Abstract
Bacillus thuringiensis is a well-known entomopathogenic bacterium used worldwide as an environmentally compatible biopesticide. During sporulation, B. thuringiensis accumulates a large number of parasporal crystals consisting of insecticidal crystal proteins (ICPs) that can account for nearly 20–30% of the cell's dry weight. However, the metabolic regulation mechanisms of ICP synthesis remain to be elucidated. In this study, the combined efforts in transcriptomics and proteomics mainly uncovered the following 6 metabolic regulation mechanisms: (1) proteases and the amino acid metabolism (particularly, the branched-chain amino acids) became more active during sporulation; (2) stored poly-β-hydroxybutyrate and acetoin, together with some low-quality substances provided considerable carbon and energy sources for sporulation and parasporal crystal formation; (3) the pentose phosphate shunt demonstrated an interesting regulation mechanism involving gluconate when CT-43 cells were grown in GYS medium; (4) the tricarboxylic acid cycle was significantly modified during sporulation; (5) an obvious increase in the quantitative levels of enzymes and cytochromes involved in energy production via the electron transport system was observed; (6) most F0F1-ATPase subunits were remarkably up-regulated during sporulation. This study, for the first time, systematically reveals the metabolic regulation mechanisms involved in the supply of amino acids, carbon substances, and energy for B. thuringiensis spore and parasporal crystal formation at both the transcriptional and translational levels.
Bacillus thuringiensis is a well-known Gram-positive, endospore-forming and entomopathogenic bacterium (1, 2) consisting of more than 71 H serotypes and 84 serovars. One of the most significant features of B. thuringiensis is the formation of parasporal crystals during sporulation, which are typically comprised of several kinds of insecticidal crystal proteins (ICPs) (3). Moreover, different strains can produce various ICPs having specific insecticidal activity. To date, B. thuringiensis has been reported to produce over 760 kinds of ICPs that are classified into 72 Cry groups (723 kinds) and 3 Cyt groups (37 kinds) (http://www.lifesci.sussex.ac.uk/Home/Neil Crickmore/Bt/; November, 2012), which endow it with an extensive spectrum of insecticidal activity (4). Moreover, many studies have confirmed that B. thuringiensis is one of the safest microbial products known (5). Therefore, it is widely used as a biopesticide for agricultural and public health applications. More recently, some ICP genes have been successfully expressed in transgenic plants, leading to a higher yield and the lowered use of chemical pesticides (6).
Strikingly, the accumulation of ICPs can account for 20–30% of the cell's dry weight. Therefore, the intrinsic mechanisms of high-level ICP syntheses and parasporal crystal formation have drawn extensive attention of biologists. The high expression levels of ICP genes appear to result from regulatory mechanisms coordinately occurring at the transcriptional (7, 8), posttranscriptional (9), translational and posttranslational levels (10). However, the regulatory mechanisms underlying the metabolic pathways that provide amino acids, carbon sources, and energy for the high-level syntheses of ICPs remain unclear.
Our laboratory isolated B. thuringiensis subsp. chinensis CT-43 from China, which is highly toxic to lepidopterous and dipterous insects. The 6.15-Mb completely sequenced genome of CT-43 contains 11 replicons: a circular chromosome (5,486,830 bp) encoding 5529 open reading frames (ORFs), and 10 circular plasmids (pCT6880, pCT8252, pCT8513, pCT9547, pCT14, pCT51, pCT72, pCT83, pCT127, and pCT281, according to their sizes ranging from 6,880 to 281,231 bp) that totally encode 737 ORFs (11). Four ICP genes, cry1Aa3, cry1Ia14, cry2Aa9, and cry2Ab1, are encoded by the largest plasmid pCT281, and another ICP gene, cry1Ba1, is located on plasmid pCT127 (11). Our previous experiments demonstrated the parasporal crystal of CT-43 contains different kinds of ICPs. To reveal the metabolic regulation mechanisms accompanying the high-level expression of ICP genes and sporulation, transcriptomics and proteomics analyses of strain CT-43 were performed in different growth phases, using the Illumina method of high throughout cDNA sequencing (RNA-seq) and isobaric tags for relative and absolute quantitation (iTRAQ)1 technique, respectively. Our results, for the first time, systematically reveal the metabolic regulation mechanisms involved in the supply of the amino acids, carbon substances, and energy for spore and parasporal crystal formation at both the transcriptional and translational levels.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions
CT-43 cells were grown at 28 °C with shaking at 200 rpm in liquid GYS medium (12), comprised of (NH4)2SO4, 2 g; MgSO4·7H2O, 0.3 g; ZnSO4·7H2O, 0.005 g; MnSO4·4H2O, 0.05 g; CaCl2, 0.08 g; CuSO4·5H2O, 0.005g; FeSO4·7H2O, 0.0005g; K2HPO4, 0.5g; glucose, 1.0 g; and yeast extract 2.0 g/L (pH 7.4). Two biological replicate cell samples were collected by centrifugation (6000 × g, 5 min, 4 °C) at 7 h, 9 h, 13 h, and 22 h. Each sample was divided into two parts for whole-genome transcriptomics and proteomics analyses.
Quantitative Transcriptomics (RNA-seq)
RNA Isolation and mRNA Purification
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA) using the standard protocol. The final total RNA was dissolved in 200 μl RNase-free water. The concentration of total RNA was determined by NanoDrop (Thermo Scientific), and the RNA integrity value (RIN) was checked using RNA 6000 Pico LabChip of Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA).
Total RNA was incubated with 10 U DNase I (Ambion, Austin, TX) at 37 °C for 1 h, and then nuclease-free water was added to bring the sample volume to 250 μl. Messenger RNA was further purified by depleting ribosomal RNA and tRNA with TerminatorTM 5′-phosphate-dependent exonuclease (Epicenter, Madison, WI) digestion. The resulting RNA samples were quantified by the spectrophotometer DU800 (Beckman Coulter, Fullerton, CA).
cDNA Synthesis and Illumina Sequencing
Double-stranded cDNA was synthesized using a RNA-Seq Library Preparation Kit (Gnome Gen) and sequenced with an Illumina Genome Analyzer IIx according to manufacturers' protocols.
Bioinformatics Analysis
The reads of each sample were mapped to reference genome using BlastN with a threshold e value of 0.00001 and the “-F F” parameter (13), which allowed mapping of reads to the genome with up to two mismatches. Reads mapped to rRNA and reads not mapped under these parameters were excluded from further analysis. The number of reads mapped to each gene was recorded. Firstly, the read number of each gene was transformed into RPKM (Reads Per Kilo bases per Million reads) (14), and then differently expressed genes were identified by the DEGseq package using the MARS (MA-plot-based method with Random Sampling model) method (15). We used FDR ≤ 0.001 and an absolute value of log2Ratio ≥ 1 as the threshold to judge the significance of gene expression difference.
Quantitative Proteomics (Isobaric Tag for Relative and Absolute Quantitation, iTRAQ)
Protein Preparation and Reductive Alkylation
The harvested cells were washed three times with ice-cold phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 10.1 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4). The supernatant was discarded after the final centrifugation at 12,000 × g for 30 min. The cells were then resuspended in lysis buffer (7 m urea, 2 m thiourea, 4% w/v CHAPS, 20 mm TBP, and 0.2% Bio-lyte (pH 3–10)), containing a protease inhibitor mixture (Sigma) with a small amount of silica beads. The cells were first mechanically disrupted with disposable tissue grinding pestles for 5 min and then treated by ultrasonication (Sonics & Materials) for 10 min. DNase I and RNase A were added to the lysate at final concentrations of 1 mg/ml and 0.25 mg/ml, respectively, and the mixture was incubated on ice for 20 min. After cell disruption, the protein solution was separated from the cell debris by centrifugation (12,000 × g, 5 min, 4°C). The crude protein extracts were further purified using the ReadyPrep 2-D Cleanup Kit (Bio-Rad Laboratories, Hercules, CA). Next, the purified proteins underwent a reductive alkylation reaction. Finally, the protein concentration was determined using a 2-D Quant Kit (GE Healthcare).
Isobaric Labeling
Proteins (100 μg) from each sample were tryptically digested and labeled with 8-plex iTRAQ reagents (Applied Biosystems, Foster City, CA) as follows: 7 h-1, 113; 7 h-2, 114; 9 h-1, 115; 9 h-2, 116; 13 h-1, 117; 13 h-2, 118; 22 h-1, 119; and 22 h-2, 121. The labeled samples were pooled and resolved into 12 fractions using an Ultremex SCX column containing 5-μm particles (Phenomenex, Torrance, CA). The eluted fractions were then desalted using a Strata X C18 column (Phenomenex) and dried under vacuum. The final average peptide concentration in each fraction was about 0.25 μg/μl. Dried peptides were stored at -80 °C before MS analysis.
LC-MS/MS Analysis
A splitless nanoACQuity (Waters, Milford, MA) system coupled with Triple TOF was used for analytical separation. Microfluidic traps and nanofluidic columns packed with Symmetry C18 (5 μm, 180 μm × 20 mm) were utilized for online trapping, desalting, and nanofluidic columns packed with BEH130 C18 (1.7 μm, 100 μm × 100 mm) were employed in analytical separation. Solvents purchased from Thermo Fisher Scientific were composed of water/acetonitrile/formic acid (A: 98/2/0.1%; B: 2/98/0.1%). A portion of a 2.25 μg (9 μl) sample was loaded, and trapping and desalting were carried out at 2 μl/min for 15 min with 99% mobile phase A. At a flow rate of 300 nL/min, analytical separation was established by maintaining 5% B for 1 min. In the following 64 min, a linear gradient to 35% B occurred in 40 min. Following the peptide elution window, the gradient was increased to 80% B in 5 min and maintained for 5 min. Initial chromatographic conditions were restored in 2 min.
Data acquisition was performed with a Triple TOF 5600 System (AB SCIEX, USA) fitted with a Nanospray III source (AB SCIEX) and a pulled quartz tip as the emitter (New Objectives). Data were acquired using an ion spray voltage of 2.5 kV, curtain gas of 30 PSI, nebulizer gas of 15 PSI, and an interface heater temperature of 150 °C. The MS was operated with a RP ≥ 30,000 FWHM for the TOF MS scans. For information dependent acquisition (IDA), survey scans were acquired in 250 ms and as many as 30 product ion scans were collected if they exceeded a threshold of 120 counts per second (counts/s) with a 2+ to 5+ charge-state. The total cycle time was fixed to 3.3 s and the Q2 transmission window was 100 Da for 100%. Four time bins were summed for each scan at a pulser frequency value of 11 kHz through monitoring of the 40 GHz multichannel TDC detector with four-anode/channel detection. A sweeping collision energy setting of 35±5 eV coupled with iTRAQ adjust rolling collision energy was applied to all precursor ions for collision-induced dissociation. Dynamic exclusion was set for 1/2 of the peak width (18 s), and the precursor was then refreshed off the exclusion list.
Database Search and Quantification
The 2.3.02 version of Mascot software (Matrix Science, Boston, MA) was used to simultaneously identify and quantify proteins. In this version, only unique peptides used for protein quantification were chosen to quantify proteins more precisely. Searches were made against the B. thuringiensis CT-43 protein database (6266 sequences, including 5529 ORFs of the chromosome and 737 ORFs of the plasmids). Spectra from the 12 fractions were combined into one MGF (Mascot generic format) file after the raw data were loaded, and the MGF file was searched. The search parameters were: i) trypsin was chosen as the enzyme with one missed cleavage allowed; ii) the fixed modifications of carbamidomethylation were set as Cys, and variable modifications of oxidation as Met; iii) peptide tolerance was set as 0.05 Da, and MS/MS tolerance was set as 0.1 Da. The peptide charge was set as Mr, and monoisotopic mass was chosen. An automatic decoy database search strategy was employed to estimate the false discovery rate (FDR). The FDR was calculated as the false positive matches divided by the total matches. In the final search results, the FDR was less than 1.5%. The iTRAQ 8-plex was chosen for quantification during the search.
The search results were passed through additional filters before data exportation. For protein identification, the filters were set as follows: significance threshold P, 0.05 (with 95% confidence) and ion score or expected cutoff less than 0.05 (with 95% confidence). For protein quantitation, the filters were set as follows: “median” was chosen for the protein ratio type (http://www.matrixscience.com/help/quant_config_help.html); the minimum precursor charge was set to 2+ and minimum peptides were set to 2; only unique peptides were used to quantify proteins. The median intensities were set as normalization, and outliers were removed automatically. The peptide threshold was set as above for identity.
Accession Number
The RNA-seq data from this article are available as raw short read data in the National Center for Biotechnology Information's Gene Expression Omnibus under accession number GSE39479.
Mass spectrometry data from this article are deposited in the mzML format in the ProteomeExchange database (http://proteomexchange.org/) under accession number PXD000020 through the PRIDE website (http://www.ebi.ac.uk/pride/).
RESULTS AND DISCUSSION
The Growth Phases of Strain CT-43 Selected for the Omics Analyses
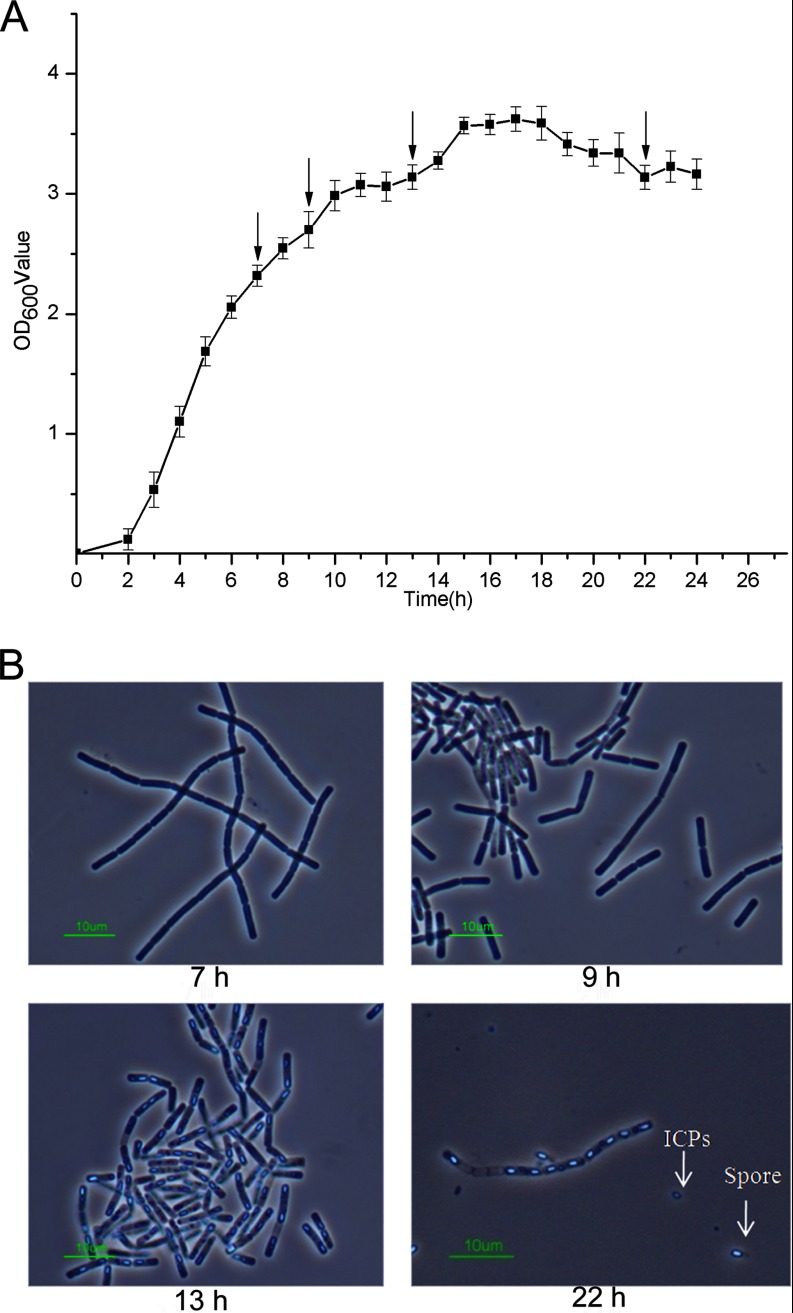
The life cycle of B. thuringiensis can be differentiated into two distinct stages: vegetative growth and sporulation. By measuring the optical density at 600 nm and observing bacterial cells under a phase contrast microscope (Nikon ECLIPSE E6000, Nikon Corp., Tokyo, Japan), the growth curve of strain CT-43 in GYS medium was obtained (Fig. 1A). The bacterial cells were collected separately at 7 h, 9 h, 13 h, and 22 h. The four time points were selected based on the following considerations: 1) 7 h represents the mid-exponential growth phase; 2) CT-43 enters the early-stationary growth phase at 9 h, and poly-β-hydroxybutyrate (PHB) particles begin to be clearly observable under a phase contrast microscope; 3) 13 h is the mid-stationary growth phase, with about 30% of cells undergoing sporulation; and 4) at 22 h, about 30% of mother cells are lysed, and some spores and parasporal crystals are released (Fig. 1B).
Fig. 1.
Growth features of strain CT-43. A, The growth curve of strain CT-43 grown in GYS. The y-axis presents the average optical densities of triplicate bacterial cultures at 600 nm at each time point. Data are averages of at least three independent experiments (error bars are S.E. from mean values).The four sampling points of 7 h, 9 h, 13 h, and 22 h for transcriptomics and proteomics are scaled out. B, Images of cell growth status at four time points of 7 h, 9 h, 13 h, and 22 h. The spore and parasporal crystal are marked. The scale bars represent 10 micrometers.
Summary of RNA-seq and iTRAQ Data
The transcriptome data were obtained by RNA-seq using the Illumina Genome Analyzer IIx sequencing platform. After omitting the low-scoring sequenced reads, the average length of the clean-reads was 110 nt, and the total numbers of clean-reads were 926,755, 1,096,665, 577,810 and 1,493,721 in the libraries at 7 h, 9 h, 13 h, and 22 h, respectively. Accordingly, the sequencing coverages of the four growth phases were 10- to 27-fold, reflecting a satisfactory sequencing depth of each sample. The clean-reads were mapped to the CT-43 genome using BlastN with a threshold e-value of 0.00001 and the “-F F” parameter. The number of unambiguously mapped reads per nucleotide was calculated and visualized by R and Origin version 8.0. The results indicated that the transcriptional percentages of the ORFs encoded by the CT-43 chromosome were 40.9%, 43.1%, 53.2%, and 17.7% for the four growth phases, respectively. These results suggested that transcriptional activity during the first three phases were more active but was severely inhibited at 22 h, which objectively reflected B. thuringiensis gene expression temporality. The gene expression level was normalized by converting the number of reads per gene into RPKM (Reads Per Kilo bases per Million reads), and the gene differential expression was analyzed using the DEGseq software package (supplemental Table S1).
A biological replicate sample was included in the iTRAQ experiment at each growth phase. After trypsinization and labeling with eight isobaric tags, the analytical separation and identification of the mixture composed of eight samples were performed by LC-MS/MS. In the eight samples, a total of 159,559 mass spectra were generated. After data filtering to exclude low-scoring spectra, 31,887 unique spectra that matched to special peptides were obtained. Through searching using Mascot version 2.3.02, a total of 8,251 peptides, 8,180 unique peptides, and the final 1,756 proteins were identified in the eight samples (supplemental Table S2). Gan et al. reported that a cutoff point at ±50% variation (±0.50) yields 88% coverage in quantification based on an analysis of biological replicates (16). In our iTRAQ data, the coverage levels of the four growth phases were between 93 and 96% when the cutoff point was at ±50% variation (supplemental Table S3). Therefore, there was better repeatability between the two biological replicates of each growth phase. Because iTRAQ quantification underestimated the amount of ‘real’ fold change between samples (17), a protein with ≥ 1.5-fold difference and a p value ≤ 0.05 was regarded as being differentially expressed in our data (18, 19) (supplemental Table S2). Our results showed that the number of differentially expressed proteins reached a peak in the temporal comparison of 7 h versus 22 h, followed by 9 h versus 22 h, 7 h versus 13 h, 13 h versus 22 h, and 7 h versus 9 h.
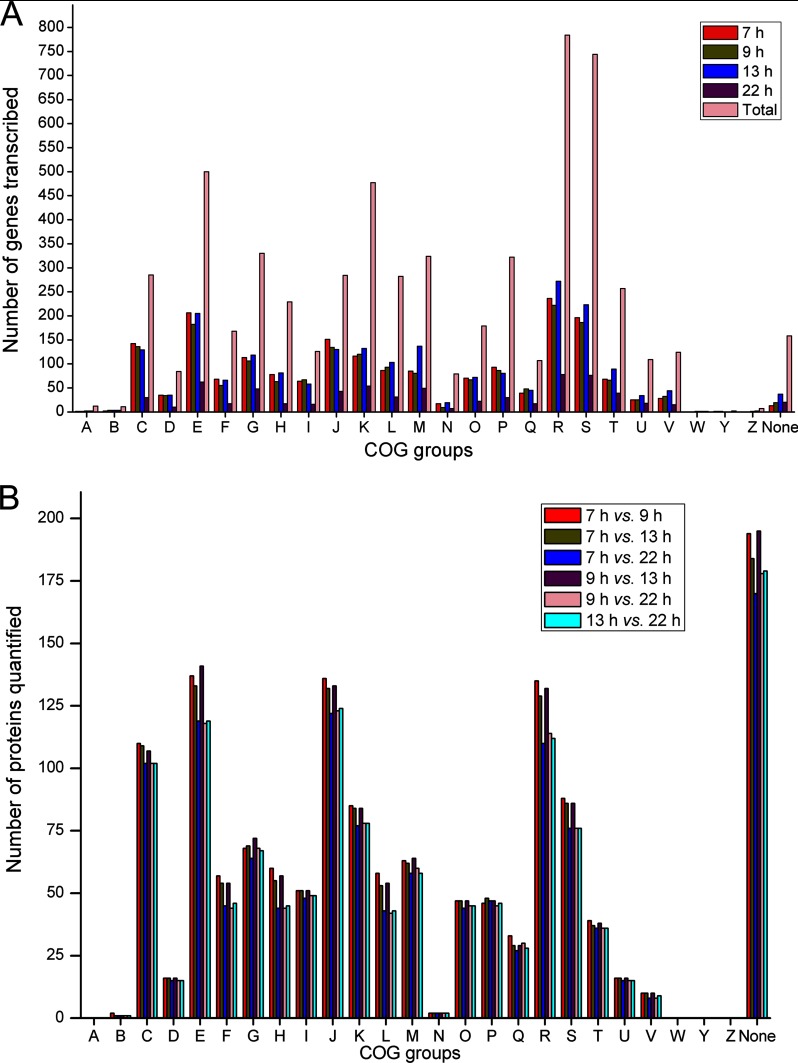
The expressed genes in each growth phase in the RNA-seq data and the quantified proteins in each temporal comparison in the iTRAQ data were subjected to Cluster of Orthologous Groups of proteins (COG) analyses. Using the results of 13 h as an example, at the transcriptional level, the order of the COG groups decided by expressed gene number was R (General function prediction only), E (Amino acid transport and metabolism), S (Function unknown), K (Transcription), J (Translation, ribosomal structure and biogenesis), G (Carbohydrate transport and metabolism), M (Cell envelope biogenesis, outer membrane), and C (Energy production and conversion) (Fig. 2A). For the quantified proteins in the temporal comparison of 7 h versus 13 h, the order was None (no COG information) R, E, J, C, K, S, and G (Fig. 2B). In terms of gene cellular functions, the two data sets were suitably consistent with each other. Collectively, these results suggested that some physiological metabolic activities, including amino acid transport and metabolism, transcription and translation, along with carbohydrate and energy metabolism, might be noticeably dynamic during sporulation, and truly reflected the regulation of the gene expression and metabolic pathways involved in sporulation and special ICP high-level expression.
Fig. 2.
COG analysis. A, Functional classification of the genes expressed at each time point in RNA-seq data. The “7 h, ” “9 h, ” “13 h, ” and “22 h ” indicate the numbers of the transcribed genes at each growth phase, whereas the “total ” represents the number of the total genes encoded by the CT-43 chromosome. B, Functional classification of the quantified proteins in each temporal comparison in iTRAQ data. The “7 h versus 9 h”, “7 h versus 13 h”, “7 h versus 22 h”, “9 h versus 13 h”, “9 h versus 22 h”, and “13 h versus 22 h ” represent the numbers of the quantified proteins in each temporal comparison. COG designations are described as follows: A, RNA processing and modification; B, Chromatin structure and dynamics; C, Energy production and conversion; D, Cell division and chromosome partitioning; E, Amino acid transport and metabolism; F, Nucleotide transport and metabolism; G, Carbohydrate transport and metabolism; H, Coenzyme metabolism; I, Lipid metabolism; J, Translation, ribosomal structure and biogenesis; K, Transcription; L, DNA replication, recombination, and repair; M, Cell envelope biogenesis, outer membrane; N, Cell motility and secretion; O, Posttranslational modification, protein turnover, chaperones; P, Inorganic ion transport and metabolism; Q, Secondary metabolite biosynthesis, transport, and catabolism; R, General function prediction only; S, Function unknown; T, Signal transduction mechanisms; U, Intracellular trafficking and secretion; V, Defense mechanisms; W, Extracellular structures; Y, Nuclear structure; Z, Cytoskeleton; None, No COG information.
The Correlation Between mRNA and Protein Expression Profiles
A coupled transcriptomics-proteomics project provides a unique opportunity to investigate how faithfully the transcriptional profile is manifested at the protein level. Therefore, we comprehensively investigated the correlation between mRNA and protein expression profiles in the temporal comparison of 7 h versus 9 h, 7 h versus 13 h, 7 h versus 22 h, 9 h versus 13 h, 9 h versus 22 h, and 13 h versus 22 h (supplemental Table S4). In each temporal comparison case, the quantified proteins could be clustered into seven groups based on the pattern of changes at the mRNA and protein levels: Group I, the mRNA and protein levels have the same change trends; Group II, the mRNA level is up-regulated but the protein level is down-regulated; Group III, the mRNA level is up-regulated but the protein level is not significantly changed; Group IV, the mRNA level remains almost unchanged while the protein level is down-regulated; Group V, the mRNA level remains almost unchanged but the protein level is up-regulated; Group VI, the mRNA level is down-regulated but the protein level is not significantly changed; Group VII, the mRNA level is down-regulated but the protein level is up-regulated (supplemental Fig. S1 and Table S4). Group I includes three subgroup: both the mRNA and protein levels are up-regulated synchronously; both the mRNA and protein levels are down-regulated synchronously; and both the mRNA and protein levels remain unchanged. Importantly, most quantified proteins belonged to the two main groups, Group I and Group VI, in all six temporal comparison cases (supplemental Fig. S1). Among the 1322 quantified proteins in the case of 7 h versus 13 h, the numbers of quantified proteins in the seven groups were: Group I, 529 (40%); Group II, 41 (3.1%); Group III, 193 (14.6%); Group IV, 25 (1.9%); Group V, 42 (3.2%); Group VI, 431 (32.6%); and Group VII, 61 (4.6%) (supplemental Fig. S1).
In principle, the more mRNA molecules are present in the cell, the more proteins can be synthesized. However, some studies in other species also found that the mRNA and protein changes of some genes were not correlated (20–22). There are several possible explanations, including: 1) although the nascent mRNA can be translated in prokaryotes, the protein production is still positively or negatively regulated at the post-transcriptional, translational, and post-translational levels; 2) generally, a protein possesses a longer half-life than mRNA; 3) the half-life of protein and/or mRNA would be different at various conditions (e.g. different growth phases). In a word, the lack of correlation between the mRNA and protein expression profiles might be attributed to the differential regulation at the mRNA and protein levels (20).
Supplement of Amino Acid Substances
Besides the myriad metabolism- and sporulation-associated proteins, many ICPs are synthesized during sporulation. Therefore, the sufficient provision of amino acids is a prerequisite for the production of these proteins. Generally, amino acids could be acquired by uptake from extracellular environment, de novo intracellular biosynthesis, and protein recycling (23, 24).
The CT-43 chromosome encodes at least 82 amino acid transport-associated genes. The CH1879∼1883 operon for branched-chain amino acid (BCAA, including isoleucine, leucine and valine) transport, the glnQHP operon for glutamine transport and the CH4808∼4810 operon for methionine transport were transcriptionally up-regulated at 13 h. Moreover, the proteins CH1879, GlnH and CH4809 were also increased by 4.2-, 7.1-, and 1.8-fold, and 9.2-, 12.3-, and 10.1-fold at 13 h and 22 h compared with 7 h, respectively (unless otherwise noted, “7 h” is hereafter used as the comparative object). Although the three genes in the CH4165∼4167 operon involved in arginine transport were markedly down-regulated at the transcriptional level at 13 h, CH4166 and CH4167 protein abundances were respectively elevated by 2.7- and 3.5-fold except that protein CH4165 could not be quantified. Two genes, CH0346 and CH0824, which encode cystine-binding proteins, displayed correspondingly increased abundances at both the mRNA and protein levels. CH0346 was transcriptionally up-regulated by 9-fold at 13 h, and its protein abundance was increased by 2.3- and 5.7-fold at 13 h and 22 h, respectively. Similarly, CH0824 was up-regulated by 7-fold at the mRNA level at 13 h and by 2.1-fold at the protein level at 22 h. Additional 14 genes for amino acid uptakes (including alanine, arginine, cystine, glutamate, lysine, proline, and threonine) were also transcriptionally up-regulated at 13 h (supplemental Table S1). Although direct amino acid uptake from the extracellular environment might be the most rapid and economical pathway, most amino acids in the GYS medium would likely be consumed during the vegetative growth phase. Accordingly, the amino acids in the extracellular environment during sporulation might come from degradation of extracellular proteins and release through cannibalism (25, 26). Furthermore, we found that CT-43 exhibited a significant ‘cannibalism’ phenomenon during sporulation (supplemental Fig. S2).
According to the KEGG Database, CT-43 carries complete biosynthesis pathways for all 20 common amino acids. Moreover, more than 300 genes participate in amino acid metabolism. Our RNA-seq results indicated that the ilvEBHC-leuABCD and ilvEBHCDA operons for BCAA biosynthesis (27) as well as the hisZGBHAFIEJ operon for histidine biosynthesis (28) were markedly up-regulated at 13 h (supplemental Table S1). Except for LeuC and LeuD, the remaining six enzymes in the ilvEBHC-leuABCD operon were increased by about 1.6- to 78.1-fold at the translational level during sporulation (supplemental Table S2). The increased expression levels of operons for both BCAA transport and biosynthesis were consistent with the fact that the BCAAs are the most abundant amino acids in both total 6266 proteins and five ICPs of CT-43 (Fig. 3). At the transcriptional level, six out of the 15 genes for lysine biosynthesis were up-regulated, and eight out of the 13 genes for methionine biosynthesis were increased at 13 h. The four asparagine synthetases (AsnA, AsnO1, AsnO2, and AsnO3) catalyzing the interconversion between aspartate and asparagine were all detected by iTRAQ. At 13 h, AsnO1 and AsnO2 were separately increased by 2.3- and 3.9-fold; AsnO3 was decreased by 2.4-fold whereas AsnA remained essentially constant. Additionally, at 13 h, transcription of some other genes for amino acid biosynthesis was specifically induced (e.g. the glutaminase gene glsA2, the pyrroline-5-carboxylate reductase gene proC1, the NADPH-dependent glutamate synthase gene CH2688, and the ferredoxin-dependent glutamate synthase gene CH3590) or up-regulated (including the threonine synthetase gene thrC, the l-serine dehydratase operon sdaAAB2, and the serine O-acetyltransferase gene cysE). Curiously, the transcripts of the trpEGDCFBA operon for tryptophan biosynthesis from chorismic acid (29) were initially detected at 13 h, perhaps because of the extremely low tryptophan content in both total 6,266 proteins and five ICPs of CT-43 (Fig. 3). These results demonstrated that some operons and genes were either induced or up-regulated in response to amino acid starvation during sporulation.
Fig. 3.
The contents of 20 common amino acids in strain CT-43. A, The contents of 20 common amino acids in a total of 6626 proteins encoded by one chromosome and 10 plasmids of CT-43. B, The contents of 20 common amino acids in five ICPs of CT-43. The number above each column represents the percentage of corresponding amino acid. The branched-chain amino acids (BCAAs), including isoleucine, leucine and valine, have very high percentages in both total 6626 proteins and five ICPs.
Nevertheless, the decreased expression of more genes involved in amino acid transport and biosynthesis may indicate that the amino acid sources from these two pathways are limited. Therefore, bacterial cells would need another way to fulfill their amino acid requirements during sporulation.
Indeed, a previous radioisotopic tracer experiment indicated that 80% of amino acids for ICP synthesis came from protein turnover (24). Accordingly, protein recycling would be the main source of amino acids during sporulation. The CT-43 chromosome encodes at least 69 proteases (not including those for spore germination such as CwlJ and SleB) and 49 peptidases (not including the 22 d-alanyl-d-alanine carboxypeptidases for peptidoglycan biosynthesis). In RNA-seq data, 42 genes encoding proteases were specifically induced or up-regulated at 13 h. Moreover, 24 proteases were detected by iTRAQ, of which 14 were up-regulated by 2.5- to 56.0-fold, and seven remained at similar levels at 13 h while the other three failed to be quantified. Among these, six ATP-dependent proteases (ClpB/C/P, HslU/U/V) were identified, of which four were up-regulated and two remained almost unchanged at 13 h (supplemental Table S2). During sporulation, some ATP-dependent proteases with high expression levels could not only rapidly degrade many abnormal polypeptides and various regulatory proteins to control protein quality and regulate many biological processes (30, 31), but could also provide a large number of amino acids. In particular, YabG (sporulation-specific protease), CH1954 (intracellular serine protease) and CH3928 (serine protease) were the most significantly up-regulated proteases, and they were increased by 16.4-, 9.5-, and 56.0-fold at 13 h, respectively. Among the 42 specifically induced or up-regulated proteases at the transcriptional level, NprB (bacillolysin), CalY (camelysin), thermitase (thermostable serine protease), and Vpr (a high-molecular-mass minor extracellular protease) were confirmed to perform important proteolysis functions (32–35). Consequently, abundant proteases with high activities could efficiently promote protein recycling to meet amino acid requirements during sporulation.
Supply of Carbon and Energy Sources
Sporulation and high-level syntheses of ICPs are biological processes that have high energy demands. Moreover, the sporulation process is initiated when nutrients are limited. Thus, where and how the energy is supplied for these biological processes is of interest.
Production and Reuse of PHB
During evolution, bacteria have developed various strategies to store carbon and energy substances. PHB is produced as an intracellular carbon and energy storage substance by a variety of bacteria (36–38) and a high PHB concentration can enhance the ICP yield (39). In a previous study, a linear relationship between the final ICP concentration and the maximum PHB concentration was observed (36). In our experiment, when CT-43 was grown in GYS medium, the intracellular PHB level began to quickly increase around 9 h and reached a maximum level at ∼17 h, whereupon it decreased rapidly (Fig. 4B). Under a phase contrast microscope, PHB granules were observable at 9 h; afterward, the sizes and numbers of these granules gradually increased in most cells and were visible in some sporulating cells even at 15 h (Fig. 4C). Recently, our laboratory found that both sporulation and parasporal crystal formation were seriously inhibited when PHB production was disturbed (40). As such, PHB metabolism plays vital roles in the processes of sporulation and ICP high-level expression in B. thuringiensis.
Fig. 4.

Biosynthesis and reuse of PHB in CT-43. A, The transcriptional level of the main genes associated with the synthesis and degradation of PHB. B, changes in the PHB level. C, PHB granules during different phases. Under a phase contrast microscope, CT-43 parasporal crystals are diamond- or spindle-shaped, whereas the PHB granules have an irregularly spherical shape. The level of intracellular PHB was measured as described previously (34). Data are averages of three independent experiments (error bars are S.E. from mean values). The photographs were obtained using a phase contrast microscope (Nikon ECLIPSE E6000). The PHB particles are marked. The scale bars represent 10 micrometers.
According to the KEGG database, two pathways are responsible for PHB synthesis from acetyl-CoA in B. thuringiensis (38, 42) (supplemental Fig. S3). In addition, some intermediates of fatty acid β-oxidation, and certain C2 and C4 compounds could flow into the PHB synthesis pathway through the three key nodal points: (R)-3-hydroxybutanoyl-CoA, acetyl-CoA, and crotonoyl-CoA, respectively (supplemental Fig. S3). The synthesized PHB is then assembled into visible PHB granules by phasins such as PhaP (38). As for the PHB degradation pathway, no ORF is annotated as a PHB depolymerase PhaZ in the genomes of B. thuringiensis strains or other sequenced Bacillus species (37). Interestingly, in vitro biochemical evidence identified PcaD (3-oxoadipate-enol-lactonase) from B. thuringiensis as a novel intracellular PHB depolymerase (37).
The RNA-seq data indicated that most PHB synthesis-associated genes were highly expressed at 7 h and 9 h, and the mRNA abundance of these genes was sharply reduced at 13 h; in contrast, transcription of the PHB degradation-associated genes pcaD (phaZ), scoT, and phbA1 was increased by about 3- to 15-fold at 13 h (Fig. 4A). At the translational level, among the three enzymes in the main PHB synthesis pathway, only PhbB was obviously decreased (3.1-fold) at 13 h, whereas PhbA (1.9-fold), PhbB (2.7-fold), and PhaC (2.0-fold) were all dramatically down-regulated at 22 h. Conversely, the protein PcaD (PhaZ) in the PHB degradation pathway was increased by 2.9-fold at 13 h. These results are in accordance with the changes in PHB concentration during the CT-43 life cycle (Figs. 4B and 4C). Notably, PhaP and PhaQ maintained high-level expression at both the transcriptional and translational levels at 13 h, indicating that these proteins possibly play important roles in both PHB granule assembly and disassembly.
Acetoin Metabolism
Acetoin (3-hydroxy-2-butanone) is produced and excreted by a number of fermentive bacteria during the exponential growth phase to prevent over-acidification of the cytoplasm and surrounding environment, and also acts as an extracellular carbon and energy store (43). Our RNA-seq data showed that the acetoin biosynthesis-associated genes alsS (α-acetolactate synthase) and alsD (α-acetolactate decarboxylase) (44) were expressed at high levels at 7 h, but were down-regulated by about threefold at 9 h, and were finally undetectable at 13 h. At the translational level, AlsS was decreased by 3.2- and 3.1-fold at 9 h and 13 h, respectively, while AlsD was detected by iTRAQ but could not be quantified. On the other hand, the acuABC operon encoding acetoin-reimporting proteins (45, 46) was markedly increased at the transcriptional level at 13 h, and another analogical operon ytrBCD was expressed slightly but still up-regulated at 13 h (supplemental Table S1). Meanwhile, the proteins AcuA/B/C were detected by iTRAQ and exhibited slight increases during the stationary growth phase. More importantly, the acoABCL operon encoding the acetoin dehydrogenase complex (47) was strongly up-regulated at both the transcriptional and translational levels at 13 h, likely to cleave reimported acetoin into acetaldehyde and acetyl-CoA (48). Among the three homologues (CH1215, CH2825 and CH3498) of dhaS (aldehyde dehydrogenase), CH1215 and CH3498 were obviously increased at the transcriptional level during the stationary growth phase, furthermore, the protein abundance of CH3498 was also increased by 3.1-fold at both 9 h and 13 h. Therefore, other than being directly converted into acetyl-CoA by ProA (bifunctional acetaldehyde-CoA/alcohol dehydrogenase), acetaldehyde would be sequentially transformed into acetyl-CoA by DhaS, AckA (acetate kinase), and EutD (phosphotransacetylase) (supplemental Fig. S3). Subsequently, the converted acetyl-CoA would enter the tricarboxylic acid (TCA) cycle for energy generation.
Low-quality Carbon and Energy Sources
In general, monosaccharides and disaccharides are preferentially utilized by bacteria as opposed to oligosaccharides and polysaccharides, which are less easily metabolized. Hence, we define the latter as low-quality carbon resources. The CT-43 chromosome encodes about 600 genes for the transport of various materials (not including 40 genes for those of oligopeptides). In RNA-seq data, many genes involved in monosaccharide and disaccharide transport were highly expressed at 7 h, and then remarkably down-regulated transcriptionally at 13 h, including ptsG (about three∼fivefold) and crr (about sevenfold) for glucose, terB (about sixfold) for trehalose, rbsB (about 31-fold) for ribose and nagE (about 33-fold) for N-acetylglucosamine. However, some genes for oligosaccharide and polysaccharide transport were specifically induced or up-regulated during sporulation (supplemental Table S1). For example, the CH2964–2961 operon for certain sugar transport was specifically induced at 13 h, and transcription of the malECD operon for cyclodextrin transport was up-regulated by about two∼sixfold at 13 h (supplemental Table S1). Particularly, the celABC3 operon (CH5241–5243) for lichenin transport was transcriptionally up-regulated by about 50∼100-fold. Moreover, the proteins CelA/B/C were increased by 4.2-, 2.2-, and 2.8-fold at 13 h, and 4.8-, 4.2-, and 2.8-fold at 22 h, respectively. Cooperatively, the amyS (cytoplasmic alpha-amylase) gene and the four glucosidase genes encoded by CT-43, including the 6-phospho-beta-glucosidase genes celF and glvA, the exo-alpha-1,4-glucosidase gene CH0357, and the oligo-1,6-glucosidase gene malL (49, 50), were transcriptionally up-regulated by about two∼threefold at 13 h. Furthermore, the proteins CelF and MalL were identified by iTRAQ, with CelF increased by 6.0- and 4.3-fold at 13 h and 22 h, respectively, whereas MalL remained almost unchanged. In addition, another 105 genes for the transport of unknown substances were specifically induced or up-regulated at 13 h at the transcriptional level (supplemental Table S1).
In addition, at the transcriptional level, the chitin transport-associated operons celABC and CH2369–2370 were specifically induced at 9 h and 13 h, respectively. Moreover, the chitin degradation-associated genes chi36 (exochitinase), CH0372 (endochitinase), CH2662 (chitosanase), and three pdaB (chitooligosaccharide deacetylase) genes (CH0148, CH1703, and CH3803) were specifically induced at 13 h. In iTRAQ data, CH2370 was increased by 4.7-fold at 13 h compared with 9 h, and CH0148 was up-regulated by 13.9-fold at 13 h compared with 7 h. Given that B. thuringiensis is an insect pathogen (1, 2) and chitin is a crucial component of insect cuticles, the expression features of these genes might reflect that the chitin can be naturally used by B. thuringiensis. Taken together, these results suggested that some low-quality carbon and energy sources that went unused during the exponential growth phase were fully used during sporulation.
Central Carbohydrate Metabolism
The glycolysis (Embden-Meyerhof-Pamas, EMP) pathway, pentose phosphate (PP) shunt, and TCA cycle constitute the central carbohydrate metabolism pathways to provide energy-yielding compounds and metabolic intermediates (51). Our results showed that a great majority of genes involved in the EMP pathway and TCA cycle were significantly down-regulated during sporulation at both the transcriptional and translational levels (supplemental Tables S1 and S2), implying an overall decrease in the activities of these pathways. Moreover, the global gene expression levels of the PP pathway were lower than those of the EMP pathway (supplemental Table S1). These results are in excellent agreement with a previous radiorespirometry experiment showing that 18 natural isolates of B. thuringiensis employed the EMP pathway (95%) almost exclusively for glucose metabolism, whereas the PP pathway played a minor role (5%) (12).
The Pentose Phosphate Shunt
The enzymes in the PP pathway (except Zwf, glucose-6-phosphate 1-dehydrogenase) were all identified by iTRAQ and remained almost unchanged during sporulation, which underscores the importance of the PP pathway in providing the reducing power (NADPH) and metabolic intermediates involved in many biosynthetic processes. In the PP pathway, the predominant route to arrive at the nodal point gluconate-6p is: 1) glucose is converted into glucose-6p; 2) the latter is converted into Glucono-1, 5-lactone-6p by Zwf; and 3) the intermediate is further transformed into gluconate-6p by 6-phosphogluconolactonase (CH3298). An alternative route is for glucose to be converted into gluconate by Gdh (glucose 1-dehydrogenase), and gluconate catalyzed into gluconate-6p by GntK (gluconokinase) (52) (supplemental Fig. S3). However, the key rate-limiting enzymes Zwf and Gdh in both routes were not detected during the exponential growth phase (7 h) at both the transcriptional and translational levels (supplemental Tables S1 and S2). The gene zwf was slightly induced at 9 h and then up-regulated at 13 h, while the gene gdh was initially induced at 13 h at the transcriptional level. Therefore, how does the PP pathway proceed during the exponential growth phase? These results could indicate a meaningful regulatory mechanism. When CT-43 cells were grown in GYS medium containing yeast extract, they selected a third route that relies on the gnt operon that participates in gluconate metabolism. In CT-43, the gnt operon (CH2189–2191) is composed of gntP (gluconate permease), gntK, and gntZ/gndA (6-phosphogluconate dehydrogenase), and lacks its own transcriptional regulator, which is comparable to the negative regulator gntR in B. subtilis (53). Our RNA-seq results showed that the gnt operon expression level reached a maximum at 7 h, and then gradually decreased. For bacteria, direct uptake of substances from the extracellular environment might be the most rapid and metabolically economical pathway. Consequently, extracellular gluconate is likely transported into the cells directly by GntP and converted into gluconate-6p by GntK, with gluconate-6p further catalyzed into ribulose-5p by GntZ/GndA (supplemental Fig. S3). Based on these results, we suggest that the zwf and gdh expression is repressed by the gluconate transported from the GYS medium, and induced when extracellular gluconate is depleted. Therefore, glucose does not enter into the PP pathway when the extracellular environment contains gluconate. Our results could thus satisfactorily explain the phenomenon observed by Nickerson and his coworkers in 1974 that 100% of glucose catabolism was through the EMP pathway when B. thurigiensis was cultured in GYS medium (although the PP pathway was not inactive), whereas the contribution of the PP pathway was still 5% in a glucose-glutamate-salts medium (12).
The EMP Pathway
During the exponential growth phase, abundant glucose flows into the EMP pathway to produce large amounts of pyruvate. According to the expression features of the associated genes revealed by the RNA-seq and iTRAQ data, pyruvate could be used to: 1) produce l-/d-alanine; 2) synthesize α-acetoacetate intermediates for BCAA and acetoin biosynthesis (41); 3) generate lactate catalyzed by three lactate dehydrogenases; and 4) yield acetyl-CoA catalyzed by the pyruvate dehydrogenase complex. Apparently, most pyruvate would be converted into acetyl-CoA. Besides entering the TCA cycle to yield energy and metabolic intermediates, quite a lot of acetyl-CoA would be utilized for fatty acid and PHB biosynthesis (36–38) (Fig. 4B and 4C, supplemental Fig. S3).
Because monosaccharides provided by the GYS medium could be exhausted during the exponential growth phase, glucose and other monosaccharides entering the EMP pathway during sporulation could come from the hydrolyzates of low-quality carbon sources, including glucosamine from chitin degradation and deacetylation, and glucose from lichenin cleavage (54, 55). Other than participating in energy metabolism and amino acid biosynthesis (particularly the BCAAs), during sporulation, pyruvate is also used for high-level synthesis of dipicolinic acid (∼25% of sporal core dry weight), which is important for spore germination and resistance (56). Consequently, pyruvate produced merely from the EMP pathway would be insufficient during sporulation. In response, the pdhABCD operon encoding the pyruvate dehydrogenase complex was transcriptionally down-regulated by more than 10-fold at 13 h. Moreover, the E1α (PdhA) and E1 β (PdhB) subunits were also translationally decreased at 13 h. On the other hand, oxaloacetate and lactate are likely important sources of pyruvate, because at 13 h: i) pckA (phosphoenolpyruvate carboxykinase) was transcriptionally increased by almost 5-fold; ii) pykA1 (an isoenzyme of pyruvate kinase) was specifically induced; and iii) an isoenzyme of lactate dehydrogenase (CH1875) was transcriptionally and translationally up-regulated by almost 6-fold and 1.5-fold, respectively. The iTRAQ data revealed an increase in the enzymes PrpB (methylisocitrate lyase), PrpC (2-methylcitrate synthase) and PrpD (2-methylcitrate dehydratase) of about 3∼9-fold at 13 h, so another source of pyruvate would be the propionyl-CoA yielded mainly from the β-oxidation of odd-chain length fatty acids via the methylcitrate cycle (57) (supplemental Fig. S3).
The TCA Cycle
Any mutant defective in the first three enzymes of the TCA cycle fails to express early sporulation genes, suggesting that the activities of these enzymes are critical for sporulation (51, 58). Conversely, α-ketoglutarate dehydrogenase, which catalyzes the fourth step of the TCA cycle, is not absolutely essential (59). During sporulation, considerable acetyl-CoA could be generated by pyruvate dehydrogenation, fatty acid β-oxidation, and the reuse of the abovementioned acetoin and PHB. Apparently, acetyl-CoA would mainly flow into the TCA cycle to yield energy. Combining our results with previous studies, we speculate that the TCA cycle is significantly modified or supplemented during sporulation as follows (supplemental Fig. S3):
The glyoxylate shunt bypasses a portion of the TCA cycle to convert isocitrate to malate (60). At 13 h, two glyoxylate shunt-specific genes aceA (isocitrate lyase) and aceB (malate synthase) were transcriptionally up-regulated by about 19- and sevenfold, respectively. Meanwhile, at the translational level, AceB was increased by 3.6- and 3.4-fold at 13 h and 22 h, respectively, whereas AceA was up-regulated by 14.1-fold at 13 h and, startlingly, by 2879.1-fold at 22 h. Therefore, these results implied that the glyoxylate shunt became more active during sporulation. As a result, the efficiencies of acetyl-CoA metabolism and energy generation would be greatly increased because the glyoxylate shunt consumed 2 molecules of acetyl-CoA per cycle.
The γ-aminobutyric acid (GABA) shunt is an additional supplement to the TCA cycle and was confirmed to be correlated with spore and parasporal crystal formation in B. thuringiensis (59, 61). GABA is synthesized through glutamate decarboxylation catalyzed by glutamate decarboxylase (59). However, we observed that the sole glutamate decarboxylase GadB (CH2716) identified so far in CT-43 was not expressed at any phase at either the mRNA or protein level, coinciding with a previous report that GABA production was relatively weak in Bacillus strains (62). Nevertheless, the mRNA of the GABA-specific permease gabP increased by about fivefold at 13 h, in agreement with an observation that the gabP gene was activated during nitrogen-limited growth (63). Meanwhile, the GABA degradation-associated enzymes GabD (succinate-semialdehyde dehydrogenase) and GabT (4-aminobutyrate aminotransferase) were transcriptionally up-regulated by about 3- and 20-fold and translationally increased by 2.9- and 3.0-fold at 13 h, respectively. These results suggest that GABA metabolism became more active during sporulation and the utilized GABA might mainly come from the extracellular environment.
The GABA shunt and the methylcitrate cycle are both interconnected with the TCA cycle at the same node point, succinate. This interconnection may lead to succinate accumulation, and therefore cause constitutive feedback inhibition of upstream reactions. Succinate could be further converted into fumarate by the succinate dehydrogenase complex SdhABC and succinyl-CoA by the succinyl-CoA synthetase complex SucCD. Of note, succinyl-CoA is the CoA donor involved in the onversion of acetoacetate to acetoacetyl-CoA in the PHB degrading pathway, and this process can accelerate PHB reuse. Indeed, SucC and SucD were just slightly decreased at 13 h as revealed by iTRAQ, which possibly implies that a significant amount of succinate could be adversely converted into succinyl-CoA during sporulation.
The final TCA cycle step is malate dehydrogenation to produce oxaloacetate. The gene citH (malate dehydrogenase) was transcriptionally down-regulated by about 142-fold, and the other two isoenzyme genes mqo (malate: quinone oxidoreductase) and malS (malate synthetase) were expressed at low levels and their transcription was down-regulated by about two and 26-fold at 13 h, respectively. Meanwhile, the enzymes Mqo and MalS were decreased by 1.9- and 2.1-fold at 13 h. Because the TCA cycle was markedly modified and supplemented (particularly, glyoxylate shunt) during sporulation, large amounts of malate would be produced. If this reaction was drastically inhibited, the energy yield and the processes of the entire TCA cycle as well as the glyoxylate and GABA shunts would have all been significantly affected. Interestingly, some studies confirmed that LeuB (3-isopropylmalate dehydrogenase) is a broad-specificity enzyme that can catalyze the oxidative decarboxylation of 3-methylmalate, as well as d- and l-malate in addition to its cognate substrate 3-isopropylmalate (64, 65). Moreover, the ilvEBHC-leuABCD operon was remarkably up-regulated at 13 h at the transcriptional level (Fig. 5A); meanwhile, the protein LeuB was also increased by 2.5-fold at 13 h. To reveal why the transcriptional level of the leuB gene was higher (about 2- to 10-fold) than those of other genes located in the operon at 13 h (Fig. 5B), the upstream regulatory region was searched visually and via DBTBS (http://dbtbs.hgc.jp/) within the region 400 nt upstream of the ORF leuB initiation codon ATG. The results showed that the recognition sequences of SigF, and SigL and its enhancer-binding protein (EBP) BkdR (66) were present within the 3′-end region of the ORF leuA located upstream of the ORF leuB (Fig. 5C). Considering that SigL was up-regulated at 13 h at both the transcriptional and translational levels (supplemental Tables S1 and S2), leuB was probably regulated by both SigL and SigF during sporulation in response to certain signals (possibly, the accumulation of malate). Collectively, LeuB would be likely used not only for BCAA biosynthesis, but also for malate dehydrogenation.
Fig. 5.

The expression features of the ilvEBHC-leuABCD operon and the leuB gene. A, The transcriptional level of the ilvEBHC-leuABCD operon at 7 h, 9 h, 13 h, and 22 h using the RPKM value of each gene as the y-axis. B, Frequencies of mapped bases of the ilvEBHC-leuABCD operon at 13 h. C, The upstream regulatory region within 400 nt upstream of the ORF leuB initiation codon ATG. The number of unambiguously mapped reads per nucleotide was calculated and visualized by R and Origin version 8.0. Each ORF is depicted with corresponding direction and length.
Oxidative Phosphorylation and Energy Generation
ATP plays a significant role in free-energy transduction in living cells. In addition to a small amount coming from substrate-level phosphorylation, most ATP molecules are synthesized by membrane-bound enzyme complexes through oxidative phosphorylation under aerobic conditions (67, 68). Complex I (NADH: quinone oxidoreductase) is the first and largest enzyme complex of the respiratory chain and plays an important role in cellular energy metabolism (69). Our results showed that the nuoA-N gene cluster encoding complex I was obviously up-regulated at the transcriptional level during sporulation. At the translational level, only the proteins NuoB/C/D/I in the nuoA-N gene cluster were detected, and NuoB and NuoC were seperately increased by 6.4- and 4.2-fold at 13 h. Complex II (succinate: quinone oxidoreductase) is the first enzyme complex of the respiratory branch chain (70). The sdhBAC operon encoding complex II was remarkably down-regulated at the transcriptional level at 13 h. However, the proteins encoded by the sdhBAC operon remained unchanged except that SdhA was decreased by 1.6-fold. The qcrABC operon (CH1449–1451) encoding complex III (menaquinol-cytochrome c reductase) reached the highest level at 9 h and maintained relatively high level throughout sporulation as revealed by RNA-seq. At the translational level, the proteins QcrA/B/C were increased by 3.0-, 2.6-, and 2.7-fold at 13 h, and 9.1-, 7.5-, and 7.2-fold at 22 h, respectively.
In Bacillus strains, the respiratory systems contain a quinol oxidase branch (with cytochrome bd, cytochrome aa3 or YthAB as its terminal oxidase) and a cytochrome oxidase branch (with cytochrome caa3 as its terminal oxidase) (71). At the transcriptional level, the qoxABCD operon encoding cytochrome aa3 was significantly down-regulated; the ythAB operon encoding a quinol oxidase was specifically expressed; and the ctaABCDEF operon encoding cytochrome caa3 reached a maximum level during the stationary growth phase. Meanwhile, the proteins CtaC/D/E/F for cytochrome caa3 were increased by 5.1-, 6.7-, 3.1-, and 7.1-fold at 13 h, respectively. Unexpectedly, the proteins QoxA/B/C for cytochrome aa3 remained almost unchanged at 13 h and were still increased by 2.2-, 1.9-, and 1.6-fold at 22 h, respectively. In addition, transctription of the cydABDC operon encoding cytochrome bd was greatly up-regulated (about two∼20-fold) at the transcriptional level at 13 h. Cytochromes P450 (P450s) are a broad class of heme b-containing mono-oxygenase enzymes involved in the oxidative degradation of various compounds (72). At the transcriptional level, two cytochrome P450 genes cypA and cypX were markedly up-regulated at 13 h and a NADPH-cytochrome P450 reductase gene cypD was specifically induced at 13 h. Moreover, the proteins CypA and CypX were separately increased by 1.6- and 1.8-fold at 13 h except that CypD could not be quantified. Integration of our results with those from previous biochemical studies in B. cereus indicated that there is a significant increase in the levels of enzymes and cytochromes involved in energy production via the electron transport system during the transition from vegetative cells to spores (73).
The F0F1-ATPase (ATP synthase) complex catalyzes ATP synthesis from ADP and Pi, driven by the proton gradient generated by the respiratory chain. However, ATP synthase is dispensable in both Escherichia coli and B. subtilis (67, 68). Within the atpIBEFHAGDC operon, atpHAGDC encodes the δ, α, γ, β and ε subunits of F1 portion, atpBEF encodes the A, C, and B subunits of F0 portion, and atpI encodes a protein with an unknown function. At the transcriptional level, the atpC gene was down-regulated by more than 20-fold and the others were decreased by about 2∼5-fold during sporulation (supplemental Table S2). At the translational level, however, the α, γ, β, and ε subunits of the F1 portion and the C and B subunits of the F0 portion were all maintained at similar levels at 13 h, and increased by more than 1.8-fold at 22 h except that the A subunit of the F0 portion failed to be quantified (supplemental Table S2). These results highlight the large energy requirement of spore and parasporal crystal formation.
CONCLUSIONS
In this study, transcriptomics and proteomics analyses that combined two high throughput and unbiased techniques, RNA-seq and iTRAQ, were used for the first time to define mechanisms that drive the high production of ICPs and sporulation in B. thuringiensis. The results revealed some important regulatory mechanisms of the metabolic pathways involved in the supply of amino acids, carbon substances, and energy for sporulation and parasporal crystal formation. (1) During sporulation, some operons and genes involved in amino acid transport and biosynthesis were either specifically induced or up-regulated in response to amino acid starvation, more importantly, abundant proteases with high activities could efficiently promote protein recycling to meet the requirements for amino acids. (2) B. thuringiensis has developed various strategies to provide carbon and energy substances for sporulation and parasporal crystal formation. When nutritional substances are rich, cells store intracellular (e.g. PHB) and extracellular (e.g. Acetoin) carbon substances that could be reused under nutrient-deficient conditions. Some low-quality carbon and energy sources that remained unused during the exponential growth phase could be fully utilized during sporulation. (3) The central carbohydrate metabolism pathways (particularly, the TCA cycle) were significantly modified during sporulation. (4) The oxidative phosphorylation-associated enzymes and cytochromes were remarkably up-regulated during sporulation.
Despite our results, it remains difficult to assess how much of the vast metabolic changes we observed are ascribed to the parasporal crystal formation. To address this question, further investigations of the metabolic changes between wild-type B. thurigiensis and a mutant strain lacking the toxin-encoded plasmids are required. Nonetheless, our study lays the foundation for metabolic engineering and industrial strain improvement of B. thurigiensis, and the construction of a heterologous gene expression system in B. thurigiensis.
Supplementary Material
Acknowledgments
We thank Chinese National Human Genome Center at Shanghai (Shanghai, China) and BGI-Shenzhen (Shenzhen, China) for the technical supports for the RNA-seq and iTRAQ, respectively.
Footnotes
* This work was supported by the National Natural Science Foundation of China (grants 30930004, 31270105 and 40830527), the National Basic Research Program of China (973 Program, grant 2013CB127504), the Fundamental Research Funds for Central Universities of China (grant 2011PY092), and the China Postdoctoral Science Foundation (20110491163).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S4.
1 The abbreviations used are:
- iTRAQ
- isobaric tags for relative and absolute quantitation
- BCAAs
- branched-chain amino acids
- BLAST
- basic local alignment search tool
- cDNA
- complementary DNA
- COG
- cluster of orthologous groups of proteins
- DPA
- dipicolinic acid
- EMP
- Embden-Meyerhof-Pamas pathway
- FDR
- false discovery rate
- GABA
- γ-aminobutyric acid
- ICPs
- insecticidal crystal proteins
- LC-MS/MS
- liquid chromatography-mass spectrometry/mass spectrometry
- ORFs
- open reading frames
- PHB
- poly-β-hydroxybutyrate
- PP
- pentose phosphate shunt
- RNA-seq
- RNA (cDNA) high throughout sequencing
- RPKM
- reads per kilo bases per million reads
- TCA
- tricarboxylic acid cycle.
REFERENCES
- 1. Raymond B., Johnston P. R., Nielsen-LeRoux C., Lereclus D., Crickmore N. (2010) Bacillus thuringiensis: an impotent pathogen? Trends Microbiol. 18, 189–194 [DOI] [PubMed] [Google Scholar]
- 2. Dubois T., Faegri K., Perchat S., Lemy C., Buisson C., Nielsen-LeRoux C., Gohar M., Jacques P., Ramarao N., Kolstø A. B., Lereclus D. (2012) Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog. 8, e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ibrahim M. A., Griko N., Junker M., Lee A. (2010) Bacillus thuringiensis: A genomics and proteomics perspective. Bioeng. Bugs 1, 31–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Frankenhuyzen K. (2009) Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 101, 1–16 [DOI] [PubMed] [Google Scholar]
- 5. Sanahuja G., Banakar R., Twyman R. M., Capell T., Christou P. (2011) Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol. J. 9, 283–300 [DOI] [PubMed] [Google Scholar]
- 6. Bravo A., Likitvivatanavong S., Gill S. S., Soberün M. (2011) Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Molec. 41, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sedlak M., Walter T., Aronson A. (2000) Regulation by overlapping promoters of the rate of synthesis and deposition into crystalline inclusions of Bacillus thuringiensis delta-endotoxins. J. Bacteriol. 182, 734–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walter T., Aronson A. (1999) Specific binding of the E2 subunit of pyruvate dehydrogenase to the upstream region of Bacillus thuringiensis protoxin genes. J. Biol. Chem. 274, 7901–7906 [DOI] [PubMed] [Google Scholar]
- 9. Agaisse H., Lereclus D. (1996) STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20, 633–643 [DOI] [PubMed] [Google Scholar]
- 10. Baum J. A., Malvar T. (1995) Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol. Microbiol. 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 11. He J., Wang J., Yin W., Shao X., Zheng H., Li M., Zhao Y., Sun M., Wang S., Yu Z. (2011) Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J. Bacteriol. 193, 3407–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nickerson K. W., St Julian G., Bulla L. A., Jr. (1974) Physiology of sporeforming bacteria associated with insects: radiorespirometric survey of carbohydrate metabolism in the 12 serotypes of Bacillus thuringiensis. Appl. Microbiol. 28, 129–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoder-Himes D. R., Chain P. S., Zhu Y., Wurtzel O., Rubin E. M., Tiedje J. M., Sorek R. (2009) Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 106, 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 15. Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 [DOI] [PubMed] [Google Scholar]
- 16. Gan C. S., Chong P. K., Pham T. K., Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 17. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J., Chen L., Wang J., Qiao J., Zhang W. (2012) Proteomic analysis reveals resistance mechanism against biofuel hexane in Synechocystis sp. PCC 6803. Biotechnol. Biofuels e5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marzinke M. A., Choi C. H., Chen L., Shih I. M., Chan D. W., Zhang H. (2012) Proteomic analysis of temporally stimulated ovarian cancer cells for biomarker discovery. Mol. Cell. Proteomics 12, 356–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fournier M. L., Paulson A., Pavelka N., Mosley A. L., Gaudenz K., Bradford W. D., Glynn E., Li H., Sardiu M. E., Fleharty B., Seidel C., Florens L., Washburn M. P. (2010) Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell. Proteomics 9, 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gedeon T., Bokes P. (2012) Delayed protein synthesis reduces the correlation between mRNA and protein fluctuations. Biophys. J. 103, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Griffin T. J., Gygi S. P., Ideker T., Rist B., Eng J., Hood L., Aebersold R. (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1, 323–333 [DOI] [PubMed] [Google Scholar]
- 23. Gong Y., Li M., Xu D., Wang H., He J., Wu D., Chen D., Qiu N., Bao Q., Sun M., Yu Z. (2012) Comparative proteomic analysis revealed metabolic changes and the translational regulation of Cry protein synthesis in Bacillus thuringiensis. J. Proteomics 75, 1235–1246 [DOI] [PubMed] [Google Scholar]
- 24. Monro R. E. (1961) Protein turnover and the formation of protein inclusions during sporulation of Bacillus thuringiensis. Biochem. J. 81, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. González-Pastor J. E., Hobbs E. C., Losick R. (2003) Cannibalism by sporulating bacteria. Science 301, 510–513 [DOI] [PubMed] [Google Scholar]
- 26. González-Pastor J. E. (2011) Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 35, 415–424 [DOI] [PubMed] [Google Scholar]
- 27. Tojo S., Satomura T., Kumamoto K., Hirooka K., Fujita Y. (2008) Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J. Bacteriol. 190, 6134–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Delorme C., Ehrlich S. D., Renault P. (1999) Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 181, 2026–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deikus G., Condon C., Bechhofer D. H. (2008) Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J. Biol. Chem. 283, 17158–17167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frees D., Savijoki K., Varmanen P., Ingmer H. (2007) Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 31. Molière N., Turgay K. (2009) Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis. Res. Microbiol. 160, 637–644 [DOI] [PubMed] [Google Scholar]
- 32. Ghosh A., Chakrabarti K., Chattopadhyay D. (2009) Cloning of feather-degrading minor extracellular protease from Bacillus cereus DCUW: dissection of the structural domains. Microbiology 155, 2049–2057 [DOI] [PubMed] [Google Scholar]
- 33. Moriyama R., Sugimoto K., Zhang H., Inoue T., Makino S. (1998) A cysteine-dependent serine protease associated with the dormant spores of Bacillus cereus: purification of the protein and cloning of the corresponding gene. Biosci. Biotechnol. Biochem. 62, 268–274 [DOI] [PubMed] [Google Scholar]
- 34. Narasaki R., Kuribayashi H., Shimizu K., Imamura D., Sato T., Hasumi K. (2005) Bacillolysin MA, a novel bacterial metalloproteinase that produces angiostatin-like fragments from plasminogen and activates protease zymogens in the coagulation and fibrinolysis systems. J. Biol. Chem. 280, 14278–14287 [DOI] [PubMed] [Google Scholar]
- 35. Nisnevitch M., Sigawi S., Cahan R., Nitzan Y. (2010) Isolation, characterization and biological role of camelysin from Bacillus thuringiensis subsp. israelensis. Curr. Microbiol. 61, 176–183 [DOI] [PubMed] [Google Scholar]
- 36. Navarro A. K., Farrera R. R., Lüpez R., Pérez-Guevara F. (2006) Relationship between poly-beta-hydroxybutyrate production and delta-endotoxin for Bacillus thuringiensis var. kurstaki. Biotechnol. Lett. 28, 641–644 [DOI] [PubMed] [Google Scholar]
- 37. Tseng C. L., Chen H. J., Shaw G. C. (2006) Identification and characterization of the Bacillus thuringiensis phaZ gene, encoding new intracellular poly-3-hydroxybutyrate depolymerase. J. Bacteriol. 188, 7592–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu D., He J., Gong Y., Chen D., Zhu X., Qiu N., Sun M., Li M., Yu Z. (2011) Proteomic analysis reveals the strategies of Bacillus thuringiensis YBT-1520 for survival under long-term heat stress. Proteomics 11, 2580–2591 [DOI] [PubMed] [Google Scholar]
- 39. Liu B. L., Tzeng Y. M. (2000) Characterization study of the sporulation kinetics of Bacillus thuringiensis. Biotechnol. Bioeng. 68, 11–17 [DOI] [PubMed] [Google Scholar]
- 40. Chen D., Xu D., Li M., He J., Gong Y., Wu D., Sun M., Yu Z. (2012) Proteomic analysis of Bacillus thuringiensis ΔphaC mutant BMB171/PHB(-1) reveals that the PHB synthetic pathway warrants normal carbon metabolism. J. Proteomics 75, 5176–5188 [DOI] [PubMed] [Google Scholar]
- 41. Law J. H., Slepecky R. A. (1961) Assay of poly-beta-hydroxybutyric acid. J. Bacteriol. 82, 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fales L., Kryszak L., Zeilstra-Ryalls J. (2001) Control of hemA expression in Rhodobacter sphaeroides 2.4.1: effect of a transposon insertion in the hbdA gene. J. Bacteriol. 183, 1568–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiao Z., Xu P. (2007) Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33, 127–140 [DOI] [PubMed] [Google Scholar]
- 44. Renna M. C., Najimudin N., Winik L. R., Zahler S. A. (1993) Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponentialphase production of acetoin. J. Bacteriol. 175, 3863–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grundy F. J., Waters D. A., Takova T. Y., Henkin T. M. (1993) Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol. Microbiol. 10, 259–271 [DOI] [PubMed] [Google Scholar]
- 46. Yoshida K. I., Fujita Y., Ehrlich S. D. (2000) An operon for a putative ATP-binding cassette transport system involved in acetoin utilization of Bacillus subtilis. J. Bacteriol. 182, 5454–5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang M., Oppermann-Sanio F. B., Steinbüchel A. (1999) Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J. Bacteriol. 181, 3837–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oppermann F. B., Steinbüchel A. (1994) Identification and molecular characterization of the aco genes encoding the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J. Bacteriol. 176, 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schönert S., Buder T., Dahl M. K. (1999) Properties of maltose-inducible alpha-glucosidase MalL (sucrase-isomaltase-maltase) in Bacillus subtilis: evidence for its contribution to maltodextrin utilization. Res. Microbiol. 150, 167–177 [DOI] [PubMed] [Google Scholar]
- 50. Yip V. L., Thompson J., Withers S. G. (2007) Mechanism of GlvA from Bacillus subtilis: a detailed kinetic analysis of a 6-phospho-alpha-glucosidase from glycoside hydrolase family 4. Biochemistry 46, 9840–9852 [DOI] [PubMed] [Google Scholar]
- 51. Ireton K., Jin S., Grossman A. D., Sonenshein A. L. (1995) Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 92, 2845–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zamboni N., Fischer E., Laudert D., Aymerich S., Hohmann H. P., Sauer U. (2004) The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J. Bacteriol. 186, 4528–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fujita Y., Fujita T. (1987) The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc. Natl. Acad. Sci. U.S.A. 84, 4524–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hsieh Y. C., Wu Y. J., Chiang T. Y., Kuo C. Y., Shrestha K. L., Chao C. F., Huang Y. C., Chuankhayan P., Wu W. G., Li Y. K., Chen C. J. (2010) Crystal structures of Bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin binding and insertion domains. J. Biol. Chem. 285, 31603–31615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Iakiviak M., Mackie R. I., Cann I. K. (2011) Functional analyses of multiple lichenin-degrading enzymes from the rumen bacterium Ruminococcus albus 8. Appl. Environ. Microbiol. 77, 7541–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Magge A., Granger A. C., Wahome P. G., Setlow B., Vepachedu V. R., Loshon C. A., Peng L., Chen D., Li Y. Q., Setlow P. (2008) Role of dipicolinic acid in the germination, stability, and viability of spores of Bacillus subtilis. J. Bacteriol. 190, 4798–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horswill A. R., Escalante-Semerena J. C. (1999) Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 181, 5615–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jin S., Sonenshein A. L. (1994) Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 176, 4669–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aronson J. N., Borris D. P., Doerner J. F., Akers E. (1975) Gamma-aminobutyric acid pathway and modified tricarboxylic acid cycle activity during growth and sporulation of Bacillus thuringiensis. Appl. Microbiol. 30, 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lohman J. R., Olson A. C., Remington S. J. (2008) Atomic resolution structures of Escherichia coli and Bacillus anthracis malate synthase A: comparison with isoform G and implications for structure-based drug discovery. Protein Sci. 17, 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu L., Peng Q., Song F., Jiang Y., Sun C., Zhang J., Huang D. (2010) Structure and regulation of the gab gene cluster, involved in the gamma-aminobutyric acid shunt, are controlled by a sigma54 factor in Bacillus thuringiensis. J. Bacteriol. 192, 346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park K. B., Oh S. H. (2006) Enhancement of gamma-aminobutyric acid production in Chungkukjang by applying a Bacillus subtilis strain expressing glutamate decarboxylase from Lactobacillus brevis. Biotechnol. Lett. 28, 1459–1463 [DOI] [PubMed] [Google Scholar]
- 63. Wray L.V., Jr., Zalieckas J. M., Ferson A. E., Fisher S. H. (1998) Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J. Bacteriol. 180, 2943–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Drevland R. M., Waheed A., Graham D. E. (2007) Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J. Bacteriol. 189, 4391–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kawaguchi H., Inagaki K., Kuwata Y., Tanaka H., Tano T. (1993) 3-Isopropylmalate dehydrogenase from chemolithoautotroph Thiobacillus ferrooxidans: DNA sequence, enzyme purification, and characterization. J. Biochem. 114, 370–377 [DOI] [PubMed] [Google Scholar]
- 66. Debarbouille M., Gardan R., Arnaud M., Rapoport G. (1999) Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181, 2059–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jensen P. R., Michelsen O. (1992) Carbon and energy metabolism of atp mutants of Escherichia coli. J. Bacteriol. 174, 7635–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Santana M., Ionescu M. S., Vertes A., Longin R., Kunst F., Danchin A., Glaser P. (1994) Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176, 6802–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Efremov R. G., Sazanov L. A. (2011) Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 [DOI] [PubMed] [Google Scholar]
- 70. Madej M. G., Müller F. G., Ploch J., Lancaster C. R. (2009) Limited reversibility of transmembrane proton transfer assisting transmembrane electron transfer in a dihaem-containing succinate: quinone oxidoreductase. Biochim. Biophys. Acta 1787, 593–600 [DOI] [PubMed] [Google Scholar]
- 71. Winstedt L., von Wachenfeldt C. (2000) Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa(3) or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182, 6557–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Munro A. W., Girvan H. M., McLean K. J. (2007) Cytochrome P450–redox partner fusion enzymes. Biochim. Biophys. Acta 1770, 345–359 [DOI] [PubMed] [Google Scholar]
- 73. Lang D. R., Felix J., Lundgren D. G. (1972) Development of a membrane-bound resiratory system prior to and during sporulation in Bacillus cereus and its relationship to membrane structure. J. Bacteriol. 110, 968–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.