest cells in prometaphase (35) and a 30 min release of this arrest to capture cells in anaphase. Each TAP-LC-MS/MS analysis was performed in duplicate.
In total, over 400 proteins were identified in at least two of the 10 Clp1 purifications by at least three unique peptides (supplemental Table S2). Proteins likely to be nonspecific (false-positive interactors) based on their identification in background or in four unrelated TAP-LC-MS/MS analyses performed in our laboratory are shaded in blue or green, respectively (supplemental Table S2). The remaining 128 Clp1-associated proteins were sorted based on the GO annotation of their function (Fig. 1C). Many of these proteins are linked to a biological process involved in cell division including cell cycle regulation, cell polarity establishment, protein degradation, chromosome segregation, cytokinesis, and cell wall biosynthesis (Fig. 1C), consistent with Clp1's known functions in anaphase (36). A large number of Clp1-associated proteins are involved in ribosome biogenesis/protein translation, or general biosynthesis and cellular metabolism. These sets of proteins have been identified frequently in other unrelated TAP-LC-MS/MS analyses performed in our laboratory, and may have been nonspecifically purified. However, given the strong functional ties between cell cycle and cell growth (37), it would not be surprising if bonafide Clp1-interacting proteins were among them. An additional GO category included several proteins involved in intracellular protein transport (Fig. 1C). Intracellular trafficking is important for delivery of materials to the cell division site (38) and so it is feasible that Cdk1/Clp1 regulates some aspect(s) of cellular trafficking. Lastly, a large group of Clp1-associated proteins were linked to transcriptional regulation, a process regulated at numerous levels by phosphorylation/dephosphorylation (Fig. 1C).
Identification of Clp1 Substrates
Because we expected Clp1 substrates to be enriched in purifications of the substrate-trapping Clp1-C286S mutant relative to wild-type Clp1, we compared the recovery of the 128 interactors from these two baits. Based on normalized spectral counts (spectral counts of the protein divided by the spectral counts of the bait Clp1, multiplied by 1000), we identified 73 proteins with at least two-fold enrichment in Clp1-C286S TAPs (supplemental Table S3). Validating this strategy and our analysis, all previously identified Clp1 substrates were recovered in this list of 73 proteins including Nsk1-Dlc1 (14), the scaffolding components of the Aurora kinase complex (INCENP/Pic1, Survivin/Bir1, and Borealin/Nbl1) (10, 39), monopolin (Mde4 and Pcs1) (13, 40), the kinesin Klp9 (12), the Cdk1 regulator Cdc25 (8, 9), and the cytokinetic ring protein Cdc15 (11) (supplemental Table S3 and Fig. 2). The remaining proteins were therefore excellent candidates for being either Clp1 substrates or the binding partners of such substrates. To distinguish Clp1 substrates, we searched our MS data set for phosphorylation sites matching the Cdk1 consensus within Clp1 interactors and found that over half (44/73) of the putative substrates contain such phosphorylation sites (Fig. 3, supplemental Tables S5 and S6), highlighting the strength of this methodology.
To better understand the cellular processes impacted by these potential Clp1 substrates, we analyzed their physical interactions annotated in BioGRID. This analysis again highlighted the connection between Clp1 and the regulation of cytokinesis and chromosome segregation (Fig. 2). As well as previously recognized Clp1 substrates involved in these processes, we found that Clp1-C286S associated with the heterodimeric kinesin-8 (Klp5-Klp6) and the DASH complex. Kinesin-8s are plus-end directed motors important for chromosome congression at the metaphase plate, conserved from yeasts through humans (reviewed by (41)). Although human kinesin-8 members, KIF18A and KIF18B, appear to be phosphorylated on Cdk1 consensus sites (42–44), to our knowledge, regulation of kinesin-8s by Cdk1 has not been reported and it will be interesting to learn whether kinesin-8s are modulated by this kinase. The DASH complex is composed of ten subunits that localize to the outer kinetochore and mitotic spindle and plays an important role in capturing and maintaining attachments of kinetochores to spindle microtubules during chromosome segregation (reviewed in (45)). Phosphorylation of multiple DASH complex components by several different protein kinases is known to control their function (45). Enrichment of DASH complex components in Clp1-C286S-TAPs suggests that phosphorylation of the DASH complex may be regulated by Clp1.
Several transcription factors (TFs) were enriched in Clp1-C286S-TAPs (Fig. 2 and supplemental Table S3). One prominent TF is Sep1, a component of pombe cell cycle box binding factor (PBF) complex, which belongs to the conserved family of forkhead transcription factors and regulates the first wave of gene transcription important for mitosis and cytokinesis (46–48). Among Sep1's targets is Ace2, another TF controlling S. pombe cell separation (48, 49). Several phosphorylation sites matching the minimal Cdk1 consensus were identified in Sep1 (Fig. 3, supplemental Tables S4–S6), and although post-transcriptional regulation of Sep1 by Cdk1 has not been reported, it is possible that Clp1 assists in reducing Sep1 activity during anaphase, fine tuning the window of its activation.
Another group of putative Clp1 substrates, Res1, Res2, and Cdc10, are all Mlu1-binding factor (MBF) TF complex subunits (Fig. 2 and supplemental Table S3). The MBF complex activates gene expression important for S phase (50). Nrm1 and Yox1, both of which are negative regulators of MBF activity and repress MBF-regulated promoters outside of G1-S phase (51, 52), are also enriched in Clp1-C286S-TAPs (supplemental Table S3). Thus, Clp1 might associate with the entire complex through an interaction with one or more of its members. Indeed, we detected Cdk1 consensus sites in Cdc10, Res1, and Nrm1 (Fig. 3) and several components of MBF have been implicated as Cdk1 targets. For example, phosphorylation of S196 on Cdc10, mediated by Cdk1, is required for formation of the Cdc10-Res1 complex (53). The cyclin Cig2 binds to Res2 and promotes Res1 phosphorylation at S130, which in turn inhibits MBF-dependent gene transcription (54). Thus, in addition to potential regulation of the M-G1 TF Mbx1 (55) and Sep1 (described above), Clp1 may also regulate the phosphorylation status and activity of the MBF TF complex.
Many other proteins involved in regulation of cytokinesis and septation were identified as potential Clp1 substrates. In addition to the F-BAR protein Cdc15, multiple cytokinetic ring proteins were identified including IQGAP Rng2, myosin II regulatory light chain Cdc4, myosin II heavy chain Myp2, and Cyk3 (Fig. 2 and supplemental Table S3). Interestingly, Cdc11, an essential scaffolding component of the Septation Initiation Network (SIN) kinase cascade that regulates the onset of cell division in S. pombe, was identified as a putative Clp1 substrate (56). Furthermore, Cdc11 is phosphorylated at Cdk1 consensus sites (reference (57), Fig. 3 and supplemental Tables S4–S6).
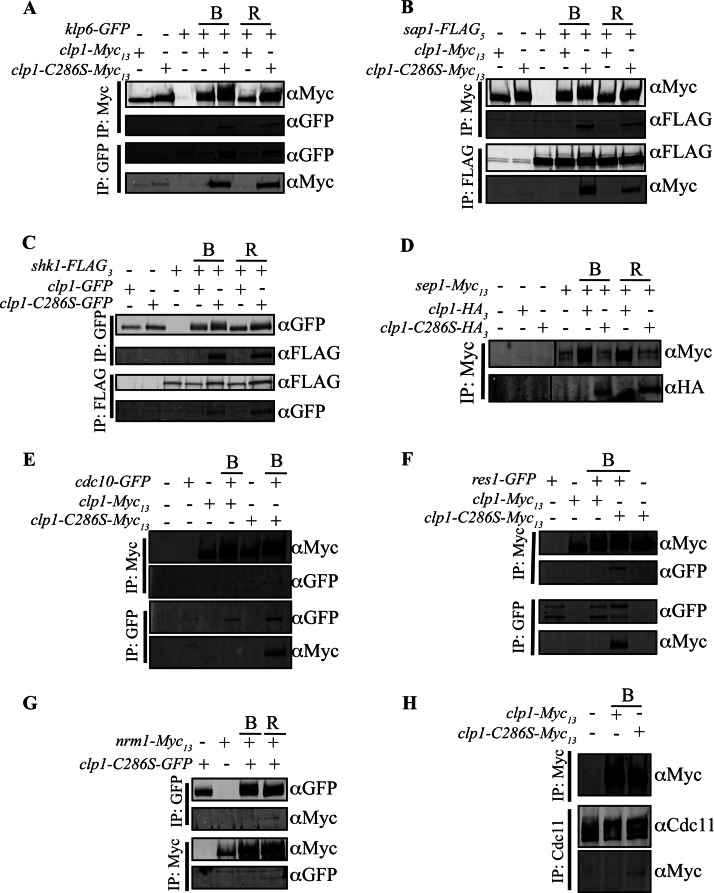
To validate our proteomic strategy, we chose an array of Clp1 interactors and confirmed their association with Clp1 by standard co-immunoprecipitation and determined if they are bona fide Clp1 substrates using phosphatase assays. In each case, Clp1-C286S co-immunoprecipitated the protein and, as expected from our MS analysis, these potential substrates co-immunoprecipitated more readily, or exclusively, with Clp1-C286S compared with wild-type Clp1 (Fig. 4). To test directly whether these proteins were Clp1 substrates, several candidates were isolated from clp1Δ cells arrested in mitosis, and treated with recombinant Clp1, catalytically inactive Clp1-C286S, or a nonspecific phosphatase. In all cases, Clp1 reduced or abolished the mobility shift due to phosphorylation (Fig. 5). In addition, the vast majority of Cdk1consensus phosphorylation sites detected in Clp1 interactors were identified in the C286S mutant TAP (supplemental Tables S4–S6), both confirming disruption of Clp1 activity in this mutant and highlighting its preference for Cdk1-phosphorylated substrates. Taken together, these results support the conclusion that the Clp1-C286S purifications are enriched in Clp1 substrates.
Fig. 4.
Co-immunoprecipitation analysis of potential Clp1 substrates. (A–H) Prior to immunoprecipitation, cells producing the indicated tagged proteins or appropriate control strains were either grown asynchronously, blocked in prometaphase (denoted B) using the cold sensitive β-tubulin mutant, nda3-KM311, or released (denoted R) from the arrest for 30 min., allowing cells to progress into anaphase when Clp1 is most active. Immunoprecipitates were then immunoblotted to detect associations with Clp1 and/or Clp1-C286S.
Fig. 5.
Clp1 dephosphorylates the candidate substrates. (A–H) The indicated substrates were immunoprecipitated from clp1Δ cells arrested in mitosis using the cold sensitive β-tubulin mutant, nda3-KM311, and then treated with H2O (mock treatment), recombinant MBP-Clp1, recombinant MBP-Clp1-C286S, or λ-phosphatase. The SDS-PAGE mobilities of the treated immunoprecipitates were then analyzed by immunoblotting. Phos-tag was incorporated into the gels to allow phosphorylation-induced mobility shifts to be detected (see Experimental Procedures).
Characterization of Cdc11 Phosphoregulation by Cdk1 and Clp1
Cdc11, a SPB protein that is required to coordinate the activities of several signaling components of the SIN, was identified among the Clp1 substrates (Figs. 4H and 5H, supplemental Table S3). The phosphorylation state of Cdc11 is postulated to regulate SIN function and cytokinesis, although the detailed mechanism is unclear (30, 57–60). Interestingly, Cdc11 was previously found to be a target of a PP2A-family phosphatase complex termed the SIN Inhibitory Phosphatase (SIP) that controls SIN function, possibly by regulating Cdc11 phosphostate (60). To explore whether Clp1 and SIP cooperate in the control of cytokinesis, we compared the effect of these two phosphatases on Cdc11 phosphostatus.
Previously, we identified phosphorylation at RXXS sites on Cdc11 catalyzed by the terminal SIN kinase Sid2, and found that they contributed to an auto-amplification loop in the SIN (57). In addition, we have identified eight phosphosites on Cdc11 matching the minimal consensus sequence of Cdk1 (S/T-P) (30, 57). Cdk1 phosphorylates MBP-Cdc11 in vitro but not when these sites are mutated to alanines indicating that these eight sites are the only Cdk1 sites in the protein (30). To examine whether Cdk1 and/or Sid2 phosphorylation sites were targeted by Clp1 and/or the SIP, we compared a cdc11-S8A allele in which the eight Cdk1 sites were mutated to alanine (generated previously (30)), a cdc11-S8D allele in which the 8 Cdk1 sites were mutated to aspartates, a cdc11-S7A allele in which the seven Sid2 sites have been mutated to alanine and a cdc11-S15A allele in which both Cdk1 and Sid2 sites were mutated to alanine. Each mutant was able to rescue cdc11::ura4+ cells indicating they retained essential function. Thus, they were integrated into the genome at the cdc11 locus by replacing cdc11::ura4+. In the following experiments, we used untagged and epitope tagged cdc11 alleles to evaluate the phosphostatus and function of the phosphomutants.
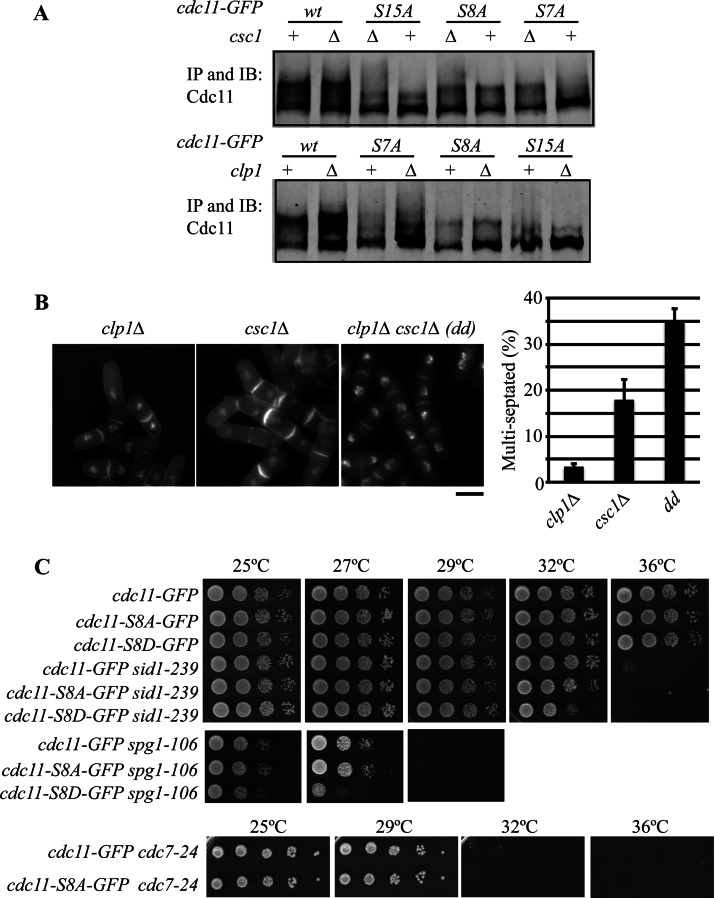
Cdc11 phosphorylation is detected as gel mobility shifts that increase during mitosis (57, 59, 60). The SDS-PAGE mobilities of Cdc11-GFP, Cdc11-S7A-GFP, Cdc11-S8A-GFP, and Cdc11-S15A-GFP were therefore examined in wild-type cells and in cells deleted for clp1 or an essential component of the SIP, csc1. As expected, mutating the Cdk1 sites, the Sid2 sites, or both, led to clear reductions in the SDS-PAGE mobility of Cdc11, consistent with these sites being phosphorylated in vivo (Fig. 6A). Ablation of Cdc11 Sid2 sites, but not the Cdk1 sites, reduced its SDS-PAGE mobility when clp1 was deleted (Fig. 6A), consistent with Clp1 specifically removing Cdk1 sites from Cdc11. In contrast, a fraction of Cdc11 migrated more slowly in the gel when the SIP component csc1 was deleted, even when both sets of phosphorylation sites were absent (Fig. 6A). These results suggest that the SIP does not remove either Cdk1 or Sid2 sites from Cdc11 and that yet another protein kinase contributes to Cdc11 phosphoregulation. Moreover, these data indicate that Clp1 affects Cdc11 function in a manner distinct from the PP2A SIP complex. Genetic evidence supporting this conclusion was obtained by examining the phenotype of the double mutant, clp1Δ csc1Δ. Whereas 3% of clp1Δ cells and 18% of csc1Δ cells failed cytokinesis, that number significantly increased to 35% in the double mutant at 36 °C (Fig. 6B).
Fig. 6.
Role of Cdc11 dephosphorylation by Clp1. A, Cdc11-GFP proteins were immunoprecipitated from the indicated strains and their SDS-PAGE mobilities were determined by immunoblotting with anti-Cdc11 serum. All retarded gel migration of Cdc11 is because of phosphorylation (57, 59). B, The indicated strains were grown at 36 °C in YE, fixed and stained with DAPI and methyl blue to visualize nuclei and cell wall, respectively (left panel). Bar, 5 μm. The number of multiseptated cells was determined and plotted (right panel). dd, double delete and error bars represent standard deviations of the mean. C, The indicated strains were grown to mid-log phase at 25 °C in YE, spotted in 10-fold serial dilutions on YE plates, and incubated at the indicated temperatures.
To assay whether Cdc11 phosphoregulation at Cdk1/Clp1 sites affects SIN signaling and cytokinesis, we took a genetic approach. We found that the cdc11-S8A-GFP mutation had no effect on the growth of sid1–239, spg1–106, and cdc7–24 mutants, which are loss of function mutants in the SIN (Fig. 6C). This is consistent with the idea that Cdk1 phosphorylation does not promote SIN signaling. In contrast, the phosphomimetic cdc11-S8D-GFP mutation exacerbated defects in SIN signaling, reducing the restrictive temperature of sid1–239 and spg1–106 and being synthetically lethal with cdc7–24 (Fig. 6C and data not shown). These results indicate that preventing Cdc11 dephosphorylation at Cdk1 sites results in a weak hypomorphic allele, consistent with the previously observed negative genetic interactions observed between clp1Δ and mutations of SIN genes (17).
Identification of Other Clp1 Interacting Proteins
We were next interested in determining whether we could identify proteins that copurified better with wild-type Clp1 than with Clp1-C286S (i.e., potential Clp1 regulators or cofactors). Based on normalized spectral counts, we identified proteins among the original 128 Clp1 interactors with more than twofold enrichment in wild-type Clp1 relative to Clp1-C286S complexes (supplemental Table S7). Although most proteins in this grouping were recovered at low levels, the top four proteins are noteworthy. First, the major hit in this category was Mid1, a protein that anchors the contractile ring in the middle of the cell (reviewed in (61)). Clp1 binds Mid1 at the medial cortex, allowing Clp1 access to a substrate at the cytokinetic ring, Cdc15 (11). Second, the transcription factor Mbx1 also fell into this category of proteins (supplemental Table S7), consistent with our previous finding that Mbx1 co-immunoprecipitated wild-type Clp1, but not Clp1-C286S (55). Lastly, the 14–3-3 proteins Rad24 and Rad25 were preferentially recovered with wild-type Clp1 compared with Clp1-C286S (supplemental Table S7). Phosphorylation of Clp1 by Sid2 or Cds1 provides docking sites for 14–3-3 proteins (RXXS), and 14–3-3 binding antagonizes Clp1 localization to the nucleolus (62–64). It is possible that because Clp1-C286S is constitutively bound to substrates, its association with Rad24 or Rad25 is partially inhibited.
Identification of the Sal3 Karopherin as the Mediator of Clp1 Nuclear Import
The regulation of Clp1 nucleoplasmic shuttling is critical for its function (64, 65) but neither the machinery at the nuclear pore, nor the sequences within Clp1 that regulate its nuclear-cytoplasmic distribution have been defined. The importin-β3, Sal3 (66) was prominent among those proteins with no apparent preference for either wild-type or Clp1- C286S (supplemental Table S8). Like other importins, Sal3 facilitates cargo transport through nuclear pore complexes (67, 68). Validating the MS results, we found that Clp1-Myc13 co-immunoprecipitated with Sal3-Flag3 (Fig. 7A) and so we pursued the implications of this association.
Fig. 7.
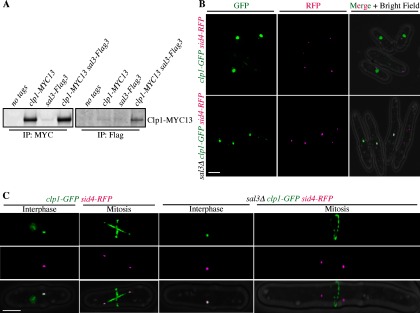
Characterization of Sal3 dependent Clp1 nuclear import. A, Clp1 and Sal3 co-immunoprecipitate. B, Live cell images of clp1-GFP sid4-RFP (Sid4 is a SPB marker, reference (83)) or clp1-GFP sid4-RFP sal3Δ cells. (C) same as (B) except single cells represent either interphase or mitosis as indicated.
Because sal3+ is not an essential gene, it was possible to evaluate Clp1-GFP localization in sal3Δ cells, although the cells are elongated because of a defect in Cdc25 nuclear import (66). Clp1-GFP was produced at normal levels in sal3Δ cells (supplemental Fig. S1) and was detected at SPBs during interphase (n = 186) and the cytokinetic ring during mitosis (n = 32) as expected (Figs. 7B and 7C). However, Clp1-GFP was absent from the nucleus in all stages of the cell cycle in sal3Δ cells, in contrast to wild-type cells (Figs. 7B and 7C). Thus, Sal3 is required for Clp1 nuclear import.
To confirm that Sal3 is the only importin involved in Clp1 nuclear import, Clp1-GFP localization was systematically examined in mutations of all other known S. pombe importins (69). There are seven importin-βs (Kap95, Kap104, Kap111, Kap113, Kap114, Kap123, and Sal3) and two importin-αs (Imp1 and Cut15) in S. pombe (69, 70). Unlike in sal3Δ cells, Clp1-GFP localized normally to the nucleus in the single deletions of the other nonessential importins (supplemental Fig. S2A), confirming that none of them affect Clp1 nucleocytoplasmic shuttling. Clp1-GFP also localized normally in the temperature-sensitive mutant of the essential importin Cut15 (supplemental Fig. S2B). Finally, because Kap95 depends on both importin-αs as adaptors for cargo selection (67), we examined Clp1-GFP localization in a double mutant, imp1Δ cut15–122, that abolishes the activity of both importin-αs at the nonpermissive temperature and found no change in Clp1 localization (supplemental Fig. S2C). Together, these data indicate that Sal3 alone controls Clp1's access to the nucleus.
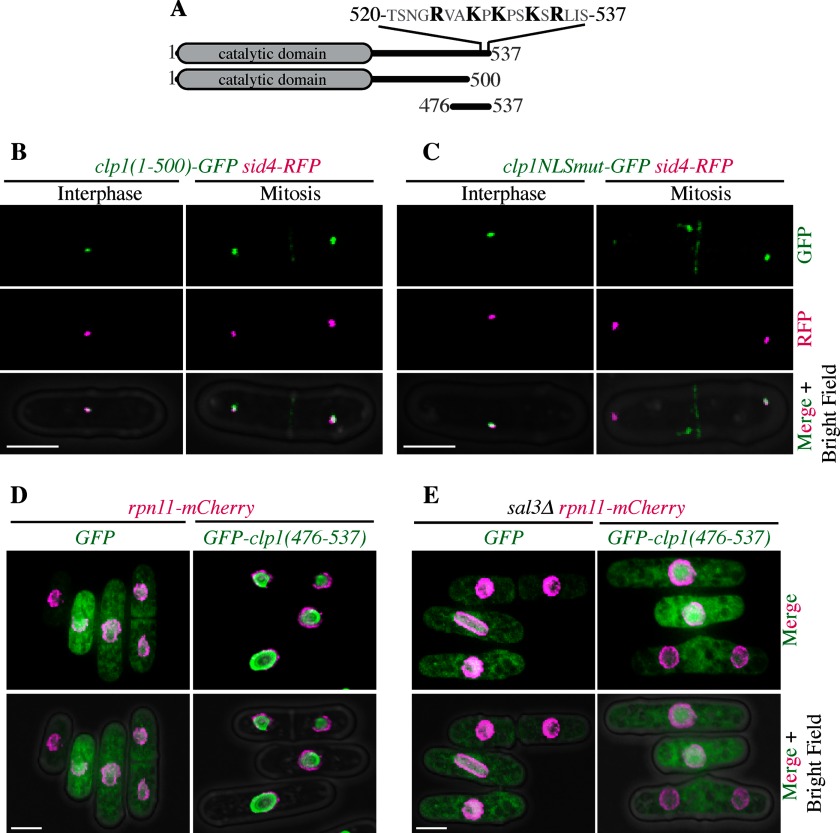
The importin-β3 family binds cargo nuclear localization sequences (NLSs) without the need for adaptor proteins (71). Thus, we next sought to identify Clp1's NLS that is recognized by Sal3. The NLS of S. cerevisiae Cdc14 is located near its C terminus (72). To determine if Clp1 shared this characteristic, we introduced a GFP::kanR cassette following amino acid 500 in the endogenous clp1+ open reading frame, eliminating the last 37 amino acids of the protein (Fig. 8A). Like Clp1-GFP in sal3Δ cells, Clp1(1–500)-GFP, was absent from the nucleus at all cell cycle stages (n = 134 interphase and 42 mitotic cells) although it maintained localization to SPBs during interphase and the cytokinetic ring during mitosis (Fig. 8B). This result suggests that Clp1's NLS is contained in the last 37 amino acids.
Fig. 8.
Identification of a functional Clp1 NLS. A, Schematic of Clp1, Clp1 truncations, and Clp1 mutations. The C-terminal 17 amino acids that contain the stretch of basic residues mutated to alanines (highlighted in black) in Clp1NLSmut are shown above full length Clp1. B, Representative live cell images of clp1(1–500)-GFP sid4-RFP cells in either interphase or mitosis. C, Same as (B) but images are of clp1NLSmut-GFP sid4-RFP cells. D, and E, Representative live cells images of rpn11-mCherry (D) (Rpn11 is a marker of the nuclear envelope (84)) or sal3Δ rpn11-mCherry (E) cells expressing GFP or GFP-clp1(476–537).
A typical NLS contains a string of basically charged amino acids (73) and we identified such a motif within amino acids 500–537 of Clp1 (Fig. 8A). To determine whether the five basic residues in this region contributed to the function of the Clp1 NLS, we replaced clp1+ with a mutant version, Clp1NLSmut-GFP, in which R524, K527, K529, K532, and R534 were mutated to alanines. Indeed, Clp1NLSmut-GFP localized to the SPBs during interphase and the cytokinetic ring during mitosis, but was not observed in the nucleus (n = 117 interphase and 38 mitotic cells) (Fig. 8C), indicating that these basic amino acids are a crucial element of the Clp1 NLS. Like wild-type clp1+ and unlike clp1Δ, the clp1NLSmut-GFP mutant opposed the function of cdc2–33, indicating that it is enzymatically active. Moreover, unlike clp1Δ, it did not negatively interact with the cytokinesis mutant, cdc3–124, signifying that its cytoplasmic functions were intact (supplemental Fig. S3).
Finally, to determine if the C terminus of Clp1 is sufficient for nuclear localization, we fused GFP to the C-terminal amino acids of Clp1, residues 476–537 (Fig. 8A), and expressed this fusion in cells. Although GFP alone localized diffusely throughout the cell, GFP-(476–537) accumulated within the nucleus (Fig. 8D), indicating that the C terminus of Clp1 is both necessary and sufficient for nuclear localization. However, GFP-(476–537) localized diffusely in the cytoplasm of sal3Δ cells (Fig. 8E), consistent with the requirement of Sal3 for Clp1 nuclear import.
DISCUSSION
Taking advantage of the nonessential nature of the S. pombe Clp1/Cdc14 phosphatase and the power of comparative proteomics yielded a wealth of information. This analysis revealed the breadth of cellular processes potentially impacted by Clp1 including vesicular trafficking, transcriptional regulation, and ribosome biogenesis in addition to its known roles in mitotic exit and cytokinesis. Many novel Clp1/Cdc14 interactors and substrates were identified, including multiple TFs, Clp1's importin and a SIN scaffolding protein, expanding our understanding of the Cdc14 phosphatase family.
Importantly, we identified a role for Clp1 in regulation of the SIN through Cdc11. Clp1-mediated dephosphorylation of Cdc11 is important for proper SIN signaling, suggesting that proper timing of SIN activation is controlled by a balance between Cdk and Clp1 activity. Clp1 is itself a target of the terminal SIN kinase Sid2 and phosphorylation of Clp1 antagonizes its nucleolar sequestration, promoting Clp1's access to substrates (62, 64). Thus, Clp1 and Cdc11 form a positive feedback loop through Sid2. That is, Clp1 dephosphorylates Cdc11, which promotes Sid2 activation and Sid2 in turn phosphorylates Clp1, prolonging its access to Cdc11. Cdc11 has emerged as a regulatory hub for SIN signaling because it is regulated by Cdk1/Clp1, Sid2, SIP and other kinases and phosphatases (i.e., Cdc11 remains phosphorylated in the absence of Cdk1 and Sid2 phosphorylation sites), adding to our understanding of the complexity of SIN phosphoregulation and regulatory feedback.
The cytokinesis-promoting SIN kinase cascade is cross-regulated by the morphogenesis-related NDR kinase network (MOR) which induces polarized growth and morphogenesis in interphase (74). Shk1, one of Clp1's putative substrates, is a PAK kinase and component of the MOR that is localized to the site of cell division. Shk1 (aka Pak1 or Orb2) phosphorylates the myosin II regulatory light chain, Rlc1 and inhibits cytokinesis (75). Dephosphorylation of Shk1 by Clp1 may result in Shk1 relocalization and/or inactivation, thereby promoting cytokinesis. Another potential Clp1 substrate, Sts5 (aka Orb4), is also involved in S. pombe cell morphology and localizes to the cell division site in close proximity to Clp1. Thus, it's possible that Clp1 is part of a redundant system of MOR inhibition during cytokinesis, contributing to the robustness of S. pombe cell morphology.
Although Clp1 has mainly been implicated in mitotic exit and cytokinesis, it appears to have a significant role in transcriptional regulation as well. This role may be related to that of human Cdc14B, which has also been implicated in the repression of cell cycle transcription (76). Clp1 may regulate Sep1 (mitosis/cytokinesis) and multiple components of the MBF transcription factor complex (50) as well as Mbx1 (55). Sep1 is essential for cell separation, and regulation by Cdk1/Clp1 may be important for its temporal activation. Similarly, perhaps dephosphorylation of MBF components or their associated inhibitors provides part of the unidirectional cell cycle engine. It will be exciting to probe the role of Clp1 in global transcriptional regulation.
Finally, a new and speculative mode of Clp1 regulation may evolve from our analysis because we identified Clp1 binding partners that appear to associate better with wild-type Clp1 than with Clp1-C286S, suggesting they are not Clp1 substrates. In budding yeast, Net1, the Cdc14 nucleolar tether, binds to and inhibits Cdc14 activity (77–79), a clear example of negative regulation by binding partners. It is possible that certain proteins bind Clp1 at or near the substrate-binding site, inhibiting substrate interactions. Our analysis revealed a few candidates—Mid1, an anillin like protein, which is necessary for Clp1 recruitment to the contractile apparatus (11), and the 14–3-3 proteins Rad24 and Rad25. Speculatively, Mid1 might recruit to and inhibit Clp1 at the cytokinetic ring and then release Clp1 allowing localized access to its ring substrates as Mid1 disappears from the ring and ring constriction proceeds. Alternatively, Mid1 may not inhibit Clp1, but simply be its tether to the cytokinetic ring. On activation of the SIN or the DNA damage checkpoint, Clp1 is released from the nucleolus and phosphorylated at RXXS sites, which facilitates Clp1's interaction with Rad24 and Rad25 and retention in the nucleoplasm (62–64, 80). Thus, 14–3-3 proteins may regulate Clp1 localization, but may also prevent its function (i.e. wild-type Clp1-TAP contains more Rad24 and Rad25 than Clp1-C286S). Future studies are needed to test these hypotheses.
Supplementary Material
Acknowledgments
We thank Drs. Paul Young and David Balasubramanian for providing the importin deletion strains used in this study, and Dr. Shelley Sazer for the cut15–122 mutant and helpful discussions.
Footnotes
* This work was supported by the Howard Hughes Medical Institute of which K.L.G. is an Investigator. M.R.B. was supported by the Biochemical and Chemical Training for Cancer Research program N.I.H. T32 CA009582. J.R.M. was supported by NCI T32CA119925.
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S8.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S8.
1 The abbreviations used are:
- SPB
- Spindle pole body
- Cdk1
- Cyclin-dependent kinase 1
- TAP
- Tandem affinity purification
- FDR
- False discovery rate
- YE
- Yeast extract
- TF
- Transcription factor
- SIN
- Septation Initiation Network
- SIP
- SIN Inhibitory Phosphatase
- PP2A
- Protein Phosphatase 2A
- MOR
- Morphogenesis-related NDR kinase network
- NDR
- Nuclear Dbf2-related kinase
- MBF
- Mlu1-binding factor
- GO
- Gene Ontology.
REFERENCES
- 1. Stegmeier F., Amon A. (2004) Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38, 203–232 [DOI] [PubMed] [Google Scholar]
- 2. Mocciaro A., Schiebel E. (2010) Cdc14: a highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 123, 2867–2876 [DOI] [PubMed] [Google Scholar]
- 3. Gray C. H., Good V. M., Tonks N. K., Barford D. (2003) The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 22, 3524–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trinkle-Mulcahy L., Lamond A. I. (2006) Mitotic phosphatases: no longer silent partners. Curr. Opin. Cell Biol. 18, 623–631 [DOI] [PubMed] [Google Scholar]
- 5. Bouchoux C., Uhlmann F. (2011) A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell 147, 803–814 [DOI] [PubMed] [Google Scholar]
- 6. Bremmer S. C., Hall H., Martinez J. S., Eissler C. L., Hinrichsen T. H., Rossie S., Parker L. L., Hall M. C., Charbonneau H. (2012) Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J. Biol. Chem. 287, 1662–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C. T., Peli-Gulli M. P., Simanis V., McCollum D. (2006) S. pombe FEAR protein orthologs are not required for release of Clp1/Flp1 phosphatase from the nucleolus during mitosis. J. Cell Sci. 119, 4462–4466 [DOI] [PubMed] [Google Scholar]
- 8. Esteban V., Blanco M., Cueille N., Simanis V., Moreno S., Bueno A. (2004) A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J. Cell Sci. 117, 2461–2468 [DOI] [PubMed] [Google Scholar]
- 9. Wolfe B. A., Gould K. L. (2004) Fission yeast Clp1p phosphatase affects G(2)/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 23, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trautmann S., Rajagopalan S., McCollum D. (2004) The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with aurora kinase. Dev. Cell. 7, 755–762 [DOI] [PubMed] [Google Scholar]
- 11. Clifford D. M., Wolfe B. A., Roberts-Galbraith R. H., McDonald W. H., Yates J. R., 3rd, Gould K. L. (2008) The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J. Cell Biol. 181, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu C., Ward J. J., Loiodice I., Velve-Casquillas G., Nedelec F. J., Tran P. T. (2009) Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell. 17, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi S. H., Péli-Gulli M. P., McLeod I., Sarkeshik A., Yates J. R., 3rd, Simanis V., McCollum D. (2009) Phosphorylation state defines discrete roles for monopolin in chromosome attachment and spindle elongation. Curr. Biol. 19, 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J. S., Lu L. X., Ohi M. D., Creamer K. M., English C., Partridge J. F., Ohi R., Gould K. L. (2011) Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation. J. Cell Biol. 195, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishra M., Karagiannis J., Trautmann S., Wang H., McCollum D., Balasubramanian M. K. (2004) The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J. Cell Sci. 117, 3897–3910 [DOI] [PubMed] [Google Scholar]
- 16. Trautmann S., Wolfe B. A., Jorgensen P., Tyers M., Gould K. L., McCollum D. (2001) Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11, 931–940 [DOI] [PubMed] [Google Scholar]
- 17. Cueille N., Salimova E., Esteban V., Blanco M., Moreno S., Bueno A., Simanis V. (2001) Flp1, a fission yeast orthologue of the s. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114, 2649–2664 [DOI] [PubMed] [Google Scholar]
- 18. Gould K. L., Ren L., Feoktistova A. S., Jennings J. L., Link A. J. (2004) Tandem affinity purification and identification of protein complex components. Methods 33, 239–244 [DOI] [PubMed] [Google Scholar]
- 19. McDonald W. H., Ohi R., Miyamoto D. T., Mitchison T. J., Yates J. R., III (2002) Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectr. 219, 245–251 [Google Scholar]
- 20. Roberts-Galbraith R. H., Chen J. S., Wang J., Gould K. L. (2009) The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J. Cell Biol. 184, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Z. Q., Tabb D. L., Burden J., Chambers M. C., Cox M. B., Cantrell M. J., Ham A. J., Litton M. D., Oreto M. R., Schultz W. C., Sobecki S. M., Tsui T. Y., Wernke G. R., Liebler D. C. (2011) Supporting tool suite for production proteomics. Bioinformatics 27, 3214–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 23. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moreno S., Klar A., Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 25. Keeney J. B., Boeke J. D. (1994) Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics 136, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast 11, 355–360 [DOI] [PubMed] [Google Scholar]
- 27. Wach A. (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265 [DOI] [PubMed] [Google Scholar]
- 28. Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127–130 [DOI] [PubMed] [Google Scholar]
- 29. McDonald W. H., Ohi R., Smelkova N., Frendewey D., Gould K. L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP- containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19, 5352–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morrell J. L., Tomlin G. C., Rajagopalan S., Venkatram S., Feoktistova A. S., Tasto J. J., Mehta S., Jennings J. L., Link A., Balasubramanian M. K., Gould K. L. (2004) Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 14, 579–584 [DOI] [PubMed] [Google Scholar]
- 31. Yoon H. J., Feoktistova A., Chen J. S., Jennings J. L., Link A. J., Gould K. L. (2006) Role of Hcn1 and its phosphorylation in fission yeast anaphase-promoting complex/cyclosome function. J. Biol. Chem. 281, 32284–32293 [DOI] [PubMed] [Google Scholar]
- 32. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (tap) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 33. Wolfe B. A., McDonald W. H., Yates J. R., 3rd, Gould K. L. (2006) Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell. 11, 423–430 [DOI] [PubMed] [Google Scholar]
- 34. Tonks N. K., Neel B. G. (1996) From form to function: signaling by protein tyrosine phosphatases. Cell 87, 365–368 [DOI] [PubMed] [Google Scholar]
- 35. Hiraoka Y., Toda T., Yanagida M. (1984) The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39, 349–358 [DOI] [PubMed] [Google Scholar]
- 36. Clifford D. M., Chen C. T., Roberts R. H., Feoktistova A., Wolfe B. A., Chen J. S., McCollum D., Gould K. L. (2008) The role of Cdc14 phosphatases in the control of cell division. Biochem. Soc. Transactions 36, 436–438 [DOI] [PubMed] [Google Scholar]
- 37. Cook M., Tyers M. (2007) Size control goes global. Curr. Opin. Biotechnol. 18, 341–350 [DOI] [PubMed] [Google Scholar]
- 38. Prekeris R., Gould G. W. (2008) Breaking up is hard to do - membrane traffic in cytokinesis. J. Cell Sci. 121, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bohnert K. A., Chen J. S., Clifford D. M., Vander Kooi C. W., Gould K. L. (2009) A Link between aurora kinase and Clp1/Cdc14 regulation uncovered by the identification of a fission yeast borealin-like protein. Mol. Biol. Cell 20, 3646–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rumpf C., Cipak L., Schleiffer A., Pidoux A., Mechtler K., Tolić-Norrelykke I. M., Gregan J. (2010) Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle 9, 3997–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gardner M. K., Odde D. J., Bloom K. (2008) Kinesin-8 molecular motors: putting the brakes on chromosome oscillations. Trends Cell Biol. 18, 307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Nat. Acad. Sci. U. S. A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cantin G. T., Yi W., Lu B., Park S. K., Xu T., Lee J. D., Yates J. R., 3rd (2008) Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. Journal of proteome research. 7, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 44. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Keri G., Stemmann O., Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 45. Buttrick G. J., Millar J. B. (2011) Ringing the changes: emerging roles for DASH at the kinetochore-microtubule Interface. Chromosome Res. 19, 393–407 [DOI] [PubMed] [Google Scholar]
- 46. Rustici G., Mata J., Kivinen K., Lio P., Penkett C. J., Burns G., Hayles J., Brazma A., Nurse P., Bähler J. (2004) Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36, 809–817 [DOI] [PubMed] [Google Scholar]
- 47. Peng X., Karuturi R. K., Miller L. D., Lin K., Jia Y., Kondu P., Wang L., Wong L. S., Liu E. T., Balasubramanian M. K., Liu J. (2005) Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell. 16, 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alonso-Nunez M. L., An H., Martin-Cuadrado A. B., Mehta S., Petit C., Sipiczki M., Del Rey F., Gould K. L., Vazquez de Aldana C. R. (2005) Ace2p Controls the Expression of Genes Required for Cell Separation in Schizosaccharomyces pombe. Mol. Biol. Cell 16, 2003–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin-Cuadrado A. B., Duenas E., Sipiczki M., De Aldana C. R., Del Rey F. (2003) The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 116, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 50. Bähler J. (2005) Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 39, 69–94 [DOI] [PubMed] [Google Scholar]
- 51. de Bruin R. A., Kalashnikova T. I., Chahwan C., McDonald W. H., Wohlschlegel J., Yates J., 3rd, Russell P., Wittenberg C. (2006) Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23, 483–496 [DOI] [PubMed] [Google Scholar]
- 52. Aligianni S., Lackner D. H., Klier S., Rustici G., Wilhelm B. T., Marguerat S., Codlin S., Brazma A., de Bruin R. A., Bähler J. (2009) The fission yeast homeodomain protein Yox1p binds to MBF and confines MBF-dependent cell-cycle transcription to G1-S via negative feedback. PLoS Gen. 5, e1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Connolly T., Caligiuri M., Beach D. (1997) The Cdc2 protein kinase controls Cdc10/Sct1 complex formation. Mol. Biol. Cell. 8, 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ayté J., Schweitzer C., Zarzov P., Nurse P., DeCaprio J. A. (2001) Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat Cell Biol. 3, 1043–1050 [DOI] [PubMed] [Google Scholar]
- 55. Papadopoulou K., Chen J. S., Mead E., Feoktistova A., Petit C., Agarwal M., Jamal M., Malik A., Spanos A., Sedgwick S. G., Karagiannis J., Balasubramanian M. K., Gould K. L., McInerny C. J. (2010) Regulation of cell cycle-specific gene expression in fission yeast by the Cdc14p-like phosphatase Clp1p. J. Cell Sci. 123, 4374–4381 [DOI] [PubMed] [Google Scholar]
- 56. Johnson A. E., McCollum D., Gould K. L. (2012) Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton 69, 686–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feoktistova A., Morrell-Falvey J., Chen J. S., Singh N. S., Balasubramanian M. K., Gould K. L. (2012) The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol. Biol. Cell 23, 1636–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krapp A., Cano E., Simanis V. (2004) Analysis of the S. pombe signalling scaffold protein Cdc11p reveals an essential role for the N-terminal domain in SIN signalling. FEBS Lett. 565, 176–180 [DOI] [PubMed] [Google Scholar]
- 59. Krapp A., Cano E., Simanis V. (2003) Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr. Biol. 13, 168–172 [DOI] [PubMed] [Google Scholar]
- 60. Singh N. S., Shao N., McLean J. R., Sevugan M., Ren L., Chew T. G., Bimbo A., Sharma R., Tang X., Gould K. L., Balasubramanian M. K. (2011) SIN-Inhibitory Phosphatase Complex Promotes Cdc11p Dephosphorylation and Propagates SIN Asymmetry in Fission Yeast. Current Biol. CB 21, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roberts-Galbraith R. H., Gould K. L. (2008) Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 22, 3082–3088 [DOI] [PubMed] [Google Scholar]
- 62. Chen C. T., Feoktistova A., Chen J. S., Shim Y. S., Clifford D. M., Gould K. L., McCollum D. (2008) The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr. Biol. 18, 1594–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Diaz-Cuervo H., Bueno A. (2008) Cds1 controls the release of Cdc14-like phosphatase Flp1 from the nucleolus to drive full activation of the checkpoint response to replication stress in fission yeast. Mol. Biol. Cell. 19, 2488–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mishra M., Karagiannis J., Sevugan M., Singh P., Balasubramanian M. K. (2005) The 14–3-3 protein rad24p modulates function of the cdc14p family phosphatase clp1p/flp1p in fission yeast. Curr. Biol. 15, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 65. Trautmann S., McCollum D. (2005) Distinct nuclear and cytoplasmic functions of the S. pombe Cdc14-like phosphatase Clp1p/Flp1p and a role for nuclear shuttling in its regulation. Curr. Biol. 15, 1384–1389 [DOI] [PubMed] [Google Scholar]
- 66. Chua G., Lingner C., Frazer C., Young P. G. (2002) The sal3(+) gene encodes an importin-beta implicated in the nuclear import of Cdc25 in Schizosaccharomyces pombe. Genetics 162, 689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chook Y. M., Blobel G. (2001) Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 [DOI] [PubMed] [Google Scholar]
- 68. Mosammaparast N., Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 69. Chen X. Q., Du X., Liu J., Balasubramanian M. K., Balasundaram D. (2004) Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast 21, 495–509 [DOI] [PubMed] [Google Scholar]
- 70. Umeda M., Izaddoost S., Cushman I., Moore M. S., Sazer S. (2005) The fission yeast Schizosaccharomyces pombe has two importin-alpha proteins, Imp1p and Cut15p, which have common and unique functions in nucleocytoplasmic transport and cell cycle progression. Genetics 171, 7–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mosammaparast N., Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 72. Mohl D. A., Huddleston M. J., Collingwood T. S., Annan R. S., Deshaies R. J. (2009) Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 184, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gupta S., McCollum D. (2011) Crosstalk between NDR kinase pathways coordinates cell cycle dependent actin rearrangements. Cell Div. 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Loo T. H., Balasubramanian M. (2008) Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J. Cell Biol. 183, 785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guillamot M., Manchado E., Chiesa M., Gomez-Lopez G., Pisano D. G., Sacristán M. P., Malumbres M. (2011) Cdc14b regulates mammalian RNA polymerase II and represses cell cycle transcription. Sci. Rep. 1, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Charbonneau H., Deshaies R. J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 97, 233–244 [DOI] [PubMed] [Google Scholar]
- 78. Traverso E. E., Baskerville C., Liu Y., Shou W., James P., Deshaies R. J., Charbonneau H. (2001) Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 276, 21924–21931 [DOI] [PubMed] [Google Scholar]
- 79. Visintin R., Hwang E. S., Amon A. (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398, 818–823 [DOI] [PubMed] [Google Scholar]
- 80. Broadus M. R., Gould K. L. (2012) Multiple protein kinases influence the redistribution of fission yeast Clp1/Cdc14 phosphatase on genotoxic stress. Mol. Biol. Cell 23, 4118–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wilson-Grady J. T., Villen J., Gygi S. P. (2008) Phosphoproteome analysis of fission yeast. J. Proteome Res. 7, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 82. Beltrao P., Trinidad J. C., Fiedler D., Roguev A., Lim W. A., Shokat K. M., Burlingame A. L., Krogan N. J. (2009) Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 7, e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chang L., Gould K. L. (2000) Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 5249–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilkinson C. R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J. P., McIntosh J. R., Gordon C. (1998) Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 17, 6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.