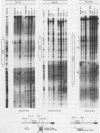
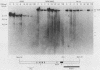
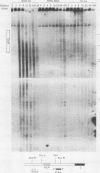
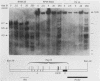
Abstract
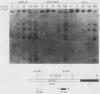
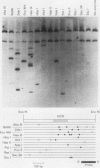
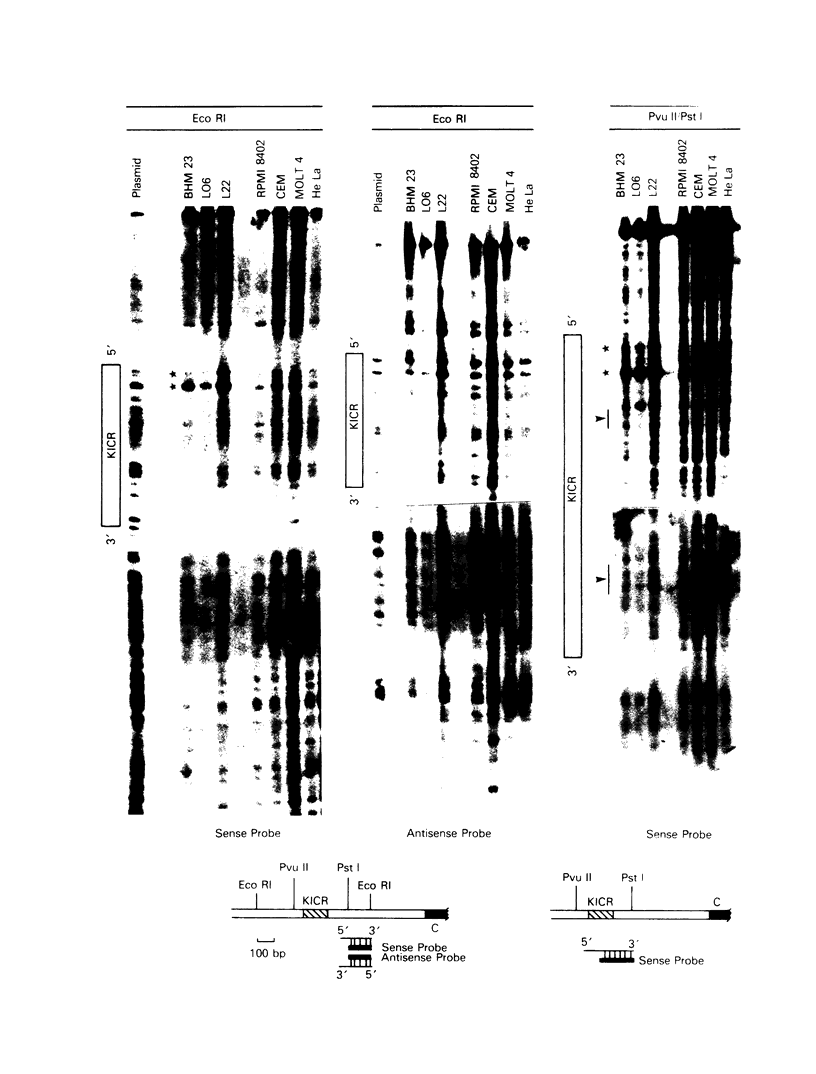
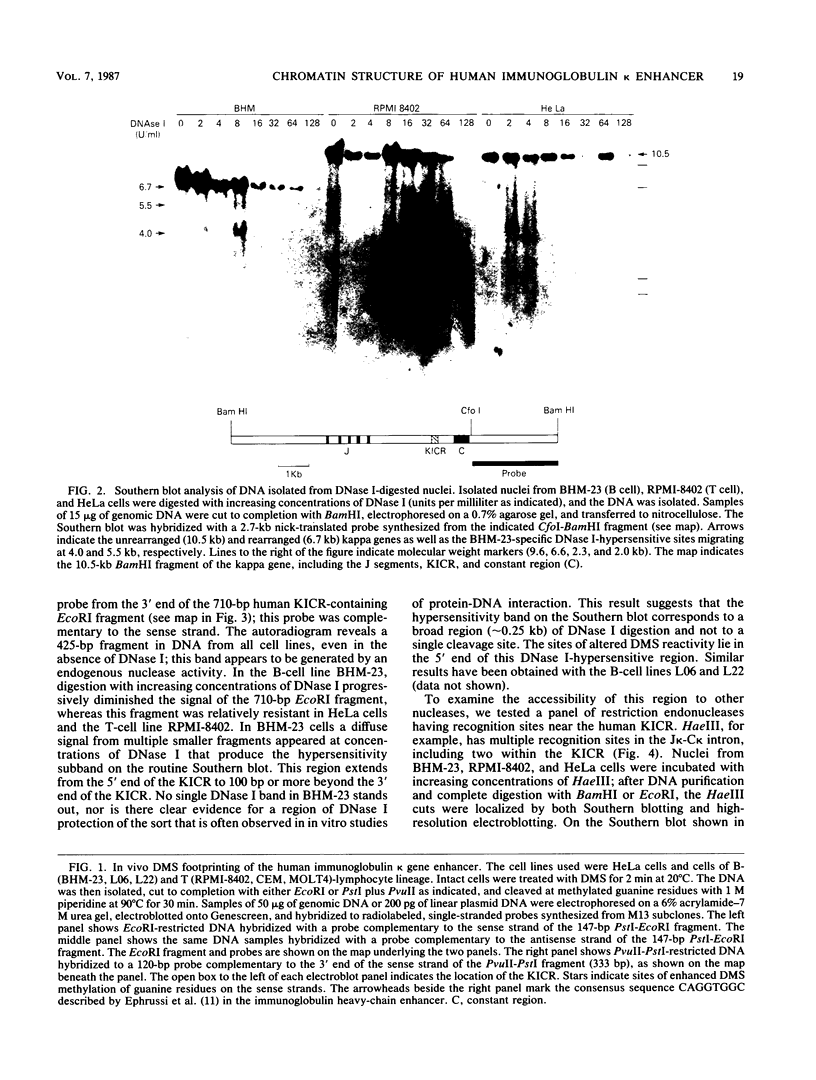
The murine immunoglobulin kappa gene enhancer has previously been found to coincide with a region of altered chromatin structure reflected in a DNase I hypersensitivity site detectable on Southern blots of B-cell DNA. We examined the chromatin structure of the homologous region of human DNA using the high-resolution electroblotting method originally developed for genomic sequence analysis by G. Church and W. Gilbert (Proc. Natl. Acad. Sci. USA 81:1991-1995, 1984). Analysis of DNA isolated from cells treated in vivo with dimethyl sulfate revealed two B-cell-specific sites of enhanced guanine methylation. Both sites are located within perfect inverted repeats theoretically capable of forming cruciform structures; one of these repeats overlaps an enhancer core sequence. No enhancement or protection of guanine methylation was observed within sequences similar to sites of altered methylation previously described in the immunoglobulin heavy-chain enhancer. Treatment of isolated nuclei with DNase I or a variety of restriction endonucleases defined a B-cell-specific approximately 0.25-kilobase region of enhanced nuclease susceptibility similar to that observed in the murine kappa enhancer. The 130-base-pair DNA segment that shows high sequence conservation between human, mouse, and rabbit DNAs lies at the 5' end of the nuclease-susceptible region.
Full text
PDF


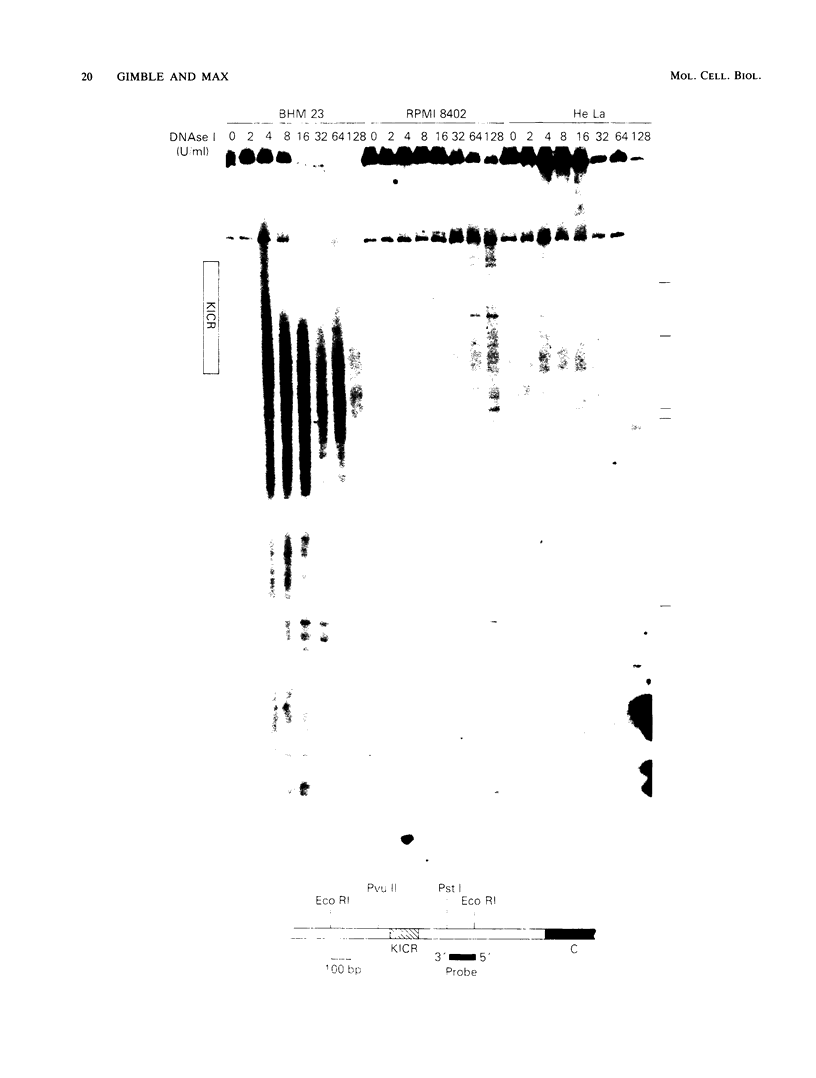

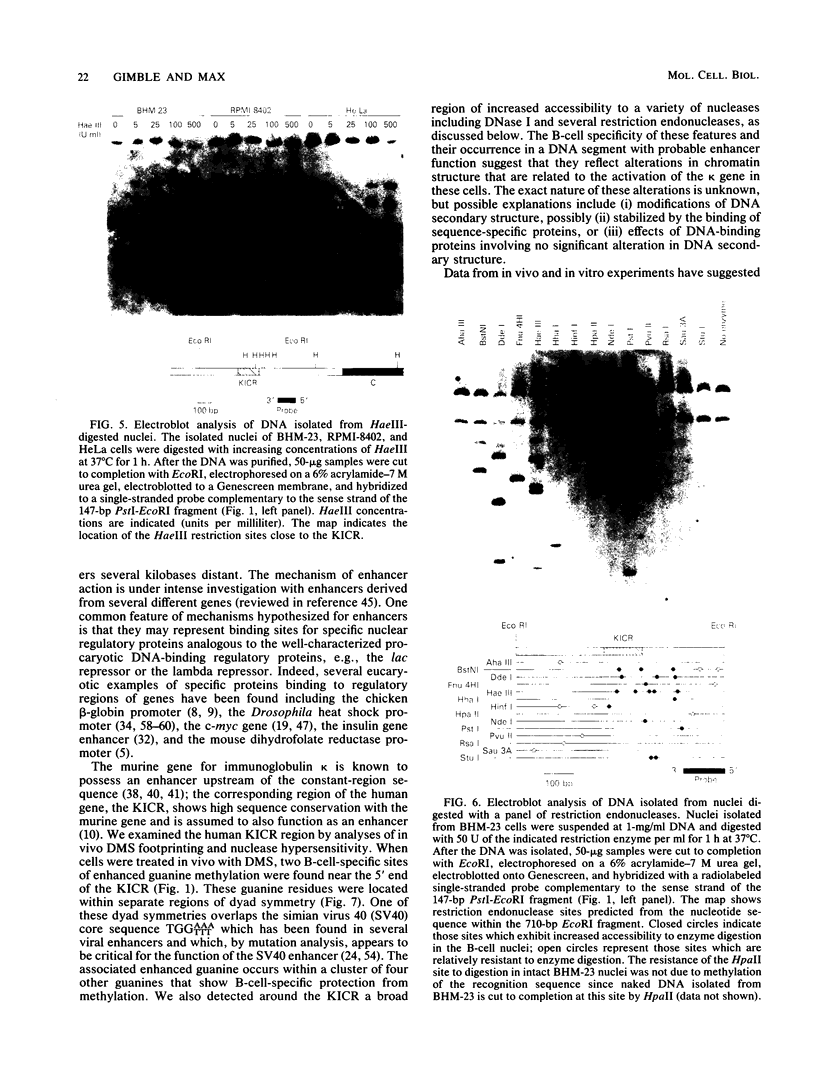
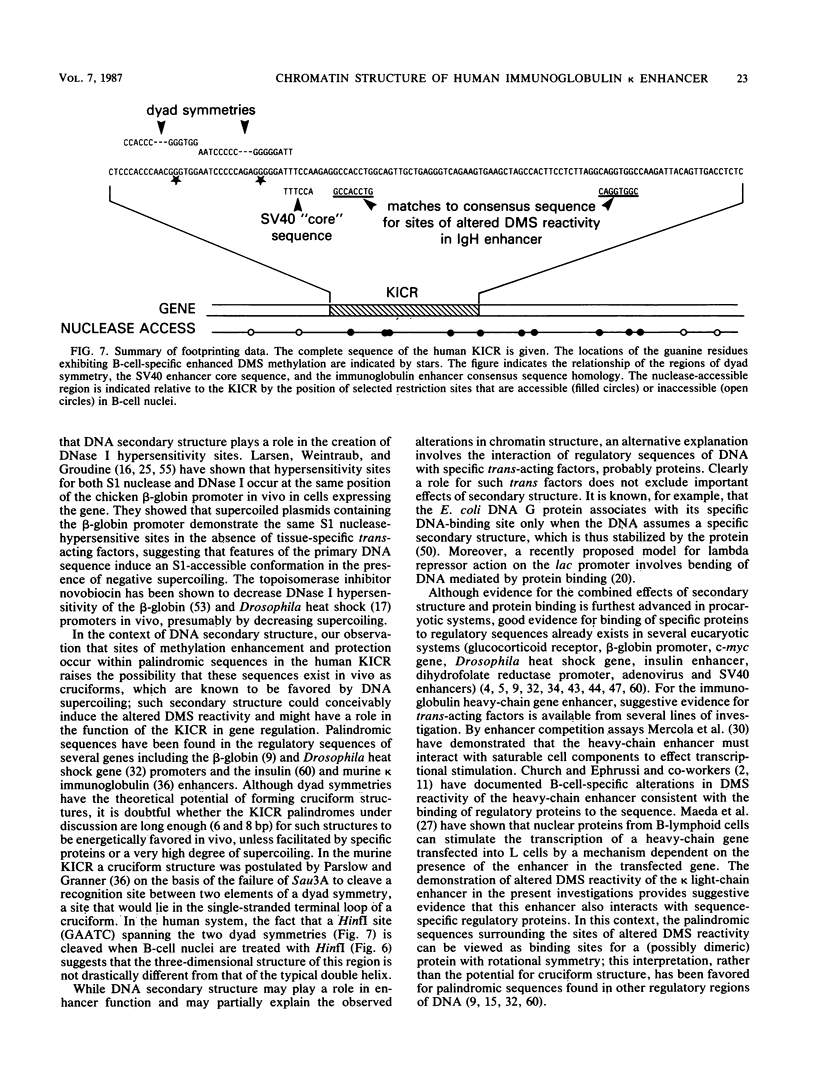


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chung S. Y., Folsom V., Wooley J. DNase I-hypersensitive sites in the chromatin of immunoglobulin kappa light chain genes. Proc Natl Acad Sci U S A. 1983 May;80(9):2427–2431. doi: 10.1073/pnas.80.9.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Ephrussi A., Gilbert W., Tonegawa S. Cell-type-specific contacts to immunoglobulin enhancers in nuclei. 1985 Feb 28-Mar 6Nature. 313(6005):798–801. doi: 10.1038/313798a0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Yamamoto K. R. Two different factors act separately or together to specify functionally distinct activities at a single transcriptional enhancer. Mol Cell Biol. 1986 Apr;6(4):993–1001. doi: 10.1128/mcb.6.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Sazer S., Tjian R., Schimke R. T. Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature. 1986 Jan 16;319(6050):246–248. doi: 10.1038/319246a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Emorine L., Kuehl M., Weir L., Leder P., Max E. E. A conserved sequence in the immunoglobulin J kappa-C kappa intron: possible enhancer element. Nature. 1983 Aug 4;304(5925):447–449. doi: 10.1038/304447a0. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell. 1983 Aug;34(1):125–134. doi: 10.1016/0092-8674(83)90142-3. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982 Aug;30(1):131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Novobiocin blocks the Drosophila heat shock response. J Mol Biol. 1985 May 5;183(1):13–29. doi: 10.1016/0022-2836(85)90277-3. [DOI] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci U S A. 1985 May;82(10):3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Siebenlist U., Danner D., Leder P., Rawlins D., Rosenfeld P., Kelly T., Jr High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985 Mar 21;314(6008):289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Jackson P. D., Felsenfeld G. A method for mapping intranuclear protein-DNA interactions and its application to a nuclease hypersensitive site. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2296–2300. doi: 10.1073/pnas.82.8.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Korsmeyer S. J., Arnold A., Bakhshi A., Ravetch J. V., Siebenlist U., Hieter P. A., Sharrow S. O., LeBien T. W., Kersey J. H., Poplack D. G. Immunoglobulin gene rearrangement and cell surface antigen expression in acute lymphocytic leukemias of T cell and B cell precursor origins. J Clin Invest. 1983 Feb;71(2):301–313. doi: 10.1172/JCI110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lohr D., Hopper J. E. The relationship of regulatory proteins and DNase I hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res. 1985 Dec 9;13(23):8409–8423. doi: 10.1093/nar/13.23.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Kitamura D., Kudo A., Araki K., Watanabe T. Trans-acting nuclear protein responsible for induction of rearranged human immunoglobulin heavy chain gene. Cell. 1986 Apr 11;45(1):25–33. doi: 10.1016/0092-8674(86)90534-9. [DOI] [PubMed] [Google Scholar]
- Max E. E., Korsmeyer S. J. Human J chain gene. Structure and expression in B lymphoid cells. J Exp Med. 1985 Apr 1;161(4):832–849. doi: 10.1084/jem.161.4.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Mercola M., Goverman J., Mirell C., Calame K. Immunoglobulin heavy-chain enhancer requires one or more tissue-specific factors. Science. 1985 Jan 18;227(4684):266–270. doi: 10.1126/science.3917575. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H., Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986 Apr 11;45(1):35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Oi V. T., Morrison S. L., Herzenberg L. A., Berg P. Immunoglobulin gene expression in transformed lymphoid cells. Proc Natl Acad Sci U S A. 1983 Feb;80(3):825–829. doi: 10.1073/pnas.80.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Structure of a nuclease-sensitive region inside the immunoglobin kappa gene: evidence for a role in gene regulation. Nucleic Acids Res. 1983 Jul 25;11(14):4775–4792. doi: 10.1093/nar/11.14.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payvar F., DeFranco D., Firestone G. L., Edgar B., Wrange O., Okret S., Gustafsson J. A., Yamamoto K. R. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983 Dec;35(2 Pt 1):381–392. doi: 10.1016/0092-8674(83)90171-x. [DOI] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Pospelov V. A., Klobeck H. G., Zachau H. G. Correlation between DNase I hypersensitive sites and putative regulatory sequences in human immunoglobulin genes of the kappa light chain type. Nucleic Acids Res. 1984 Sep 25;12(18):7007–7021. doi: 10.1093/nar/12.18.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Queen C., Stafford J. Fine mapping of an immunoglobulin gene activator. Mol Cell Biol. 1984 Jun;4(6):1042–1049. doi: 10.1128/mcb.4.6.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Sundaresan V., Ausubel F. M. Specific protection of nucleotides in the lac operator from DMS methylation and DNase I nicking by crude bacterial cell extracts. Gene. 1982 Jun;18(3):247–255. doi: 10.1016/0378-1119(82)90162-7. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P., Wildeman A., Chambon P. A trans-acting factor is responsible for the simian virus 40 enhancer activity in vitro. Nature. 1985 Feb 7;313(6002):458–463. doi: 10.1038/313458a0. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Sims J., Benz E. W., Jr Initiation of DNA replication by the Escherichia coli dnaG protein: evidence that tertiary structure is involved. Proc Natl Acad Sci U S A. 1980 Feb;77(2):900–904. doi: 10.1073/pnas.77.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984 Sep 6;311(5981):81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- Wu C. An exonuclease protection assay reveals heat-shock element and TATA box DNA-binding proteins in crude nuclear extracts. Nature. 1985 Sep 5;317(6032):84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]