Abstract
RepoMan is a protein phosphatase 1 (PP1) regulatory subunit that targets the phosphatase to key substrates throughout the cell cycle. Most work to date has focused on the mitotic roles of RepoMan/PP1, although equally important interphase role(s) have been demonstrated. Initial mapping of the interactome of nuclear RepoMan, both endogenous and tagged, was complicated by various factors, including antibody cross-reactivity and low sensitivity of the detection of chromatin-associated partners above the high background of proteins that bind nonspecifically to affinity matrices. We therefore adapted the powerful combination of fluorescence imaging with labeling-based quantitative proteomics to map the “fragmentomes” of specific regions of RepoMan. These regions demonstrate distinct localization patterns and turnover dynamics that reflect underlying binding events. The increased sensitivity and signal-to-noise ratio provided by this unique approach facilitated identification of a large number of novel RepoMan interactors, several of which were rigorously validated in follow-up experiments, including the association of RepoMan/PP1 with a specific PP2A-B56γ complex, interaction with ribosomal proteins and import factors involved in their nucleocytoplasmic transport and interaction with proteins involved in the response to DNA damage. This same strategy can be used to investigate the cellular roles of other modular proteins.
Reversible protein phosphorylation is the major general mechanism regulating most physiological processes in eukaryotic cells, with protein kinases and protein phosphatases playing key roles in the control of cell proliferation, differentiation, and a host of other critical events. A common theme in phosphatase regulation is a mechanism whereby localization of the enzyme determines its access to substrates. In the case of the ubiquitous serine/threonine protein phosphatase 1 (PP1)1, this is mediated by interaction of the catalytic subunit with a panel of regulatory proteins termed “targeting subunits” to generate a range of holoenzyme complexes with distinct subcellular roles. Using a novel combination of live cell imaging with Stable Isotope Labeling by Amino acids in cell Culture (SILAC)-based quantitative proteomics, we identified several known and novel nuclear PP1 targeting subunits and determined their specificity for two of the three mammalian PP1 isoforms, α and γ (1). We further demonstrated that one of the novel γ-specific interactors, RepoMan, is a targeting subunit that recruits a pool of PP1 to chromatin initially at anaphase (Recruits PP1 to Mitotic chromatin at ANaphase) and retains it there throughout the following interphase until its release at the entry into mitosis.
Not surprisingly, this anaphase recruitment of a pool of RepoMan/PP1 has since been linked to regulation of several mitotic exit events, including chromosome segregation (2, 3), chromosomal Aurora B kinase targeting (4) and nuclear envelope reassembly (5). Persistence on chromatin throughout interphase suggests equally important nonmitotic roles for this phosphatase complex, however. PP1 is known to function in regulation of transcription and chromatin remodeling (for review see 6, 7), and RepoMan/PP1 may therefore contribute to modulation of the activity or localization of specific transcription factors, or chromatin accessibility in general. Consistent with our initial observation that RNA interference-induced knockdown of RepoMan in HeLa cells causes large-scale cell death by apoptosis (i.e., the protein promotes cell survival;1), it was recently shown that increased levels of cellular RepoMan increase the activation threshold of a DNA damage checkpoint, whereas knockdown restores normal DNA damage response (8).
To identify binding partners for RepoMan that would provide clues to the specific chromatin-associated regulatory pathway(s) in which this PP1 complex is involved, we initially applied our optimized quantitative labeling-based affinity purification/mass spectrometry (AP/MS) approach (9, 10) to assess the interactome of nuclear fluorescence protein (FP)-tagged RepoMan. We identified and validated several known and novel interactors, however none provided obvious links to the interphase chromatin association of RepoMan. The SILAC isotope labeling strategy, with its built-in negative control, provides great help in separating specific interactors from nonspecific interactors, of which the latter can account for up to 95% of identified proteins in an AP/MS experiment (11, 9). However, not all specific interactions can be unambiguously determined, particularly near the threshold level where signal-to-noise ratios are close to background. A large number of chromatin-associated proteins bind nonspecifically to affinity matrices (9), reducing the ability to distinguish genuine protein–protein interactions. We therefore needed a way to increase the signal, that is, the abundance of chromatin-associated RepoMan complexes.
We show here that photokinetic analyses of FP-tagged RepoMan and truncated fragments spanning predicted globular domains in the protein provides important clues to the specific regions involved both in chromatin association and in association with other nuclear structures and complexes. Fragments with overlapping regions show similar localization patterns and turnover dynamics, reflecting conserved underlying binding events. When we extended these analyses to interactome mapping in a quantitative “fragmentome” approach, we identified a large number of previously undetected RepoMan interactions mediated by distinct regions within this modular protein. They include the first complex detected between a PP1 holoenzyme (RepoMan/PP1) and a PP2A holoenzyme (PP2Ac/R1A/B56γ), association of RepoMan with ribosomal proteins and import factors involved in their nucleocytoplasmic trafficking and an interaction between RepoMan and the catalytic subunit of the DNA damage response protein kinase DNA-PK. With the exception of 14-3-3 proteins, which may represent a cell cycle- or perturbation-specific interaction, all of these associations, uncovered by our unique fragmentome approach, validated with full-length RepoMan. This confirms the increased sensitivity of the fragmentome approach and suggests previously unknown roles for RepoMan in a diverse range of intracellular pathways.
EXPERIMENTAL PROCEDURES
Cloning and Plasmid Constructs
Previously described EGFP- and mCherry (mCh)-tagged RepoMan constructs (1) were used as PCR templates to create the RepoMan truncation mutants used in this study, a full list of which can be found in supplemental Table S1. Phospho-mimic (Ser/Thr to Asp/Glu) and nonphosphorylatable (Ser/Thr to Ala) mutants were generated using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). EGFP-RPL27, EGFP-RPL5, and GFP-RPS6 were described previously (12), as was pEYFP-NLS (13). mCh-KPNA2 and mCh-KPNB1 were generous gifts from Dr. Saskia Hutten, EGFP-Caprin from Dr. Ben Wang and the R1A coding sequence from Dr. Anne-Claude Gingras.
Cell Culture
HeLa cells (ATCC, Manassas, VA) were grown in Dulbecco's modified Eagles' medium (DMEM) supplemented with 10% fetal bovine serum and 100 U/ml penicillin and streptomycin (Wisent, St. Bruno, QC). HeLa/EGFP, HeLa/EGFP-PP1α, and HeLa/EGFP-PP1γ stable cell lines were described previously (1). Cells were transiently transfected using either the Effectene transfection reagent (Qiagen, Valencia, CA) or polyethylenimine (Polysciences, Inc., Warrington, PA). FACS analyses were performed as previously described (14). For mitotic arrest experiments, cells were treated with either 0.1 μg/ml nocodazole, 10 nm taxol, or 5 μm S-trityl-l-cysteine (STLC) for 18 h before harvesting by mitotic shake-off. S-phase arrest treatments included: 0.5 mm l-mimosine for 22 h, 2 mm hydroxyurea for 16 h (which was then removed for 2 h before harvest by changing the media) or a double thymidine block in which cells were treated with 2 mm thymidine for 14 h, the thymidine removed for 18 h and then 2 mm thymidine added for another 14 h before harvest.
Fluorescence Imaging and Photokinetic Experiments
Live cell imaging experiments were carried out as described previously, using a wide-field deconvolution-based fluorescence microscope system (DeltaVision CORE; Applied Precision, Issaquah, WA) equipped with a three-dimensional motorized stage, temperature-and gas-controlled environmental chamber, Xenon light source, and quantifiable laser module (13). Images were collected using a 60 × NA 1.4 Plan-Apochromat objective and recorded with a CoolSNAP coupled-charge device (CCD) camera (Roper Scientific, Trenton, NJ). The microscope was controlled by SoftWorX acquisition and deconvolution software (Applied Precision, Seattle, WA). For the permeabilization assay, cells were imaged before and after 5 min incubation with 0.1% Triton X-100 in ASE buffer (20 mm Tris pH 7.5, 5 mm MgCl2, 0.5 mm EGTA) at 37 °C.
For FRAP experiments, a single section was imaged before photobleaching, a region of interest (ROI) bleached to 50–60% of its original intensity using a 488-nm laser, and a series of images acquired after the photobleaching period. Images were analyzed and recovery half-times and mobile fractions calculated using SoftWorX or GraphPad Prism. Recovery curves were plotted using Microsoft Excel. For FLIP experiments, different areas of the cell were monitored while a specific region was repeatedly photobleached over time.
Cell Extracts and Immunoblotting
Whole-cell extracts and cytoplasmic, nucleoplasmic and nucleolar fractions were prepared as previously described (9, 15). The final buffer for each extract was RIPA (50 mm Tris pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and protease inhibitors), and all extracts were cleared by centrifuging at 2800 × g for 10 min at 4 °C. Nuclear extracts were prepared from purified nuclei as previously described (1). Total protein concentrations were measured using a Qubit Fluorometer (Invitrogen, Carlsbad, CA) or Pierce® BCA Protein Assay Kit (Thermo Scientific, Rockford, IL).
For affinity purification of GFP- and mCh-tagged proteins, extracts were incubated with either GFP-Trap_A™ or RFP-Trap_A™, respectively (Chromotek, Martinsried, Germany). For immunoprecipitation (IP) of endogenous proteins, affinity purified antibodies were covalently conjugated to protein G Sepharose (GE Healthcare, Mississauga, ON) at a concentration of 1 mg/ml. Covalently-conjugated affinity purified IgG from the same species was used for control IPs. Beads were incubated with extracts for 1 h at 4 °C, after which the extracts were removed, the beads washed several times with ice-cold RIPA buffer and bound proteins eluted by heating in LDS sample buffer (Invitrogen). Lysate and IP samples were separated on 4–12% Novex Nu-PAGE bis-Tris polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes for immunoblotting.
To demonstrate the mobility shift of the phosphorylated proteins, Phos-tag™ ligand (final concentration 50 μm) and MnCl2 (final concentration 100 μm) were added to the 10% polyacrylamide separating gels before polymerization. After electrophoresis the gel was soaked in 2 mm EDTA, 25 mm Tris, and 192 mm glycine for 15 min to eliminate manganese ions from the gel and then equilibrated in transfer buffer (25 mm Tris,192 mm glycine, and 20% methanol) for another 15 min. Proteins were transferred to nitrocellulose membranes at 4 °C for 400 Vh.
A full list of all of the primary antibodies used for immunoblotting can be found in supplemental Table S2. To reduce the antibody heavy chain signal and improve resolution of the B56γ band, the ReliaBLOT® Western blot kit (Bethyl Laboratories, Inc., Montgomery, TX) was used according to the manufacturer's instructions. Primary antibodies were detected via HRP-conjugated secondary antibodies (Thermo Scientific) and ECL Plus Western blotting substrate (GE Healthcare). Full Western blots are provided (Supplemental Figs. S6–S8) for all cropped blots presented in Figs. 1, 4, 5, 6 and 7.
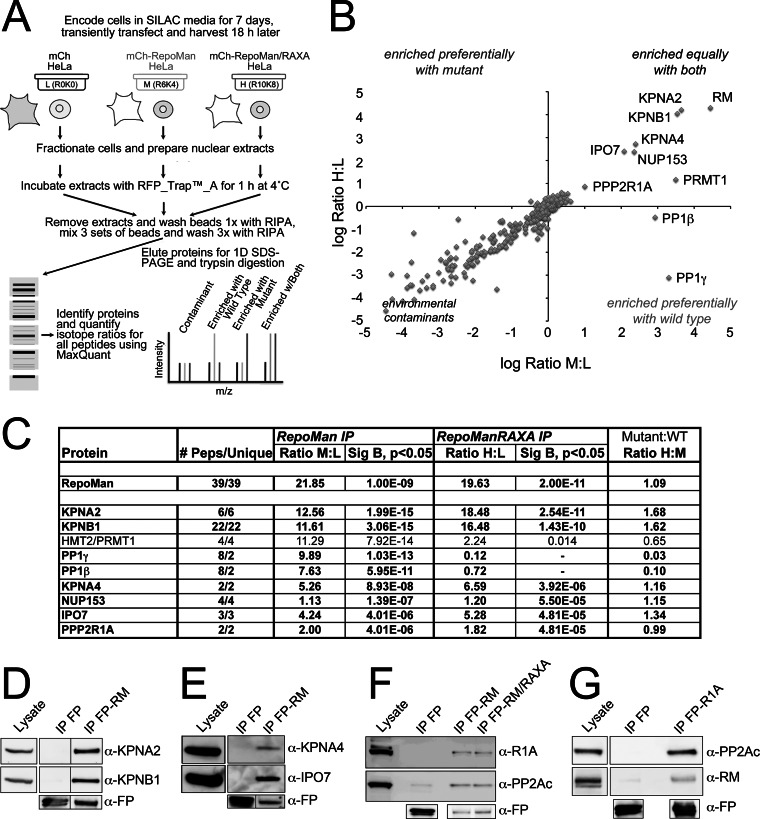
Fig. 1.
Quantitative analysis of the tagged RepoMan interactome. A, Design of the quantitative IP experiment comparing mCh-RepoMan (M), the non-PP1 binding mCh-RepoMan/RAXA mutant (H), and the mCh tag alone (L). B, Plot of log H:L ratio (enrichment in mCh-RM/RAXA IP) versus log M/L ratio (enrichment in mCh-RM IP). Contaminants cluster around a log ratio 0 (SILAC ratio 1:1), whereas proteins enriched equally with both fall in the upper right quadrant. C, Table listing # peptides (total and unique), ratio M/L (enrichment with mCh-RepoMan), ratio H/L (enrichment with mCh-RepoMan/RAXA) and ratio H/M (enrichment with mutant relative to wild type) for proteins with M/L ratios > 2 identified with two or more peptides. Significance B (Sig B) values, which are weighted by relative intensity, are shown for those proteins with ratios significantly different from the mean (p < 0.05). Proteins highlighted in bold have been validated as bona fide RepoMan interactors by IP/WB analysis. D, Validation of the copurification of KPNB1 and KPNA2 with tagged, full-length RepoMan (RM) from asynchronous cell extracts. E, Co-IP/WB validation of novel interactions of RepoMan with two additional importins, KPNA4 and IPO7. F, Copurification of PPP2R1A with FP-tagged wild type and RAXA mutant RepoMan. G, Reciprocal validation of the R1A/RepoMan interaction demonstrated by copurification of endogenous RepoMan with FP-tagged R1A. As a control, PP2Ac was also shown to copurify with FP-R1A. Anti-FP antibodies demonstrate the amount of fusion protein (or free FP) recovered in each IP.
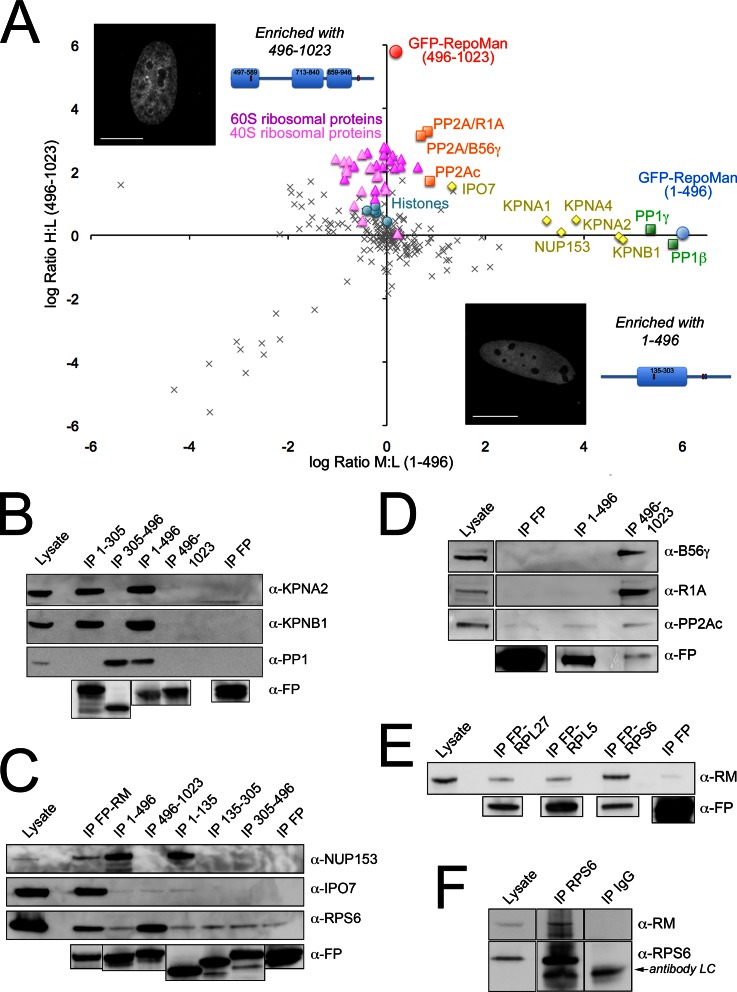
Fig. 4.
N- and C-terminal halves of RepoMan bind distinct pools of interactors. A, Plot of log H:L ratio (enrichment in GFP-RepoMan/496-1023 IP) versus log M:L ratio (enrichment in GFP-RepoMan/1–496 IP). Contaminants cluster around a log ratio 0 (SILAC ratio 1:1). The N-terminal fragment (blue circle) enriched a cluster of nuclear import factors (yellow triangles) and two isoforms of PP1 (green squares), whereas the C-terminal fragment (red circle) enriched PP2Ac/R1A/B56γ (orange squares) and ribosomal proteins (RPLs, dark pink triangles; RPSs, light pink triangles). Histones (turquoise circles) are close to threshold, albeit slightly enriched with the C terminus. Inset images are single optical sections of live HeLa cells expressing FP-RepoMan/1–496 or FP-RepoMan/496-1023. Both fragments are predominantly nuclear. Scale bars are 5 μm. B, Validation and further mapping of N-terminally mediated interactions with KPNA2, KPNB1 and PP1. C, Validation of NUP153, IPO7 and RPS6 association with full-length FP-tagged RepoMan (FP-RM). Interaction of NUP153 with the N-terminal half of RepoMan was further mapped to the 1–135 domain, whereas RPS6 enrichment was confirmed to be mediated by the C-terminal half of RepoMan. IPO7 was weakly enriched with both halves and with the 1–135 domain. D, Mapping of the PP2Ac/R1A/B56γ association with RepoMan to the C-terminal half of the protein was confirmed by IP/WB analysis. E, Reciprocal validation of the RepoMan/ribosomal protein interaction was demonstrated by copurification of endogenous RepoMan (RM) with a range of FP-tagged ribosomal proteins (RPL27, RPL5 and RPS6). Anti-FP antibodies were used to demonstrate the amount of fusion protein (or free FP) recovered in each IP. F, Endogenous RPS6, immunoprecipitated using anti-RPS6 antibodies (compared with an IgG control IP), copurifies endogenous RepoMan. RPS6 runs just above the 25 kDa antibody light chains (LC), which are indicated (arrow) on the WB.
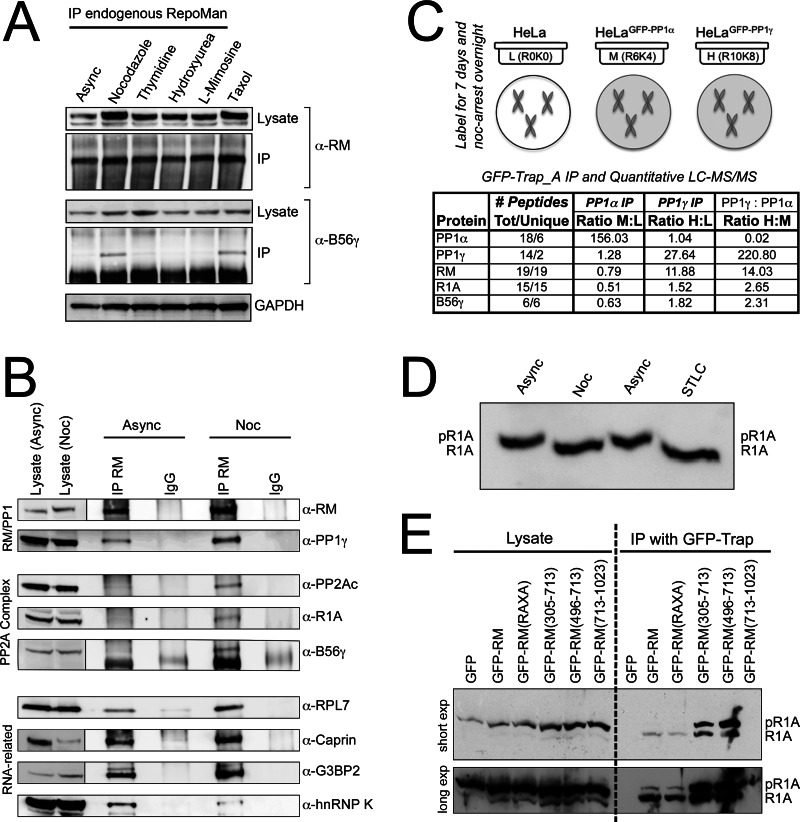
Fig. 5.
Mitosis-specific association of RepoMan/PP1 and PP2A/R1A/B56γ. A, IP/WB analysis demonstrating copurification of B56γ with endogenous RepoMan (RM) predominantly from extracts prepared from cells arrested in mitosis (using nocodazole or taxol), as compared with asynchronous cells or cells arrested in S-phase (using thymidine, hydroxyurea or l-mimosine). Lysate lanes demonstrating the relative amounts of RepoMan and B56γ (and the loading control GAPDH) before IP are shown below the IP lanes. B, IP/WB analysis of endogenous RepoMan immunoprecipitated from either asynchronous or nocodazole (noc)-arrested cell extracts, compared with an IgG control IP. The WB was probed with the antibodies indicated. C, Quantitative comparison of GFP-PP1α and GFP-PP1γ interactomes in nocodazole-arrested cells. Specific enrichment of RepoMan and PP2Ac/R1A/B56γ with the γ isoform is indicated by high H:L and H:M ratios and low M:L ratios. PP1α is shown for comparison (high M:L ratio, and low H:L and H:M ratios). D, Phos-tag gel/WB analysis of R1A in asynchronous cell extracts versus extracts from cells arrested in early mitosis (using nocodazole or the Eg5 inhibitor STLC). The Phos-tag amplifies phosphorylation-induced band shifts. E, Phos-tag gel/WB analysis of R1A copurified with FP-tagged wild type RepoMan, the RAXA mutant and the 305–713 and 713–1023 fragments. Control GFP alone and RepoMan/713-1023 IPs are shown for comparison.
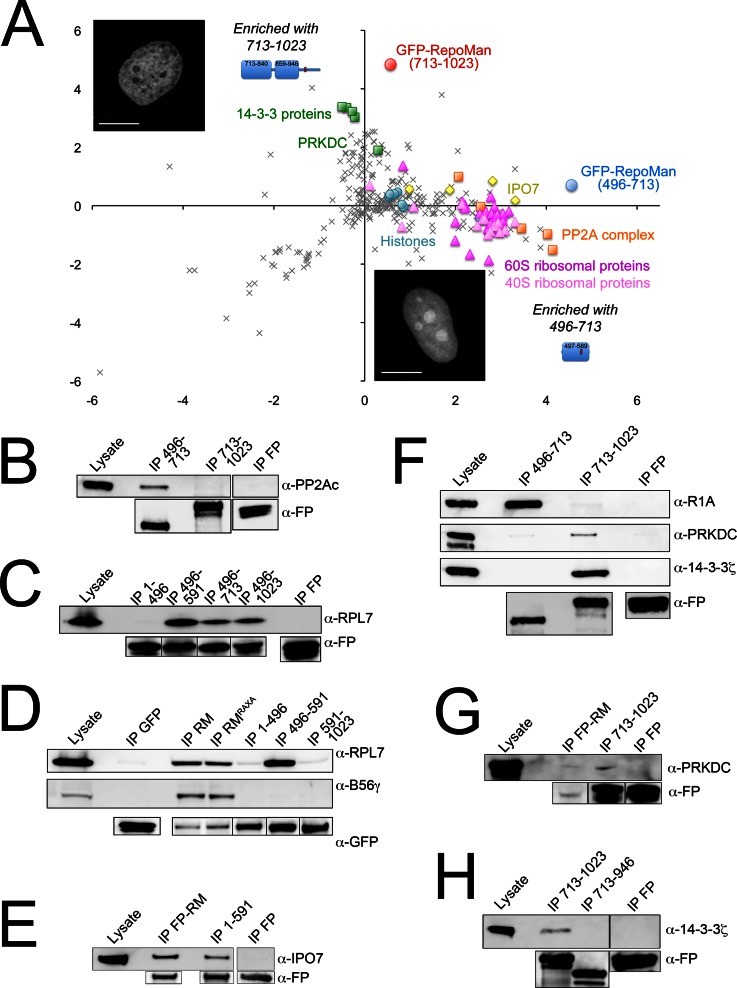
Fig. 6.
C-terminal half of RepoMan contains distinct bindings domains for diverse interactors. A, Plot of log H:L ratio (enrichment in GFP-RepoMan/713–1023 IP) versus log M:L ratio (enrichment in GFP-RepoMan/496–713 IP). Contaminants cluster around a log ratio 0 (SILAC ratio 1:1). The 496–713 fragment (blue circle) enriched a cluster of ribosomal proteins (RPLs, dark pink triangles; RPSs, light pink triangles), PP2A complex members (orange squares) and importins (yellow diamonds), whereas the 713–1023 fragment (red circle) enriched several 14-3-3 proteins and the DNA-dependent protein kinase PRKDC (green squares). Inset images are single optical sections of live HeLa cells expressing FP-RepoMan/496–713 or FP-RepoMan/713–1023. Both fragments are predominantly nuclear, with 496–713 showing additional accumulation within nucleoli. Scale bars are 5 μm. B, Validation of the mapping of PP2Ac association to the 496–713 domain. C, Interaction of the ribosomal protein RPL7 with the 496–713 domain of RepoMan was validated, and binding further mapped to the 496–591 domain. D, RPL7 and B56γ associate equally with wild type RepoMan (RM) and the non-PP1 binding RAXA mutant. Although RPL7 association mapped to the 496–591 domain, splitting the C terminus of RepoMan at this site disrupts association with PP2A complex members. E, Consistent with the weak interaction observed for IPO7 with regions in both the N- (1–135) and C- (496–1023, 496–591) terminus, the 1–591 fragment that spans both of these regions copurifies IPO7 nearly as efficiently as full-length FP-RepoMan. F, Validation of the domain-specific interactions of R1A (496–713), PRKDC (713–1023) and 14-3-3ζ (713–1023). G, Validation of the interaction of PRKDC with both the 713–1023 fragment and with full-length FP-RepoMan. H, Validation of the interaction of 14-3-3ζ with the 713–1023 fragment. Removal of the extreme C terminus (947–1023) from this fragment abrogated 14-3-3ζ binding. Anti-FP antibodies were used to demonstrate the amount of fusion protein (or free FP) recovered in each IP.
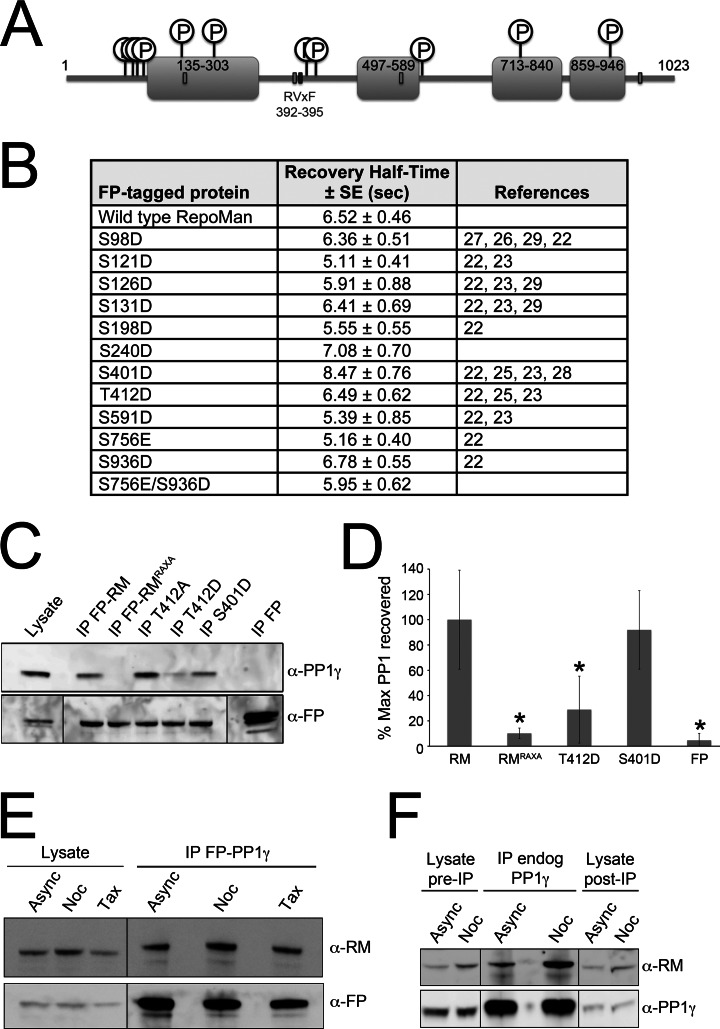
Fig. 7.
Effect of early mitotic phosphorylation on RepoMan dynamics and PP1 binding. A, Diagram noting the location of 11 phosphoresidues mapped in endogenous RepoMan immunoprecipitated from nocodazole-arrested cells. Pale gray boxes indicate the positions of predicted NLSs whereas the dark gray box marks the PP1-binding RVxF motif. B, Summary of FRAP experiments comparing the recovery rates of single phosphomimic mutants (S/T to D/E) to that of wild type RepoMan. A double phosphomimic mutant of the two sites (S756E/S936D) mapped in the chromatin-associated 713–1023 domain was also tested. Data are mean ± S.E. (n = 5–25). C, IP/WB experiment demonstrating the loss of PP1 binding induced by the RAXA and T412D phosphomimic mutations compared with wild type FP-RepoMan (FP-RM). The T412A nonphosphorylatable mutant retains PP1 binding, as does the S401D phosphomimic mutant. The results of 4 quantified experiments are summarized in (D). Data are mean ± S.E. Asterisks indicate a significant reduction in associated PP1 compared with wild type FP-RepoMan (p < 0.05 in an unpaired Student's t test). E, Endogenous RepoMan (RM) copurifies with FP-tagged PP1γ from HeLa/GFP-PP1γ cell extracts (both asynchronous and arrested in early mitosis using nocodazole or taxol). F, Endogenous RepoMan copurifies with endogenous PP1γ immunopurified from both asynchronous and nocodazole-arrested HeLa cell extracts.
Quantitative SILAC IP
Quantitative SILAC-based affinity purification experiments were carried out as previously described (9). Cells were encoded in media containing either “light” l-arginine and l-lysine (Sigma-Aldrich, Oakville, ON), “medium” l-arginine 13C and l-lysine 4,4,5,5-D4 or “heavy” l-arginine 13C/15N and l-lysine 13C/15N (Cambridge Isotope Laboratories, Andover, MA) and extracts prepared as described above. Control (tag alone) and fusion protein purifications were carried out separately, the extracts removed and the beads washed once with ice-cold RIPA buffer, and then the control and experimental beads carefully combined and washed an additional three times with ice-cold RIPA buffer. Proteins were eluted, separated by 1D SDS-PAGE, and trypsin-digested for MS analysis as described previously (9).
Mass Spectrometry and Data Analysis
An aliquot of the tryptic digest (prepared in 5% acetonitrile/0.1% trifluoroacetic acid in water) was analyzed by LC-MS on an LTQ-Orbitrap mass spectrometer system (ThermoElectron, Rockford, IL) as previously described (9). The Orbitrap was set to analyze the survey scans at 60,000 resolution and the top five ions in each duty cycle selected for MS/MS in the LTQ linear ion trap. Database searching (against the human IPI database v3.68; 87,061 entries) and quantitation were performed using MaxQuant software v1.1.1.14 and the Andromeda search engine (16, 17). The following criteria were used: peptide tolerance = 10 ppm, trypsin as the enzyme (2 missed cleavages allowed) and carboxyamidomethylation of cysteine as a fixed modification. Variable modifications were oxidation of methionine and N-terminal acetylation. Medium SILAC labels were Arg6 and Lys4 and heavy SILAC labels were Arg10 and Lys8. Minimum ratio count was 2 and quantitation was based on razor and unique peptides. Peptide and protein FDR was 0.01. The Perseus data mining and management module of MaxQuant was used to calculate Significance B values. For quantitation of specific PP1 isoforms and RepoMan fragments, only unique SILAC ratios were averaged. All data sets (minus common environmental contaminants as per http://maxquant.org and proteins identified via the decoy database) are provided as Supplemental Data.
Phosphopeptide Mapping
For phosphopeptide mapping of RepoMan in nocodazole-arrested HeLa cells, the endogenous protein was immunopurified from cell extracts using a covalently conjugated polyclonal RepoMan antibody (Abcam, Cambridge, MA). Proteins were separated by SDS-PAGE and the region containing the phosphorylated RepoMan band was excised from the gel (as determined by alignment with a Western blot run concurrently to highlight the position of phospho-RepoMan). The excised band was incubated with 50 mm iodoacetamide to alkylate cysteine residues, washed and digested with trypsin for 16 h. Tryptic digests were analyzed by LC-MS/MS using a Proxeon Easy-LC connected to an LTQ orbitrap velos system via a Proxeon nanospray source fitted with a New Objective FS360–20-20 uncoated emitter. An aliquot of 18/100 μl of the tryptic digest was injected onto 20 × 0.1 mm C18 guard column equilibrated in buffer A (2% acetonitrile/0.1% formic acid in water) at 7 μl/min. After washing the injector loop with 30 μl buffer A, the guard column was switched in line with a 150 × 0.075 mm PepMap C18 column (Dionex) equilibrated in buffer A at 300 nl/min. The column was developed with a 60 min discontinuous gradient of buffer B (80% acetonitrile/0.1% formic acid in water). Nanoelectrospray was performed by applying a voltage of 1.6 kV to the emitter and the orbitrap was set to perform a survey scan of m/z 350–1800 at a resolution of 60000 and the top 10 multiply charged ions (minimum intensity 10000 cps) were selected for collision induced dissociation in the LTQ and then excluded for 30 s after two occurrences. The raw data file was converted to a Mascot Generic File (mgf) and searched using Mascot 2.4 and Proteome Discoverer 1.3. The following criteria were applied: Database, SwissProt; Species, Human;Enzyme, Trypsin (1 missed cleavage permitted); Fixed modification, Carboxyamidomethylation of cysteine; Variable Modification, Oxidation of Methionine, phosphorylation (STY), Gln>pyroglu; Precursor mass tolerance, 10 ppm, MS/MS mass tolerance, 0.8 Da. Phosphopeptides were assigned using phosphoRS and validated by searching a reverse database with the FDR set at 0.01 (Strict) and 0.05 (relaxed) Supplemental Fig. S9.
RESULTS
Quantitative SILAC IP Identifies Novel RepoMan Complexes
RepoMan is predominantly nuclear in interphase cells (1), and we therefore combined our quantitative SILAC IP approach (9) with cellular fractionation to reduce background contaminants and increase sensitivity of our nuclear RepoMan interactome mapping. Two sets of isotopic labels (R6K4 and R10K8) were used to directly compare the wild type RepoMan interactome to that of a non-PP1 binding mutant (RepoMan/RAXA;1), to determine whether the ability to recruit PP1 affects the molecular complexes with which RepoMan associates. Isotopically labeled HeLa cells were transiently transfected with either mCh alone (control) or mCh-tagged RepoMan fusion proteins, harvested and fractionated into cytoplasm and nuclei. Pull-down experiments were performed on nuclear extracts using the red fluorescent protein (RFP)-Trap_A™ affinity matrix and combined before mass spectrometric analysis (Fig. 1A).
As shown in the graph in Fig. 1B, although the majority of identified proteins represent common bead contaminants, as expected (9), a subset was enriched with tagged RepoMan. This group includes both known and novel interactors. With the exception of the PP1 isoforms, all of the detected interactors were enriched equally with wild type and mutant RepoMan. Fig. 1C summarizes the top hits (log2 M: L ratios > 1), all of which were identified by two or more unique peptides and enriched significantly above background (see Supplemental Data for full data set). Proteins shown in bold were validated as bona fide RepoMan interactors. Although the protein arginine N-methyltransferase PRMT1 appeared enriched with both wild type and mutant RepoMan in the SILAC experiment, we were unable to convincingly validate this interaction by IP/WB analysis (data not shown).
Previously identified interactors include NUP153, KPNA2, and KPNB1, believed to link RepoMan to regulation of nuclear envelope formation at mitotic exit (5). We confirmed interaction of KPNA2 and KPNB1 with FP-RepoMan in asynchronous HeLa cell extracts by immunoprecipitation and Western blot (IP/WB) analysis (Fig. 1D), and demonstrated that FP-tagged KPNA2 and KPNB1 copurify endogenous RepoMan (supplemental Fig. S4D). We also identified and validated novel interactions between FP-RepoMan and two additional importins, KPNA4 and IPO7 (Fig. 1E).
A surprising result was enrichment of the PP2A regulatory subunit R1A (a.k.a. PR65A, α-isoform of the 65 kDa scaffolding A subunit), which suggests association of a PP1 holoenzyme complex with a PP2A holoenzyme complex. To validate this interaction, we first demonstrated copurification of endogenous R1A (and the PP2A catalytic subunit PP2Ac) with tagged RepoMan (wild type and RAXA mutant) by IP/WB analysis (Fig. 1F). We then cloned R1A and demonstrated copurification of endogenous RepoMan (and PP2Ac) with tagged R1A (Fig. 1G). This confirms that RepoMan can complex with the core PP2Ac/R1A holoenzyme. No PP2A regulatory B subunits were identified in the SILAC screen.
Fluorescence Imaging-based Assays of GFP-tagged RepoMan Fragments Reveal Distinct Subnuclear Localization and Binding Affinity Regions within the Full-Length Protein
Although our quantitative IP analysis of tagged full-length RepoMan identified several interaction partners, there was a distinct absence of chromatin-associated proteins. RepoMan is recruited to and persists on chromatin during mitotic exit, and its localization and dynamics in interphase are consistent with maintenance of this chromatin association (1). Because RepoMan itself does not contain any known DNA binding domains, it is assumed that this association is indirect, with RepoMan binding chromatin-associated proteins. The predominance of importins in the SILAC experiment suggested that repeated identification of these highly abundant interactors may have precluded identification of less abundant binding partners. With evidence that importin β association maps to the extreme N terminus (5) and chromatin association to the C-terminal half (8), we hypothesized that splitting RepoMan into distinct domains with different underlying binding events would increase signal/noise and facilitate identification of lower abundance binding partners. We therefore combined a microscopy-based characterization of RepoMan truncation mutants with our quantitative proteomic approach in a unique “fragmentome” strategy.
Although the three-dimensional structure of RepoMan has not yet been solved, structural algorithms such as GlobPlot (18) can be used to predict likely globular versus disordered regions within the protein based on amino acid sequence (Fig. 2A). We used these predicted regions to design a RepoMan fragment cloning strategy that would maximize proper folding of the expressed fragments, with all fragments being fused to an N-terminal GFP tag.
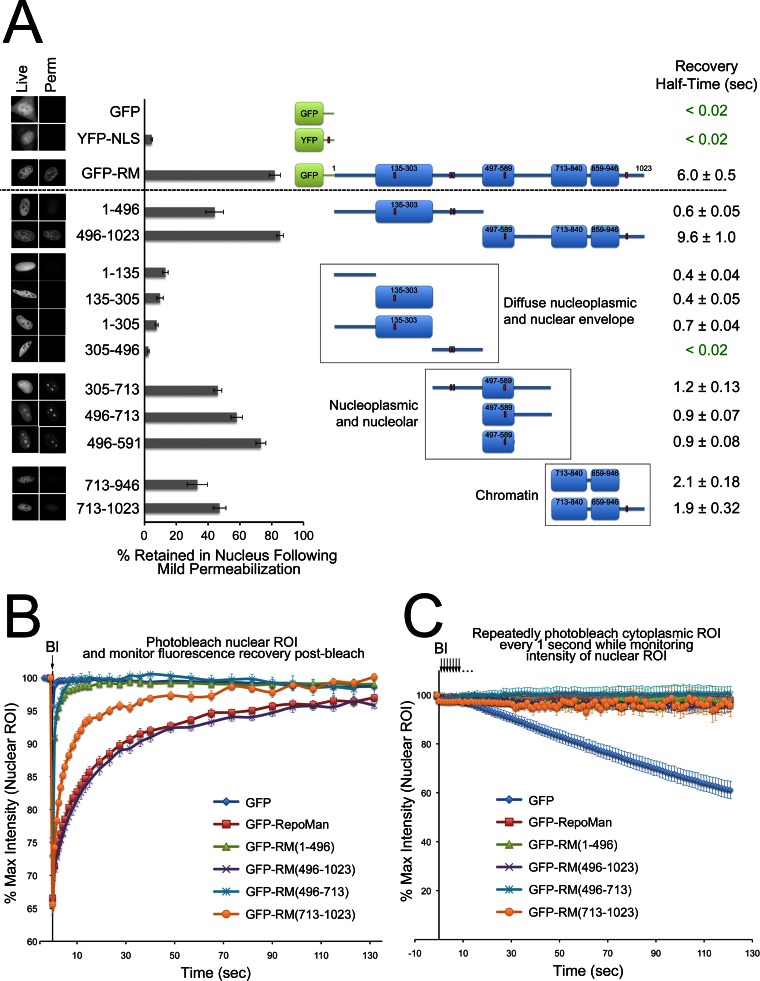
Fig. 2.
Design of truncation mutants for the analysis of domain-specific RepoMan interactomes. A, Localization of GFP-RepoMan (GFP-RM) in live HeLa cells before (“Live” panel) and after (“Perm” panel) mild permeabilization with 0.1% Triton X-100. Localization of free GFP and YFP-NLS (nuclear localization signal added to enhance nuclear and nucleolar accumulation) in the cell is shown for comparison. Scale bars are 5 μm. The diagram in the center shows predicted globular (box) and disordered (line) regions in RepoMan and highlights the PP1-binding RVxF motif (blue box) and predicted NLSs (red boxes). Localization in live cells before and after mild permeabilization is shown for each fragment and summarized on the graph as % retained in nucleus. Data are mean ± S.E. (n = 13–40, 3–4 separate experiments). B, FRAP (Fluorescence Recovery After Photobleaching) experiment comparing the recovery rates of the photobleached N-terminal (1–496, green triangles) and C-terminal (496–1023, purple crosses) halves of RepoMan within a nucleoplasmic region of interest (ROI) to that of the full-length protein (red squares) and free GFP (blue diamonds). Also shown are the recovery curves for the split C-terminal halves: 496–713 (turquoise crosses) and 713–1023 (orange circles). A pre-bleach image was acquired, a nucleoplasmic ROI photobleached at the 0 time point (arrow) and the GFP signal within that ROI monitored over time. Data are mean ± S.E. (n = 7–31, three separate experiments). Recovery half-times for nucleoplasmic pools of all fragments tested are summarized in the far right column in (A) as mean ± S.E. C, FLIP (Fluorescence Loss In Photobleaching) experiment assessing nucleocytoplasmic shuttling. A cytoplasmic region of interest was continually photobleached every 1 s (arrows) for a total of 126 s, with images taken after each bleach event. Total intensity over time for a nuclear ROI was normalized for photobleaching because of image acquisition. Data are mean ± S.E. (n = 4–7, 3 separate experiments). GFP, which shuttles freely between the nucleus and cytoplasm (nuclear pool is continually depleted as the protein trafficks through the cytoplasmic region of interest and is irreversibly photobleached) was used as a positive control.
Using a range of in vivo fluorescence imaging approaches, we compared the behavior of the various RepoMan fragments to that of the full-length protein, starting with analysis of localization in live cells. Cells were imaged before and after mild permeabilization with 0.1% Triton X-100, to compare nuclear retention. Freely soluble proteins are rapidly lost from the nucleus in response to this treatment (see GFP and YFP-NLS in Fig. 2A), whereas proteins that form more stable associations with other nuclear proteins or structures are retained. Binding events also affect the turnover of GFP-tagged proteins in fluorescence recovery after photobleaching (FRAP) experiments, in which a pool of GFP-protein is irreversibly photobleached by exposure to high intensity 488 nm light and recovery of the fluorescence signal within this region of interest (which represents turnover of the protein on binding sites) monitored over time. Lastly, fluorescence loss in photobleaching (FLIP) experiments were used to test nuclear/cytoplasmic shuttling. Repeated photobleaching of a region of interest in the cytoplasm of a cell leads to an eventual loss of nuclear fluorescence if the GFP-tagged protein actively shuttles or freely distributes between the nucleus and cytoplasm (and thus passes through the photobleached region at some point).
Like endogenous RepoMan, GFP-RepoMan is predominantly nuclear in interphase cells, with a localization pattern similar to that of DNA binding dyes such as DAPI and Hoechst 33342 (Fig. 2A,1). The protein contains four predicted nuclear localization signals (NLSs) and no nuclear export signals (NESs). Consistent with this, once RepoMan is recruited to the nucleus it remains there and does not shuttle between the nucleus and cytoplasm, as demonstrated by FLIP experiments (Fig. 2C). Compared with the rapid loss of free GFP following mild permeabilization of the cells, ∼80% of GFP-RepoMan is retained in the nucleus (Fig. 2A), reflecting participation in multiprotein complexes and/or association with nuclear structures.
All of the FP-tagged RepoMan fragments tested here are also predominantly nuclear, which is not entirely surprising given that nearly all of them contain a predicted NLS (Fig. 2A). Two exceptions are the 1–135 fragment, previously shown to bind directly to KPNB1 in vitro (5), and the C-terminal 713–946 fragment, which is likely recruited via association with other nuclear proteins. The majority of the fragments were also retained in the nucleus to varying extents following mild permeabilization, although differences in subnuclear localization patterns and turnover dynamics indicate that distinct binding events are associated with each of these regions.
When RepoMan was split into N-terminal (1–496) and C-terminal (496–1023) halves, the C-terminal half showed a similar localization pattern to that of full-length RepoMan, both before and after permeabilization, whereas the N-terminal half displayed a diffuse nucleoplasmic pattern and increased loss from permeabilized cells. Consistent with these observations, the N-terminal 1–496 fragment also demonstrated a more rapid recovery rate in FRAP experiments, whereas the turnover dynamics of the C-terminal half (496–1023) were similar to that of the full-length protein (Figs. 2A, 2B). Neither fragment was lost from the nucleus in FLIP experiments (Fig. 2C), indicating that they were retained in the nucleus once they were imported. This is consistent with the fact that neither has an export signal nor is small enough to be lost by free diffusion through nuclear pores.
Truncated fragments of the N-terminal half of RepoMan all showed predominantly diffuse nucleoplasmic localization patterns, although we did observe colocalization of a small pool of FP-RepoMan (and the 1–305 and 1–496 fragments) with nuclear pore complex markers at the nuclear envelope (data not shown), consistent with a previous study (5). These smaller N-terminal fragments were not efficiently retained in the nucleus following permeabilization (Fig. 2A) and showed similar and more rapid recovery rates in FRAP experiments (Fig. 2A, supplemental Fig. S1), suggesting that they do form larger complexes, but that these complexes are predominantly soluble and not as stably associated with nuclear structures as the full-length protein.
Splitting the C-terminal half of RepoMan into two fragments (496–713 and 713–1023) uncovered a surprising nucleolar accumulation of the 496–713 fragment that mapped to a minimal predicted globular region of 496–591 (Fig. 2A and supplemental Fig. S5C). This region contains a bipartite NLS, and sequence analysis using the Nucleolar Localization Sequence Detector (NoD;19) identified a predicted nucleolar localization signal (NoLS) overlapping this NLS (Fig. 8B). Although full-length RepoMan is predominantly nucleoplasmic, as judged both by imaging of endogenous and GFP-tagged RepoMan in cells and by cell fractionation/WB analysis (1, supplemental Fig. S4A), this simply indicates that there is no accumulation of the protein in this subnuclear structure under steady-state conditions. FLIP analysis revealed that the nuclear pool of GFP-RepoMan does in fact shuttle through nucleoli (supplemental Fig. S5D). The NoLS may therefore be important for one or more of RepoMan's roles within the nucleus. The 3 fragments that span this NoLS-containing region (305–713, 496–713 and 496–591) are all retained to a greater extent in the nucleus (and nucleolus) following mild permeabilization than the N-terminal fragments and also have slower turnover dynamics, albeit faster than that of full-length RepoMan (Figs. 2A, 2B, supplemental Fig. S1).
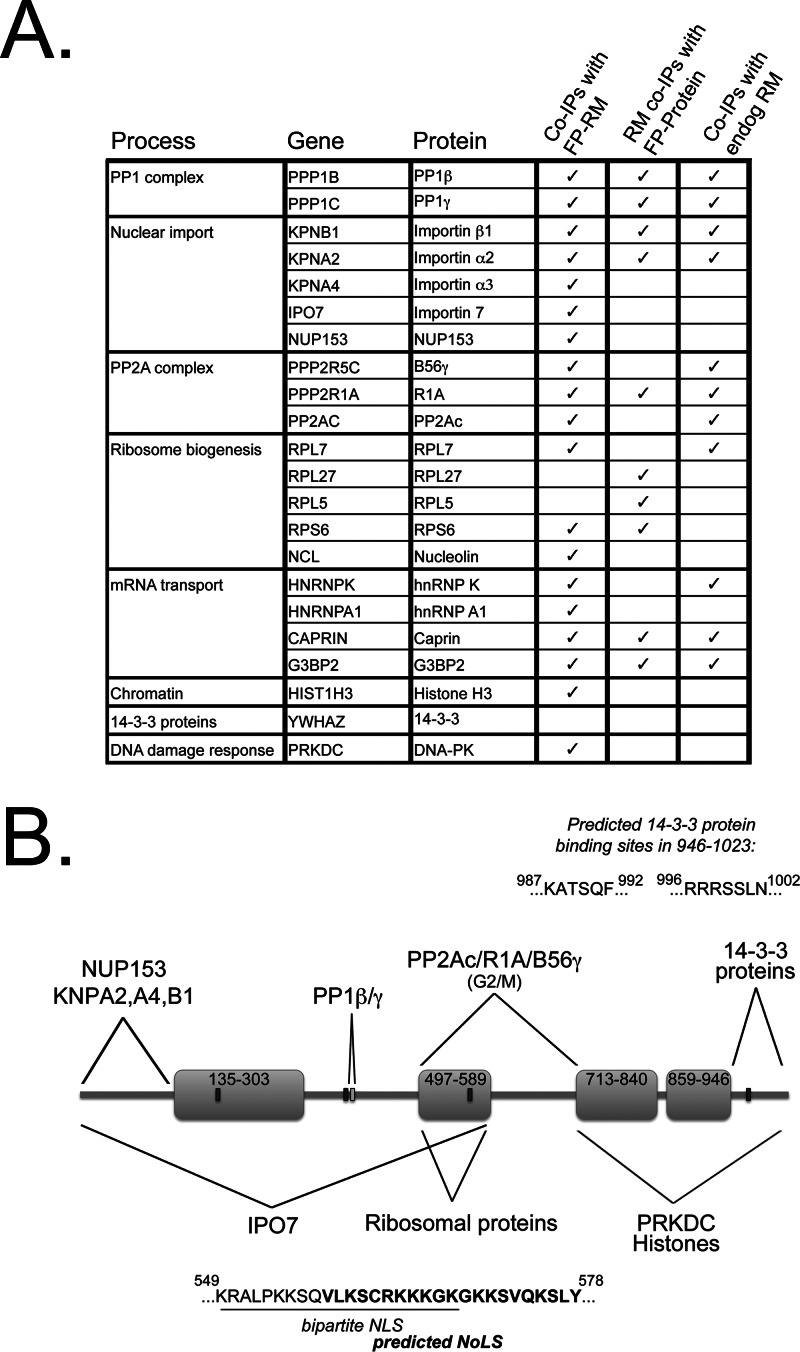
Fig. 8.
Validation summary and model of the modular interaction events mediated by specific domains of RepoMan. A, Table summarizing the validation of the RepoMan interactors discussed in this study. Where indicated, endogenous interactors were detected in pulldowns of full-length FP-tagged RepoMan (Co-IPs with FP-RM), endogenous RepoMan was detected in pulldowns of FP-tagged interactors (RM co-IPs with FP-Protein) and/or endogenous interactors were detected in pulldowns of endogenous RepoMan (Co-IPs with endog RM). With the exception of 14-3-3ζ, all of the interactors shown here validated with full-length RepoMan. B, Diagram summarizing the domain-specificity of the interactions uncovered using the “fragmentome” mapping approach. Although the extreme N terminus mediates most interactions with importins and NUPs, IPO7 (and other nonclassic importins) binds weakly to both the 1–135 and 496–591 regions of RepoMan, and more efficiently to the 1–591 fragment encompassing both of these regions. The PP1-binding RVxF domain in the N terminus mediates interaction with PP1β and PP1γ. Region 496–591, which mediates interaction with ribosomal proteins and shows additional accumulation in nucleoli, contains both a predicted bipartite nuclear localization signal (NLS, underlined) and a predicted nucleolar localization signal (NoLS, bold text). The longer 496–713 domain is required for association with PP2Ac/R1A/B56γ, and this association occurs predominantly in early mitosis. The chromatin localization pattern observed for RepoMan in interphase cells is maintained by the extreme C terminus (713–1023), which also mediates association with histone H3 and with the DNA damage response-related PRKDC kinase and 14-3-3 proteins. Deletion of the extreme C terminus (947–1023), which contains 2 predicted 14-3-3 binding sites, results in loss of 14-3-3 binding.
Although the 713–1023 fragment showed a similar steady-state nuclear localization pattern to that of full-length RepoMan (and the 496–1023 C-terminal half), it was not retained in the nucleus to the same extent following permeabilization (Fig. 2A) and its turnover dynamics as measured by FRAP were faster, albeit not as fast as the 1–496 N-terminal half or the 496–713 fragment (Figs. 2A, 2B). This highlights the importance of assessing both localization and dynamics, as the latter can detect changes in underlying binding events that are not necessarily reflected in steady-state localization patterns.
Further analysis of the recovery curve of full-length GFP-RepoMan revealed that the data are better fit by a two-phase association consisting of a fast recovery pool (t1/2 = 1.6 s) and a slow recovery pool (t1/2 = 18.9 s). This suggests that, under steady-state conditions in interphase nuclei, RepoMan is found in more than one complex. Our inability to detect chromatin-associated interactors above the highly abundant importins in our intial interactome screen of full-length RepoMan (Fig. 1) may reflect differences in the relative levels of these complexes. We therefore set out to map the “fragmentome” of the C-terminal half expressed on its own, compared it to that of the dynamic N-terminal half that contains PP1 (1) and importin β (5) association domains. Our hypothesis was that this reduced complexity, along with the higher expression levels that we observe for RepoMan fragments in cells compared with the FP-tagged full-length protein (supplemental Fig. S2D), should substantially increase our signal-to-noise ratio in interactome experiments.
RepoMan Fragmentome Identifies Distinct Pools of Interactors for the N- and C-Terminal Halves of the Protein
We used our optimized quantitative SILAC IP approach (9) to compare the fragmentomes of the N-terminal (1–496) and C-terminal (496–1023) halves of RepoMan, as outlined in Fig. 3A. HeLa cells were isotopically labeled for 7 days before transient expression of either GFP alone or the two GFP-tagged RepoMan fragments. Because the fragments vary in their nuclear retention in response to mild permeabilization, we tested their nuclear retention during cell fractionation and found that a pool of the 1–496 fragment does end up in the cytoplasmic fraction (supplemental Fig. S4C), despite the fusion protein being predominantly nuclear in live cells (Fig. 2A; Fig. 4A). For that reason we decided to prepare whole-cell rather than nuclear extracts. As previously noted, we observed both an increased transfection efficiency and higher level of fusion protein expressed in cells for the fragments compared with full-length tagged RepoMan, which was reflected in a 50% increase in the relative amount of total protein recovered in the SILAC IP as judged by relative abundance in the MS analysis (supplemental Fig. S3).
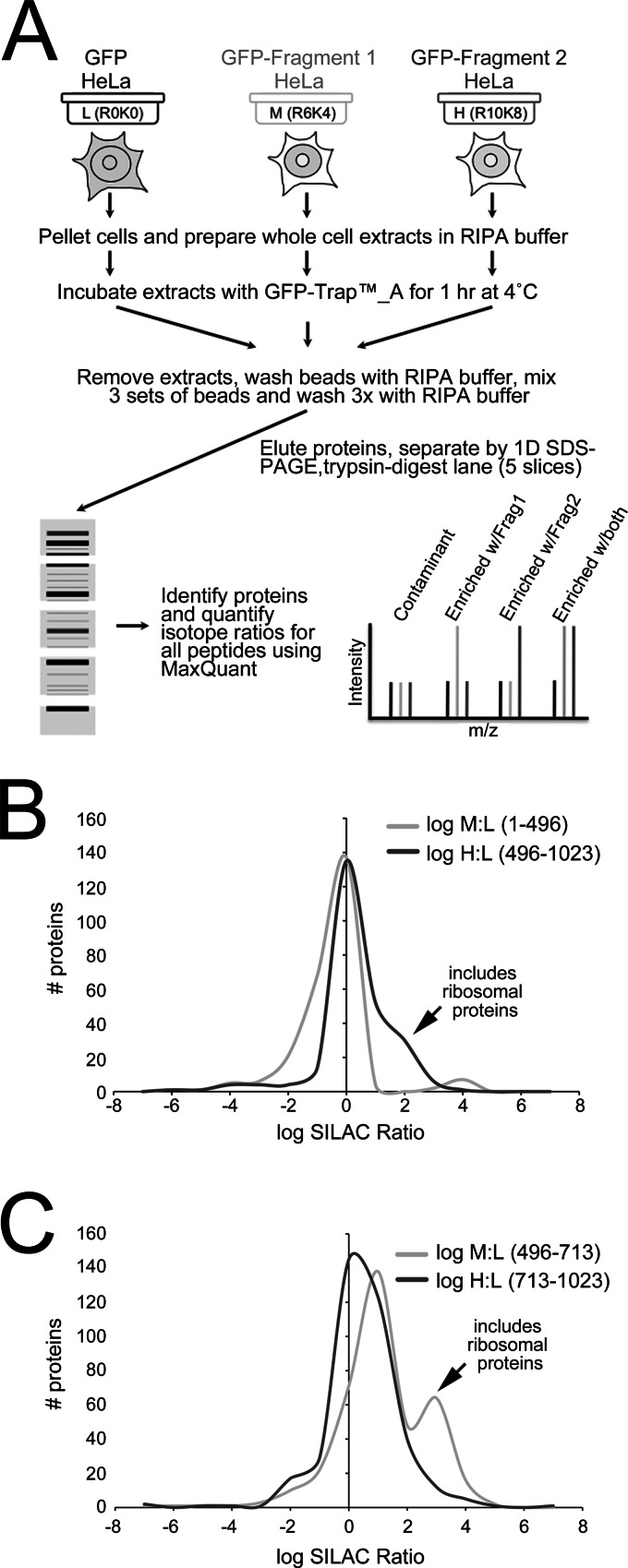
Fig. 3.
Quantitative mapping of RepoMan “fragmentomes.” A, Design of the quantitative IP experiments comparing the interactomes of RepoMan fragments. B, Distribution of log SILAC (M:L and H:L) ratios for the fragmentome experiment comparing GFP-RepoMan/1–496 (M) to GFP-RepoMan/496-1023 (H), with free GFP as the built-in negative control (L). The small peak (arrow) of log H:L ratios (i.e. enriched with 496–1023) that separates from the contaminants (peak over log ratio 0, i.e. ratio 1:1) contains enriched ribosomal proteins. C, Distribution of log SILAC (M/L and H/L) ratios for the fragmentome experiment comparing GFP-RepoMan/496–713 (M) to GFP-RepoMan/713–1023 (H), with free GFP as the built-in negative control (L). The peak (arrow) of log M:L ratios (i.e. enriched with 496–713) that separates from the contaminants (peak over log ratio 0, i.e. ratio 1:1) contains enriched ribosomal proteins.
The samples were analyzed twice by LC-MS/MS, with a total of 238 and 277 proteins identified and quantified, respectively, in each run (full data sets provided as Supplemental Data). The top hits are summarized both in supplemental Table S3 and in Fig. 4A, which shows a plot of the log H:L ratios (enrichment with 496–1023) versus log M:L ratios (enrichment with 1–496). As expected, PP1 copurified with the 1–496 fragment containing the PP1-binding RVxF motif (residues 392–395). WB/IP analysis confirmed association of PP1γ both with this fragment and with the 305–496 disordered region (Fig. 4B). NUP153, identified in our original nuclear RepoMan interactome screen (Fig. 1), also copurified with the 1–496 fragment. We confirmed interaction of NUP153 with both full-length RepoMan and the 1–496 fragment by IP/WB, and mapped it to the same minimal 1–135 region previously shown to mediate KPNB1 binding (Fig. 4C).
KPNB1, along with the importins KPNA1, A2 and A4, was also enriched with the N terminus of RepoMan (Figs. 4A, 4B). Interestingly, the importin IPO7 did not map specifically to this region in the proteomic screen, but instead was enriched equally with both the N- and C-terminal halves of RepoMan (Fig. 4A). We had previously identified and validated IPO7 as a RepoMan interactor in our nuclear interactome screen (Fig. 1). IP/WB analysis revealed that efficient copurification of IPO7 is disrupted by splitting the protein in half, although slight enrichment is observed with both halves (fragments 1–496 and 496–1023), consistent with the proteomic results (Fig. 4C). This suggests that association of IPO7 with RepoMan involves domains in both the N- and C-terminal halves of the protein.
Also consistent with its identification in our original full-length RepoMan interactome, the PP2A scaffolding subunit R1A was found to be enriched with the C-terminal fragment, along with the catalytic subunit PP2Ac and the regulatory B subunit B56γ (Fig. 4A, supplemental Table S3). IP/WB analysis confirmed both the association of full-length RepoMan with this PP2A complex (supplemental Fig. S4G) and specific mapping of the interaction to the C-terminal half of RepoMan (Fig. 4D).
As shown in Fig. 4A, there was a surprising enrichment of a large number of ribosomal proteins with the C-terminal half of RepoMan. Although these are common contaminants known to bind nonspecifically to affinity matrices (9, 10), genuine enrichment in a quantitative SILAC IP will shift their isotope ratios above those of nonspecific contaminant proteins that fall in a distribution over a ratio of 1:1 (log ratio 0). We demonstrated this previously for the nucleolar protein RRP1B, which targets PP1 to pre-60S subunits and specifically enriches RPL proteins (13). When we plotted the distribution of ratios for the FP-RepoMan/1–496 (log M:L) and FP-RepoMan/496-1023 (log H:L) fragments, we saw a normal distribution for the log M:L ratios, that is, the majority of proteins are bead contaminants enriched equally under all conditions and thus distributed over a ratio of 1:1 (log ratio 0), whereas a small pool of enriched proteins is shifted to higher ratios. The log H:L ratio distribution, however, included a substantial “bump” that was shifted to the right (> 1:1) of the contaminant ratio peak (Fig. 3B). This group includes a large number of ribosomal proteins (supplemental Table S3) that appear to be enriched with the C-terminal half of RepoMan.
Given the increased and variable background caused by nonspecific binding of ribosomal proteins in control IPs, enrichment above background must be rigorously and reproducibly demonstrated to claim validation of the interaction. We first quantified four separate IP/WB experiments, demonstrating significant enrichment of representative 60S and 40S subunit ribosomal proteins (RPL7 and RPS6) with full-length FP-RepoMan above the background nonspecific binding observed in control FP IPs (supplemental Fig. S5B). Overexpression of RepoMan (or the C-terminal fragment) does not affect total cellular levels of ribosomal proteins (data not shown), verifying that the enrichment is specific to the tagged protein and not simply because of the potential for higher levels of nonspecific binding to the affinity matrix. We next demonstrated reciprocal copurification of endogenous RepoMan with three different FP-tagged ribosomal proteins (RPL27, RPL5, and RPS6) identified in the fragmentome screen (Fig. 4E). Lastly, we demonstrated copurification of endogenous RepoMan with endogenous RPS6 (Fig. 4F).
This stringent approach validates ribosomal proteins as genuine RepoMan interactors and confirms that the C-terminal half of RepoMan mediates the association. Although we have looked at a subset of ribosomal proteins here, the SILAC screen identified enrichment of a much larger number (Fig. 4A, supplemental Table S3, supplemental Fig. S3C), suggesting that the interaction is related to a more general role in ribosome biogenesis, such as ribosomal protein import or ribosome subunit export.
RepoMan Forms a Cell Cycle-dependent Complex with PP2A/R1A/B56γ
The identification of a PP2A complex copurifying with RepoMan was intriguing not only because it is the first demonstration of an interaction between two specific PP1 and PP2A holoenzyme complexes, but also because RepoMan is predominantly nuclear in interphase cells whereas PP2Ac, R1A and B56γ are predominantly cytoplasmic (supplemental Fig. S4B). We initially uncovered the interaction in our nuclear FP-RepoMan SILAC IP, but only R1A was detected (and identified by only two peptides, Fig. 1C), suggesting that the complex was either at a very low abundance in the nucleus or represented trace amounts of cytoplasmic contamination during fractionation. Identification of B56γ as a member of the complex in the fragmentome experiment suggested another possibility. This specific B56 isoform is one of two that have been proposed to have a nuclear pool (20), and more recently a PP2Ac/B56γ complex was shown to accumulate in nuclei during S phase (21). To test the possibility of a nuclear, albeit cell cycle-specific, interaction with RepoMan, we assessed the copurification of B56γ with RepoMan in asynchronous cells compared with cells arrested either in S phase or in early mitosis. When endogenous RepoMan pulldowns were probed with anti-B56γ antibodies, significant enrichment was only observed in cells arrested in early mitosis (Fig. 5A). We then confirmed the mitosis-specific interaction of R1A and PP2Ac with RepoMan (Fig. 5B).
In comparison, PP1γ is found to associate with RepoMan under both conditions, along with a representative ribosomal protein (RPL7) and three additional interactors identified in the fragmentome screen (Caprin, G3BP2 and hnRNP K). Although RepoMan is expressed at all stages of the cell cycle, protein levels are higher in mitotic cells (see RepoMan lysate lanes in Fig. 5B) and consequently more RepoMan is recovered from mitotic cells. When quantified as amount interactor detected/amount RepoMan recovered, most showed only slight differences, however the amount of B56γ recovered from nocodazole-arrested cells with RepoMan was 2-fold higher than that recovered from asynchronous cells. In comparison, the hnRNP K interaction is more predominant (twofold higher amount recovered) in asynchronous cells.
In parallel with our RepoMan interactome screen, we carried out a quantitative comparison of mitotic PP1α and PP1γ complexes, by nocodazole arrest of our HeLa/GFP, HeLa/GFP-PP1α and HeLa/GFP-PP1γ stable cell lines. As shown in Fig. 5C, we confirmed the specific enrichment of Repoman with PP1γ, and detected significant enrichment of both R1A and B56γ with this isoform. This provides further evidence that a pool of RepoMan/PP1 associates with a PP2Ac/R1A/B56γ complex in early mitosis.
The non-PP1 binding mutant of RepoMan copurifies the complex as efficiently as the wild type protein (Figs. 1F, 6E), and we found no evidence that RepoMan/PP1 regulates phosphorylation of any of the complex members. R1A does show a shift in phosphorylation on entry into early mitosis, however, as highlighted by Phos-tag gel/WB analysis of asynchronous versus nocodazole (G2/M) or STLC (metaphase) arrested cell extracts (Fig. 5D). The Phos-tag binds phosphate groups on proteins and amplifies the apparent molecular mass shift (22). In asynchronous cells, FP-RepoMan (and the RAXA mutant) copurify the less abundant, lower (or un-) phosphorylated form of R1A that likely represents the small proportion (∼20% of the total) of mitotic cells (Fig. 5E). Interestingly, although the 305–713 (contains PP1 binding site) and 496–713 (no PP1 binding site) fragments of RepoMan both copurify R1A, the specificity for the less phosphorylated form observed with full-length RepoMan is lost (Fig. 5E). Overexpression of these fragments does not affect cell cycle distribution (supplemental Fig. S2), and this result therefore suggests that they can associate with R1A in interphase as well as in mitosis. No differentially phosphorylated forms of PP2Ac or B56γ were identified in these experiments (data not shown).
Further Fragmentation of the C Terminus of RepoMan Reveals Distinct Domains Mediating Association with Ribosomal Proteins, IPO7, the PP2A Complex and Proteins Related to DNA Damage Response
Similar to our concern that interactions with abundant importin proteins masked our ability to identify lower abundance interactors in our nuclear RepoMan interactome screen, interaction of the C-terminal (496–1023) half of RepoMan with abundant ribosomal proteins has the potential to mask lower abundance chromatin-associated interactors. Fluorescence imaging-based analysis indicated that chromatin association mapped predominantly to the 713–1023 region (Fig. 2A), and we therefore used our quantitative AP/MS approach (Fig. 3A) to map the fragmentomes of FP-RepoMan/496–713 and FP-RepoMan/713–1023. The samples were analyzed twice by LC-MS/MS, with a total of 378 and 309 proteins identified and quantified, respectively. The top hits are summarized both in supplemental Tables S4, S5 and in the graph in Fig. 6A, which plots log H:L ratios (enrichment with 713–1023) versus log M:L ratios (enrichment with 496–713). Interaction with the PP2A complex (PP2Ac, R1A, and B56γ) mapped to the 496–713 domain of RepoMan, although specificity for the B56 subunit isoform appeared to be lost, with additional enrichment of B56ε, B56α, and B56δ (supplemental Table S4). This could either reflect increased sensitivity of detection or indicate that additional regions of RepoMan are required to confer specificity for a particular PP2A complex.
Association of ribosomal proteins with RepoMan also mapped to the 496–713 region, with an even more dramatic shift of the isotope ratios of these proteins away from the contaminants peak in the FP-RepoMan/496–713 IP (Fig. 3C). Fig. 6A and supplemental Table S4 also highlight the fact that ribosomal proteins represent a large fraction of the proteins enriched with FP-RepoMan/496–713. Interaction with ribosomal proteins was further mapped to the minimal 496–591 domain (Fig. 6C, supplemental Fig. S5A), which, like the 496–713 fragment (Fig. 6A), is predominantly nucleoplasmic with additional accumulation in nucleoli (supplemental Fig. S5C). The nucleolar pool accounts for ∼20% of the total nuclear pool of FP-RepoMan/496–591, as measured by quantitative WB analysis of subcellular fractions (supplemental Fig. S5C). To demonstrate that increased nucleolar accumulation alone cannot account for increased enrichment of ribosomal proteins, we confirmed that both FP-RepoMan/496–591 and FP-RepoMan enrich significantly more RPL7 than either free FP or FP tagged with a short NLS (RKKR) that leads to accumulation in nucleoli (supplemental Figs. S5A, 5B).
Although full-length RepoMan does not show a steady-state accumulation in nucleoli (Fig. 2A and supplemental Fig. S4A), it does traffic through them (supplemental Fig. S5D), and therefore has the potential to come in contact with ribosomal proteins either in the nucleoplasm, the nucleolus or both. IP/WB results showing that RPL7 copurifies predominantly with the nucleoplasmic pool of FP-RM/496–591 (supplemental Fig. S5C), suggest that the interaction occurs primarily in this region.
Another RepoMan interactor that mapped to the 496–713 fragment is IPO7 (Fig. 6A, supplemental Table S4). As previously noted, IPO7 binds full-length RepoMan most efficiently, and the SILAC IP results and weak binding observed for the fragments in IP/WB experiments suggest that both N- and C-terminal domains are important for this binding. Having observed a weak interaction of IPO7 with both the 1–135 and 496–591 fragments of RepoMan in IP/WB experiments (data not shown), we tested binding to a 1–591 fragment that includes both these domains. Increased enrichment of IPO7 with this fragment (Fig. 6E) confirms that these two regions are required for efficient binding, and explains the loss of binding when the protein is split in half at residue 496.
Although association of ribosomal proteins with RepoMan could be mapped to the predicted globular domain spanning residues 496–591, association with the PP2A complex was lost when RepoMan was split in half at residue 591 (Fig. 6D). Phosphorylation of residue S591 in nocodazole-arrested cells has been mapped both by our lab (Fig. 7A) and others (23, 24), and we therefore tested whether the phospho-status of this residue could affect association of RepoMan with the PP2A complex. As shown in supplemental Fig. S4G, there is no significant difference in the amount of PP2A complex members copurified with a nonphosphorylatable (S591A) or phosphomimic (S591D) mutant compared with full-length FP-RepoMan. It is more likely that splitting at this particular site disrupts binding properties, as the C-terminal 591–1023 fragment showed very different nuclear retention and dynamics compared with other C-terminal fragments (supplemental Figs. S1A, S1B).
The most striking observation for the 713–1023 fragmentome was the significant enrichment of four 14-3-3 protein family members (Fig. 6A, supplemental Table S5), known to bind and regulate various chromatin-associated proteins. This association was validated for 14-3-3ζ by co-IP/WB analysis (Fig. 6F). The extreme C terminus of RepoMan (residues 946–1023) contains two predicted 14-3-3 binding sites (Fig. 8B). We observed that 14-3-3 protein binding is lost when this region is deleted from FP-RepoMan/713-1023, suggesting that it is involved in the interaction (Fig. 6H).
Although we could validate the interaction of 14-3-3ζ with FP-RepoMan/713–1023, we could not detect copurification of 14-3-3 proteins with full-length FP-RepoMan. It is not yet clear whether this is a sensitivity issue, or whether the interaction normally occurs with full-length RepoMan under very specific conditions that we have not yet identified. These proteins have been shown to bind nonspecifically to affinity matrices (9), which can complicate validation. Direct comparison of the distribution of identified proteins between the N- (1–496) and C-terminal (496–1023) halves of RepoMan in the initial fragmentome experiment revealed that 14-3-3 proteins, like histones, are slightly enriched with the C-terminal fragment, albeit buried in the threshold (supplemental Fig. S3C). This suggests that sensitivity of detection of specific binding above nonspecific background binding may be a factor in our inability to validate the interaction with full-length RepoMan.
Another chromatin-associated protein found to be enriched with the 713–1023 fragment was PRKDC, the catalytic subunit of the protein kinase DNA-PK. Although this protein is known to bind nonspecifically to affinity matrices (9), we demonstrated that it is enriched above background with both full length FP-RepoMan and the 713–1023 fragment (Figs. 6F, 6G).
We did not observe significant enrichment of histones above background in any of our SILAC IP experiments, however they did show a slight enrichment with the 496–1023 fragment (Fig. 4A, supplemental Fig. S3C). We carried out a series of IP/WBs of full-length RepoMan and the fragments used in our SILAC experiments, assessing copurification of histone H3 (supplemental Fig. S5E), which is a known RepoMan/PP1 substrate (4). The results, summarized in supplemental Fig. S5F, indicate that although H3 is not significantly enriched above background with full-length FP-RepoMan, significant enrichment is observed with both the 496–1023 and the 713–1023 fragments. This increased histone association may represent a direct interaction and/or a higher degree of association of RepoMan with chromatin complexes that include histones. Interestingly, the histone H1 family, and H1X in particular, were enriched with the 496–713 fragment (supplemental Table S4), and we validated this increased association by IP/WB analysis for both the 496–713 and 496–1023 fragments (data not shown). Although the specific function of this H1 variant is not yet clear, it was shown to exhibit a G1 phase-dependent nucleolar accumulation (25), which may relate to its association with the nucleolar-targeted 496–713 fragment.
Other putative RepoMan interactors that were detected close to threshold in our fragmentome experiments and validated by IP/WB analysis include Nucleolin (NCL), Caprin, G3BP2, hnRNP K, and hnRNP A1. Fig. 8 summarizes the validation experiments that were carried out for these and other interactors identified in the RepoMan fragmentome experiments, and the minimal association domains to which they were mapped.
Effects of Mitotic Phosphorylation of RepoMan on Turnover Dynamics and Protein Binding
Having previously observed a high level of RepoMan phosphorylation in early mitosis, coincident with release of the protein from chromatin (2), we set out to identify the specific residues by phosphopeptide mapping to test the effect of mimicking phosphorylation (using S/T - D/E phosphomimic mutants) on localization and turnover dynamics in interphase cells. Endogenous RepoMan was affinity purified from nocodazole-arrested HeLa cell extracts, separated by SDS-PAGE and trypsin-digested, and phosphopeptides enriched using IMAC resin. Mass spectrometric analysis identified 11 phosphopeptides (see Supplemental Data set), highlighted on the RepoMan diagram in Fig. 7A. Most have also been identified in one or more large-scale phosphoproteomic screens (23, 24, 26–30), although this is the first detection of phospho-S240. We mutated each of these residues singly to acidic D or E residues and compared their turnover dynamics to that of wild-type RepoMan by FRAP analysis. Our prediction was that phosphorylation of one or more of these sites would increase the turnover dynamics of nucleoplasmic RepoMan by weakening association with chromatin. None of the single phosphomutants increased the turnover rate significantly (Fig. 7B), nor did a double phosphomutant mimicking phosphorylation of both residues (S756 and S936) mapped in the C-terminal 713–1023 domain. It is therefore unlikely that phosphorylation of RepoMan at the onset of mitosis, in the region that we have shown to mediate interphase chromatin association, is sufficient to induce release from chromatin.
A previous study had mapped T412 as the predominant site phosphorylated by CDK1 in vitro, and suggested that this phosphorylation results in a loss of PP1 binding in early mitosis (5). As this was inconsistent with our evidence, from both RepoMan and PP1 interactome studies, that RepoMan/PP1 complexes persist in cells arrested in early mitosis (Figs. 5B, 7E, 7F), we directly assessed the interaction of endogenous PP1 with FP-tagged RepoMan/T412 mutants. As shown in Fig. 7C, mimicking phosphorylation at this site with a T412D mutant resulted in a significant, but not complete, loss of PP1 binding, whereas our previously described RAXA mutant (1) abolished PP1 binding entirely. The nonphosphorylatable T412A mutant retained PP1 binding, as did the S401D mutant (which is even closer to the PP1 binding motif found at residues 392–395). This experiment (with the exception of the T412A mutant) was repeated three times and the results summarized in Fig. 7D. The persistence of a RepoMan/PP1 complex in early mitosis is likely because of two factors: 1. the incomplete loss of PP1 binding when this site is phosphorylated and 2. the existence of a pool of RepoMan that is not phosphorylated at this site. In support of the latter, we found that a relatively efficient depletion (∼70%) of endogenous PP1γ from nocodazole-arrested cells copurified RepoMan but had only a minimal effect on the total level of RepoMan in the extract (Fig. 7F). Although this could be accounted for in part by RepoMan persisting in complexes with the β isoform of PP1, it also suggests that there are pools of RepoMan in the cell that are bound to PP1 and pools that are not bound to PP1. Because nobody has yet quantified the total amount of RepoMan that is phosphorylated on T412 (or any of the other residues), we can assume that there is a mixed population in the cell. Phosphorylation of this residue was also detected in asynchronous T cells in which the ERK pathway was inhibited (26), suggesting that it is not necessarily mitosis-specific.
DISCUSSION
We initiated this study with the intention of mapping interphase chromatin-associated RepoMan complexes, using transiently expressed FP-tagged fusion protein as bait in our optimized quantitative SILAC IP workflow (9). Although it is possible to affinity purify endogenous RepoMan using our in-house or commercially available polyclonal antibodies, the depletion efficiency varies. More importantly, all of these antibodies detect unrelated bands in cell lysates in addition to RepoMan, suggesting a high potential for cross-reactivity that would complicate a nonbiased AP/MS screen (10). We confirmed this by attempting such a screen with the most commonly used commercial RepoMan antibody and identifying/validating strong cross-reactivity with at least one other cellular protein (DBC1/KIAA1967; data not shown). Although our endogenous RepoMan interactome screen did identify several top hits that we later identified/validated in our tagged RepoMan screens (PP1β and γ, KPNB1, KPNA4, Caprin, G3BP2, hnRNP K and several ribosomal proteins), other hits did not validate as RepoMan interactors, suggesting copurification with DBC1 or another protein with which the antibody cross-reacted.
In contrast, FP-tagged proteins can be rapidly and reproducibly purified from extracts using high affinity/specificity single chain llama antibodies, and the powerful combination of fluorescence imaging with quantitative proteomics has been used by our lab and others to dissect diverse cellular pathways including protein phosphatase targeting (1), ribosomal protein dynamics (12), and composition of the anaphase promoting complex (31, 9). When applied here to the analysis of FP-tagged RepoMan complexes in the nucleus, we identified and validated a handful of known (PP1, KPNB1, KPNA2, NUP153) and novel (KPNA4, IPO7, PP2R1A) interactors, all of which (with the exception of PP1) were enriched equally with the wild type protein and non-PP1-binding RAXA mutant. Detection of a RepoMan/R1A/PP2A complex was particularly intriguing because it suggested a stable association of two different serine/threonine phosphatase complexes. When the increased sensitivity of our fragmentome approach uncovered an additional member of this PP2A complex, the regulatory subunit B56γ, we explored the interaction further and discovered that it predominantly occurs between the endogenous proteins in mitosis. This was consistent with our observed copurification of RepoMan, R1A, and B56γ with FP-PP1γ from nocodazole-arrested HeLa/GFP-PP1γ cell extracts. Interestingly, we and others have shown phosphorylation of RepoMan on residue T412 (which can affect interaction with PP1) in early mitosis, although the stoichiometry of this phosphorylation has not yet been quantified. We demonstrate here that mimicking phosphorylation at this site disrupts but not does completely abrogate PP1 binding. Taken together with our finding that a substantial fraction of RepoMan remains following near complete depletion of endogenous PP1γ from nocodazole-arrested cell extracts, we suggest that RepoMan is present in more than one complex under these conditions (i.e. with and without PP1), making mitotic regulation of this particular PP1 complex more complicated than previously thought (5).
B56-PP2A complexes have been implicated both in the protection of centromeric cohesion before the metaphase-anaphase transition (32) and in the regulation of the stable attachment of kinetochores and microtubules (33). Although several distinct roles for RepoMan/PP1 have been identified at mitotic exit, including counteraction of Aurora B activity on anaphase kinetochores to stabilize chromosome segregation (3) and coordination of nuclear envelope reassembly (5), the complex has also been shown to oppose the Haspin kinase-mediated spreading of histone H3T3 phosphorylation to chromosome arms before metaphase, before mediating net dephosphorylation of this residue at mitotic exit (4). Future studies will address the functional significance of the PP1/PP2A complex association that we identified here.
We found no evidence that RepoMan/PP1 regulates net phosphorylation of any of the PP2A complex subunits (or vice-versa), although we did demonstrate a previously unreported decrease in net R1A phosphorylation in cells entering mitosis. This was consistent with our observation that RepoMan, which interacts with R1A predominantly in mitosis, associates with the less phosphorylated form of the protein. Interestingly, although overexpression of smaller fragments of RepoMan (305–713 and 496–713) that bind R1A had no effect on cell cycle distribution (i.e., did not increase the fraction of cells in mitosis), these fragments were able to copurify both the low/unphosphorylated (mitosis) and the more phosphorylated (interphase) forms of the protein. This suggests that additional regions in RepoMan mediate the mitotic specificity of the interaction.
We also noted a loss of B56 isoform specificity when we screened the 496–1023 C-terminal half of RepoMan versus the smaller 496–713 region, with the latter enriching not only B56γ, but also B56ε, δ, and α. We do not know yet whether this is simply a detection issue, or whether it suggests that additional regions in RepoMan contribute to binding specificity. Mass spectrometric analysis of the protein Shugoshin, shown to associate with PP2Ac/B56, also detected interaction with numerous isoforms of the B56 subunit (32), suggesting that they may be interchangeable in certain complexes or under certain conditions. In addition, a recent study that appeared while this work was under review detected copurification of RepoMan/CDCA2 with tagged R1A (confirming our findings) and with both tagged B56γ and B56ε (34). Although the only interactions that have been confirmed between the endogenous proteins so far are the copurifications of PP2Ac, R1A, and B56γ with RepoMan presented here, we cannot rule out the possibility that other B56 isoforms interact with RepoMan in vivo.
Another surprising finding uncovered by our fragmentome analysis is the presence of a functional nucleolar localization signal within RepoMan. We have shown here that although RepoMan does not accumulate within this nuclear structure under steady-state conditions, it does traffic through it. This trafficking may be related to the equally surprising finding that RepoMan associates with ribosomal proteins, which we went to great lengths in this study to rigorously validate. These abundant proteins are common contaminants in AP/MS experiments, however it is important in a nonbiased screen to not simply subtract them from data sets because there are circumstances in which they are genuinely enriched with the protein of interest (13). When our fragmentome screens revealed significant enrichment of ribosomal proteins with the C-terminal half of RepoMan (further mapped to the 496–713 and then 496–591 domain), we set out to validate interaction with full-length RepoMan, demonstrating significant enrichment with both the FP-tagged and endogenous protein. This highlights the power of the fragmentome approach, which can uncover interactions hidden by the lower sensitivity and signal-to-noise ratio of more complex full-length protein interactome screens.
Taken together with our data showing that the same domain within RepoMan mediates association with numerous import factors involved in a range of ribosomal protein import pathways (IPO7, IPO5, KPNB2, KPNB1, and IPO4), interaction with ribosomal proteins suggests the possibility of a role for RepoMan in regulation of ribosome biogenesis. With RepoMan found at the nuclear pore complex, in the nucleoplasm, and trafficking through nucleoli, it is not unfeasible to suggest some type of chaperone role in nuclear import and/or nucleolar targeting of ribosomal proteins. There is a wealth of literature linking reversible phosphorylation events to regulation of nucleocytoplasmic transport (see (35) for review), including enhanced recognition or binding affinity for importins on phosphorylation of residues within (36) or near (37) classic NLSs. Phosphorylation events have also been shown to activate noncanonical transport signals mediating nuclear import, as in the case of enhanced recognition of phosphorylated ERK1/2 by IPO7. Nuclear accumulation of active ERK5 is triggered when phosphorylation unmasks an NLS by disrupting an NES (38), the consequences of which can include increased activation of the AP-1 transcription factor that controls proliferation, differentiation and apoptosis (39). Treatment with the PP1/PP2A inhibitors okadaic acid and microcystin has been shown to inhibit nuclear import pathways mediated by KPNB1 and KPNB2/transportin (40), whereas in yeast, PP1 activity has been shown to antagonize PKA-dependent phosphorylation at the nuclear import domain of Msn2, a transcription factor involved in stress response (41).
Although IPO7 can act as an adaptor-like protein in association with KPNB1, mediating nuclear import of proteins such as histone H1 (another protein enriched with the 496–713 fragment), it can also function as an autonomous nuclear transport receptor. With respect to this latter role it has been shown, along with IPO5 and KPNB2, to mediate nuclear import of ribosomal proteins including RPL23A, RPS7, and RPL5 (42). Assembly of the 40S and 60S ribosomal subunits take place in the nucleolus, and the ∼75 ribosomal proteins required for this assembly and maturation that are synthesized in the cytoplasm are rapidly and actively imported into the nucleus via diverse import pathways (43) and targeted to the nucleolus (12). The redundancy of import pathways used for ribosomal proteins is likely required to support the high production rate of ribosomes in proliferating cells, but complicates regulation of import. A regulatory mechanism linked to all of these pathways would therefore provide a means of rapidly switching import on or off as required. We will continue to explore the potential for such a regulatory role for RepoMan in ribosomal protein transport, and whether it is mediated via PP1.
With respect to chromatin association, we showed that the extreme C terminus of RepoMan (residues 713–1023) confers the predominant interphase chromatin localization pattern, although its more rapid turnover dynamics compared with full-length RepoMan and the larger 496–1023 C-terminal half suggest that certain binding events may have been disrupted or weakened. Of the top hits in the FP-RepoMan/713-1023 fragmentome, several have been linked to DNA damage response pathways, including the 14-3-3 proteins gamma, epsilon, zeta, and theta (for review see (44), the catalytic subunit of the DNA damage sensor kinase DNA-PK (PRKDC; 45), the DNA mismatch repair protein MSH6 (46) and TRIP13, which is involved in DNA double-strand break repair (47).
PRKDC, which we confirmed copurifies with full-length FP-RepoMan, had previously been shown to interact with both PP2A and the PP2A-like protein phosphatase 6 (PP6;48). PRKDC recruits the PP6 complex to sites of DNA damage, where it dephosphorylates γ-H2AX. This results in dissolution of γ irradiation-induced foci and release of cells from the G2/M checkpoint. A PRKDC/RepoMan complex would likely feed into the damage sensing/repair pathway at another point. Although previous work by another lab has shown an association between RepoMan and the ATM kinase (8), we did not identify ATM in our interactome screens and could not detect an interaction by IP/WB analysis of endogenous RepoMan, full-length FP-RepoMan or any of our fragments. This may be because of differences in cell lines or experimental conditions.
In summary, by employing fluorescence imaging techniques to monitor localization, nuclear retention and turnover dynamics of specific regions of RepoMan, we identified specific domains involved in association with the nuclear pore complex, trafficking through nucleoli and association with chromatin. We then directly extended these observations, via quantitative interactome screens, to identify the underlying protein-protein binding events. The results highlight the fact that RepoMan is a modular protein that participates in more than one regulatory pathway and likely exists in multiple complexes throughout the cell at any given time. Importantly, this powerful technique can be extended to other modular proteins to add both increased sensitivity of detection and domain specificity to interactome analyses.
Supplementary Material
Acknowledgments
We thank Lawrence Puente at the Ottawa Hospital Research Institute Proteomics Core Facility for technical support. We also thank Dr. Judith Sleeman for assistance with FRAP analysis and Drs. Jocelyn Côté, Anne-Claude Gingras, Saskia Hutten, Angus Lamond and Ben Wang for providing plasmids and antibodies used in this study.
Footnotes
* This work was supported by a Canadian Cancer Research/Terry Fox Foundation New Investigator Award (Ref: 20148). LTM holds a CIHR New Investigator Salary Support Award.
 This article contains supplemental Tables S1 to S5 and Figs. S1 to S9.
This article contains supplemental Tables S1 to S5 and Figs. S1 to S9.
1 The abbreviations used are:
- PP1
- protein phosphatase 1
- AP/MS
- affinity purification/mass spectrometry
- FLIP
- fluorescence loss in photobleaching
- FRAP
- fluorescence recovery after photobleaching
- GFP
- green fluorescent protein
- IP/WB
- immunoprecipitation/western blot
- mCh
- mCherry
- NES
- nuclear export signal
- NLS
- nuclear localization signal
- NoLS
- nucleolar localization signal
- SILAC
- stable isotope labeling by amino acids in cell culture
- STLC
- S-trityl-L-cysteine.
REFERENCES
- 1. Trinkle-Mulcahy L., Andersen J., Lam Y. W., Moorhead G., Mann M., Lamond A. I. (2006) Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 172, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vagnarelli P., Hudson D. F., Ribeiro S. A., Trinkle-Mulcahy L., Spence J. M., Lai F., Farr C. J., Lamond A. I., Earnshaw W. C. (2006) Condensin and Repo-Man–PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 8, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wurzenberger C., Held M., Lampson M. A., Poser I., Hyman A. A., Gerlich D. W. (2012) Sds22 and Repo-Man stabilize chromosome segregation by counteracting Aurora B on anaphase kinetochores. J. Cell Biol. 198, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M. (2011) PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Current Biol. 21, 766–773 [DOI] [PubMed] [Google Scholar]
- 5. Vagnarelli P., Ribeiro S., Sennels L., Sanchez-Pulido L., de Lima Alves F., Verheyen T., Kelly D. A., Ponting C. P., Rappsilber J., Earnshaw W. C. (2011) Repo-man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Cell 21, 328–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moorhead G. B., Trinkle-Mulcahy L., Ulke-Lemée A. (2007) Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8, 234–244 [DOI] [PubMed] [Google Scholar]
- 7. Ceulemans H., Bollen M. (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 8. Peng A., Lewellyn A. L., Schiemann W. P., Maller J. L. (2010) Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Current Biol. 20, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trinkle-Mulcahy L. (2012) Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry. Proteomics 12, 1623–1638 [DOI] [PubMed] [Google Scholar]
- 11. Selbach M., Mann M. (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat Meth 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 12. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Current Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chamousset D., De Wever V., Moorhead G. B., Chen Y., Boisvert F. M., Lamond A. I., Trinkle-Mulcahy L. (2010) RRP1B targets PP1 to mammalian cell nucleoli and is associated with Pre-60S ribosomal subunits. Mol. Biol. Cell 21, 4212–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trinkle-Mulcahy L., Sleeman J. E., Lamond A. I. (2001) Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 114, 4219–4228 [DOI] [PubMed] [Google Scholar]
- 15. Chamousset D., Mamane S., Boisvert F. M., Trinkle-Mulcahy L. (2010) Efficient extraction of nucleolar proteins for interactome analyses. Proteomics 10, 3045–3050 [DOI] [PubMed] [Google Scholar]
- 16. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protocols 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 17. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 18. Linding R. (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scott M. S., Boisvert F. M., McDowall M. D., Lamond A. I., Barton G. J. (2010) Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 38, 7388–7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCright B., Rivers A. M., Audlin S., Virshup D. M. (1996) The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm*. J. Biol. Chem. 271, 22081–22089 [DOI] [PubMed] [Google Scholar]
- 21. Lee T. Y., Lai T. Y., Lin S. C., Wu C. W., Ni I. F., Yang Y. S., Hung L. Y., Law B. K., Chiang C. W. (2010) The B56γ3 regulatory subunit of protein phosphatase 2A (PP2A) regulates S phase-specific nuclear accumulation of PP2A and the G1 to S transition. J. Biol. Chem. 285, 21567–21580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kinoshita E. (2005) Phosphate-binding Tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 23. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of aurora and polo-like kinase activities in mitotic cells. Sci. Signal. 4, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dephoure N., Zhou C., Villen J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stoldt S., Wenzel D., Schulze E., Doenecke D., Happel N. (2007) G1 phase-dependent nucleolar accumulation of human histone H1x. Biol. Cell 99, 541–552 [DOI] [PubMed] [Google Scholar]
- 26. Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Science Signaling 2:ra46. [DOI] [PubMed] [Google Scholar]
- 27. Wang B., Malik R., Nigg E. A., Körner R. (2008) Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal. Chem. 80, 9526–9533 [DOI] [PubMed] [Google Scholar]
- 28. Yu L. R., Zhu Z., Chan K. C., Issaq H. J., Dimitrov D. S., Veenstra T. D. (2007) Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J. Proteome Res. 6, 4150–4162 [DOI] [PubMed] [Google Scholar]
- 29. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 30. Malik R., Lenobel R., Santamaria A., Ries A., Nigg E. A., Körner R. (2009) Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 8, 4553–4563 [DOI] [PubMed] [Google Scholar]
- 31. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol.189, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kitajima T. S., Sakuno T., Ishiguro K. I., Iemura S. I., Natsume T., Kawashima S. A., Watanabe Y. (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. EMBO J. 441, 46–52 [DOI] [PubMed] [Google Scholar]
- 33. Foley E. A., Maldonado M., Kapoor T. M. (2011) Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. EMBO J. 13, 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmstrom L., Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 [DOI] [PubMed] [Google Scholar]
- 35. Nardozzi JD, Lott K, Cingolani G. (2010) Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kitamura R., Sekimoto T., Ito S., Harada S., Yamagata H., Masai H., Yoneda Y., Yanagi K. (2006) Nuclear import of Epstein-Barr virus nuclear antigen 1 mediated by NPI-1 (Importin α5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential. J. Virol. 80, 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao C. Y. (1997) SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J. Biol. Chem. 272, 22191–22198 [DOI] [PubMed] [Google Scholar]
- 38. Kondoh K., Terasawa K., Morimoto H., Nishida E. (2006) Regulation of nuclear translocation of extracellular signal-regulated kinase 5 by active nuclear import and export mechanisms. Mol. Cell. Biol. 26, 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morimoto H., Kondoh K., Nishimoto S., Terasawa K., Nishida E. (2007) Activation of a C-terminal transcriptional activation domain of ERK5 by autophosphorylation. J. Biol. Chem. 282, 35449–35456 [DOI] [PubMed] [Google Scholar]
- 40. Kehlenbach R. H. (2000) Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J. Biol. Chem. 275, 17848–17856 [DOI] [PubMed] [Google Scholar]
- 41. De Wever V., Reiter W., Ballarini A., Ammerer G., Brocard C. (2005) A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 24, 4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jäkel S., Albig W., Kutay U., Bischoff F. R., Schwamborn K., Doenecke D., Görlich D. (1999) The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plafker S. M., Macara I. G. (2002) Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Mol. Cell. Biol. 22, 1266–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bridges D., Moorhead G. B. G. (2004) 14-3-3 Proteins: a number of functions for a numbered protein. Sci. Signal. 242:re10. [DOI] [PubMed] [Google Scholar]
- 45. Hill R., Lee P. W. (2010) The DNA-dependent protein kinase (DNA-PK): More than just a case of making ends meet? Cell Cycle 9, 3460–3469 [DOI] [PubMed] [Google Scholar]
- 46. Kunkel T. A., Erie D. A. (2005) DNA Mismatch Repair*. Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 47. Li X., andSchimenti J. C. (2007) Mouse pachytene checkpoint 2 (Trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Douglas P., Zhong J., Ye R., Moorhead G. B., Xu X., Lees-Miller S. P. (2010) Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates γ-H2AX. Mol. Cell. Biol. 30, 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.