Background: Human topoisomerase IIα unlinks catenated chromosomes and preferentially relaxes positive supercoils.
Results: Supercoil chirality, twist density, and tension determine topoisomerase IIα relaxation rate and processivity.
Conclusion: Strand passage rate is determined by the efficiency of transfer segment capture that is modulated by the topoisomerase C-terminal domains.
Significance: Single-molecule measurements reveal the mechanism of chiral discrimination and tension dependence of supercoil relaxation by human topoisomerase IIα.
Keywords: Biophysics, DNA Replication, DNA Topoisomerase, DNA Topology, Single Molecule Biophysics, Chiral Discrimination, Magnetic Tweezers
Abstract
Type IIA topoisomerases (Topo IIA) are essential enzymes that relax DNA supercoils and remove links joining replicated chromosomes. Human topoisomerase IIα (htopo IIα), one of two human isoforms, preferentially relaxes positive supercoils, a feature shared with Escherichia coli topoisomerase IV (Topo IV). The mechanistic basis of this chiral discrimination remains unresolved. To address this important issue, we measured the relaxation of individual supercoiled and “braided” DNA molecules by htopo IIα using a magnetic tweezers-based single-molecule assay. Our study confirmed the chiral discrimination activity of htopo IIα and revealed that the strand passage rate depends on DNA twist, tension on the DNA, and the C-terminal domain (CTD). Similar to Topo IV, chiral discrimination by htopo IIα results from chiral interactions of the CTDs with DNA writhe. In contrast to Topo IV, however, these interactions lead to chiral differences in relaxation rate rather than processivity. Increasing tension or twist disrupts the CTD-DNA interactions with a subsequent loss of chiral discrimination. Together, these results suggest that transfer segment (T-segment) capture is the rate-limiting step in the strand passage cycle. We propose a model for T-segment capture that provides a mechanistic basis for chiral discrimination and provides a coherent explanation for the effects of DNA twist and tension on eukaryotic type IIA topoisomerases.
Introduction
The duplex state of DNA provides stability and protection, in addition to a robust duplication mechanism, for genetic information. However, the double helical structure creates topological complications because DNA metabolic processes including replication, transcription, and repair require DNA strand separation (1). These topological complications are resolved by topoisomerases, ubiquitous enzymes found in all phyla that regulate DNA topology and maintain topological homeostasis (2–7). Topoisomerases are characterized as type I or II based on the number of DNA strands cleaved and dependence on ATP hydrolysis (2, 4, 5, 7). Topo IIA3 enzymes share similar core domains and the corresponding catalytic “two-gate” mechanism: passing one segment of duplex DNA (the transfer or T-segment) through a transient double-stranded break in a second segment of DNA (the gate or G-segment) in an ATP-dependent process (5, 6, 8–10). Catalytically dispensable C-terminal domains (CTDs) are structurally diverse among Topo IIA enzymes, but are important for cellular function as they contain phosphorylation sites and nuclear localization signals (11–16). Growing evidence suggests that CTD variations among Topo IIA enzymes also alter the topological specificity and activity of the otherwise similar core domain functions of prokaryotic and eukaryotic Topo IIA enzymes (17–23). For example, Escherichia coli topoisomerase IV (Topo IV) preferentially relaxes positive supercoils in a chiral discrimination process mediated by the CTDs (5, 24, 25). The molecular mechanisms underlying chiral discrimination by Topo IV have not been entirely elucidated, although it was shown to arise from chirality-dependent differences in enzymatic processivity, i.e. the number of strand passages catalyzed per binding event (25). To date, an atomic structure of a eukaryotic Topo IIA CTD has not been solved, precluding a detailed structural comparison with prokaryotic CTDs. Nonetheless, it has been suggested that eukaryotic Topo IIA CTDs differ from bacterial CTDs in that they are likely unstructured and lack a globular DNA binding domain (5). Furthermore, most eukaryote Topo IIA enzymes studied to date lack chirality-dependent differences in supercoil relaxation (24, 26). Interestingly, htopo IIα, but not the other human Topo II isoform, htopo IIβ, preferentially relaxes positive supercoils (20, 21). Deletion of the htopo IIα CTD or replacement with the htopo IIβ CTD abolishes chiral discrimination, whereas a chimera of the htopo IIβ core and htopo IIα CTD exhibits robust chiral discrimination (20, 21). These findings establish that chiral discrimination is mediated by the CTDs; however, the mechanistic basis of chiral discrimination by htopo IIα and its relationship to chiral discrimination by Topo IV remain unclear. Furthermore, a recent single-molecule measurement of DNA unlinking by htopo IIα failed to observe chiral discrimination (27). Here we used a magnetic tweezers-based single-molecule assay to investigate supercoil relaxation and DNA unlinking activities of wild type (WT) htopo IIα and a CTD deletion mutant of htopo IIα (ΔCTD; deletion from 1175 to 1531) (20).
Chiral discrimination by htopo IIα could result from either twist-dependent or writhe-dependent differences in relaxation (24, 28). Two single-molecule DNA geometries were employed to distinguish between these possibilities (Fig. 1, A and B). The first geometry, the single-molecule supercoil relaxation assay, is based on a single rotationally constrained DNA molecule in which supercoils are generated by rotating a magnetic bead tethered to the surface by the DNA. This geometry affords precise control over twist and writhe while providing high-resolution measurements of relaxation rate and processivity (29). The second geometry, the braiding assay, in which two DNA strands are wound around one another, allows creation of DNA links with well defined writhe in the absence of twist (29). DNA tension, which is likely substantial during DNA processing in vivo, can also be varied in this geometry.
FIGURE 1.
DNA experimental geometry and supercoil relaxation data trace. A, DNA extension as a function of turns (linking number difference, ΔLk). Supercoiling was achieved by rotating a magnetic bead (green) attached to rotationally constrained DNA (blue) (inset graphic). Supercoiling twists the DNA until a buckling transition, past which the DNA twist remains constant, and further rotation of the bead is converted to writhe (plectonemes) that reduces the extension of the bead (49). Thus, DNA twist can be controlled by applied tension because the buckling transition occurs at higher twist density as the tension increases (50). At F = 0.2 pN (red circles), DNA extension decreases symmetrically for both positive and negative turns. At F = 2.0 (green circles), DNA extension decreases only for positive turns as negative turns lead to DNA melting, rather than plectoneme formation. B, a DNA braid is formed by wrapping two DNA molecules tethered to a bead around one another by rotating the bead. At 0.5 turns, the DNA extension (z) decreases sharply by an amount dependent on the distance between the two DNA molecules (ρ) and the length of the DNA (z0). At nbuckle > n > 0.5, the range of bead rotations (n) over which our measurements were made, the extension decreases as: z(n) = √ z02 − (2ρ + 2πR(n − 0.5))2, where R is the effective electrostatic diameter of the DNA molecule (∼11 nm under our conditions) (24, 37). At nbuckle < n (black dashed lines indicate nbuckle), the DNA braid forms a second order plectonemic structure. nbuckle is determined by the condition z(n)/z0 = 1/√(2) (37). C, trace of a htopo IIα supercoil relaxation experiment. Supercoils are introduced by rotating the magnets (pink arrows), and the DNA extension decreases as the DNA buckles and forms plectonemes. DNA extension increases when htopo IIα removes supercoils (green dashed lines). The magnets automatically rotate to reintroduce a fixed number of supercoils when the DNA extension reaches a threshold value (red dashed line), thereby creating a pseudo-infinite substrate for htopo IIα. Twait is defined by the time duration required for htopo IIα to complete N cycles (N is the half of the magnetic turns introduced each time the DNA extension reaches the threshold). The enzyme works processively until it falls off. The processivity was determined by counting the number of cycles completed during a processive burst (red shaded region).
Relaxation measurements of supercoiled DNA revealed that WT and ΔCTD htopo IIα displayed opposite chiral preferences at low DNA twist density, yet they both relaxed negative and positive supercoils with comparable processivity. Unlinking experiments of “braided” DNA revealed preferential relaxation of positive writhe by WT htopo IIα and negative writhe by the ΔCTD, consistent with chiral discrimination arising from selection of a preferred crossing geometry by the CTDs. These results further imply that T-segment capture is the rate-determining step in the strand passage reaction during processive relaxation, which we argue is likely the case for the majority of eukaryotic type IIA topoisomerases. We propose a DNA topology-dependent model for htopo IIα activity that accounts for CTD-dependent chirality sensing and the effects of DNA twist and tension on supercoil relaxation. More generally, this work contributes to our understanding of the complex interplay between DNA topology and the activity of topoisomerases that modify and control topology.
EXPERIMENTAL PROCEDURES
Coilable DNA Preparation
DNA templates with multiple biotin moieties on one extremity and multiple digoxigenin moieties on the other were made by ligating two 500-bp DNA handles labeled with either biotin or digoxigenin on the ends of a 5-kb dsDNA as described previously (30, 31). Briefly, the 5-kb DNA was made by PCR from pET28 plasmid (EMD4 Biosciences) with primers (forward, 5′-GGACCTGCTTTCCAACGCCATATTCAACGGGAAACG-3′ at position 3999; and reverse, 5′GGGTCTCGACCAAACAGCTGATTGCCCTTCAC-3′ at position 1728, Invitrogen) that were both extended to include nonpalindromic restriction sites at their 5′ ends to generate ssDNA regions complementary to the DNA handles after digestion. The two handles were made by PCR using pBluescript II (Stratagene) as a template and primers also containing nonpalindromic restriction sites (forward, 5′-GGACCTGCTTTCGTTGTGGCGTAATCATGGTCATAG-3′; and reverse, 5′-GGGTCTCGTGGTTTATAGTCCTGTCGGGTTTC-3′, Invitrogen). For multiple labeling with biotin or digoxigenin, 10–20% of appropriately labeled dUTP (Roche Applied Science) was added to the PCR reaction. After PCR, the products were purified (Qiagen PCR purification kit) and cut with restriction endonucleases BsaI (New England Biolabs) for the digoxigenin-labeled DNA handle, BfuAI (New England Biolabs) for the biotin-labeled DNA handle, and both enzymes for the 5-kb DNA fragment. After restriction enzyme digestion, the biotin- and digoxigenin-labeled DNA handles were ligated to the main DNA segment at room temperature for 3 h (30, 31).
Braided DNA Preparation
12-kb DNA labeled with a single biotin and digoxigenin at each 5′ end, respectively, was made by PCR employing 48-kb λ phage DNA (New England Biolabs) as a template with a biotin-labeled forward primer and a digoxigenin-labeled reverse primer (Eurofins MWG Operon). PCR products were purified as above.
Protein Preparation
WT and ΔCTD htopo IIα were prepared as described previously (32, 33).
Single-molecule Uncoiling Experiments
To prepare torsionally constrained (coilable) DNA tethers, 0.3 nm of multiple digoxigenin and biotin end-labeled DNA prepared as described above was mixed with 32 ng of anti-digoxigenin (Roche Applied Science) in 50 μl of 1× PBS buffer for 1 h at room temperature. This mixture was introduced into a sample cell (∼30 μl total volume) coated with a low concentration of stuck beads and incubated overnight at 4 °C (30). Unbound DNA was washed out with 200 μl of wash buffer (WB; 1× PBS, 0.04% Tween 20, 0.3% w/v BSA). 20 μl of streptavidin-coated magnetic beads (Dynabeads MyOne, Invitrogen) was then introduced in wash buffer and allowed to tether for 1 h and then washed with 1 ml of WB. Once a coilable DNA substrate was located, 200 μl of topoisomerase buffer (10 mm Tris-HCl (pH 8.0), 0.1 mm EDTA, 5 mm MgCl2, 175 mm KCl, 0.01% v/v Tween 20, and 0.3% w/v BSA) was washed through the flow cell, and topoisomerase was added at a concentration of 50–500 pm in 200 μl of topoisomerase buffer supplemented with 1 mm ATP. Topoisomerase activity was measured by tracking the height of the tethered bead in real time from CCD images captured at 100 Hz. The position of the sample cell with respect to the objective was actively stabilized by tracking the position of a fiducial bead affixed to the surface of the flow cell and moving the three-dimensional piezoelectric stage to compensate for the drift (31). To accurately measure the number of stand passages per enzyme binding event, i.e. the processivity, which could be much larger than the total writhe in the DNA molecule, we generated an “infinite” supercoiled substrate (34). As supercoils were removed, the accompanying increase in the height of the bead was recorded in real time (Fig. 1). When the height of the bead exceeded a threshold value, corresponding to the removal of all but three crossings, a fixed number of supercoils was automatically reintroduced by rotating the magnets faster (>10 turns × s−1) than the average enzyme relaxation rate (∼3 cycles × s−1). In this manner, processive bursts consisting of hundreds of strand passages were accurately measured.
Single-molecule Unlinking Experiments
To prepare double DNA tethers for braiding experiments, 1 μl of 60 pm 12-kb dsDNA singly biotin- and digoxigenin-labeled at each end was incubated with 10 μl of magnetic beads (1% w/v, Dynabeads MyOne, Invitrogen) in 200 μl of WB overnight (3:1 DNA to bead ratio). The sample cell (∼30 μl total volume) was incubated with 30 μl of 10 μg/ml anti-digoxigenin in 1× PBS for 1 h at room temperature followed by a wash with 200 μl of WB. The DNA-bead mixture was incubated in the anti-digoxigenin-coated sample cell for 30 min before washing with 400 μl of WB. Topoisomerase activity was measured as described above.
Catalytic Rate Analysis
Data traces were analyzed with custom software to identify the change in DNA extension, the relaxation rate, and the duration of each processive relaxation event (30, 31). To determine the mean relaxation or unlinking rate, the waiting times, Twait, between the imposition and relaxation of a fixed number of supercoils or links were measured during processive htopo IIα relaxation events (Fig. 1C). Under the assumption that strand passage is governed by a single rate-determining step, i.e. a Poisson process, Twait is a convolution of N exponential steps that is described by a Gamma distribution (35). Because the mean relaxation rate per N enzymatic cycles is N/Twait, the distribution of the mean rates was fitted to an inverse Gamma distribution to obtain the average rate
 |
where v is the velocity measured over N cycles, each occurring with rate k and P0 is a scaling factor.
RESULTS
Chirality-dependent Relaxation Rates at Low DNA Twist Density
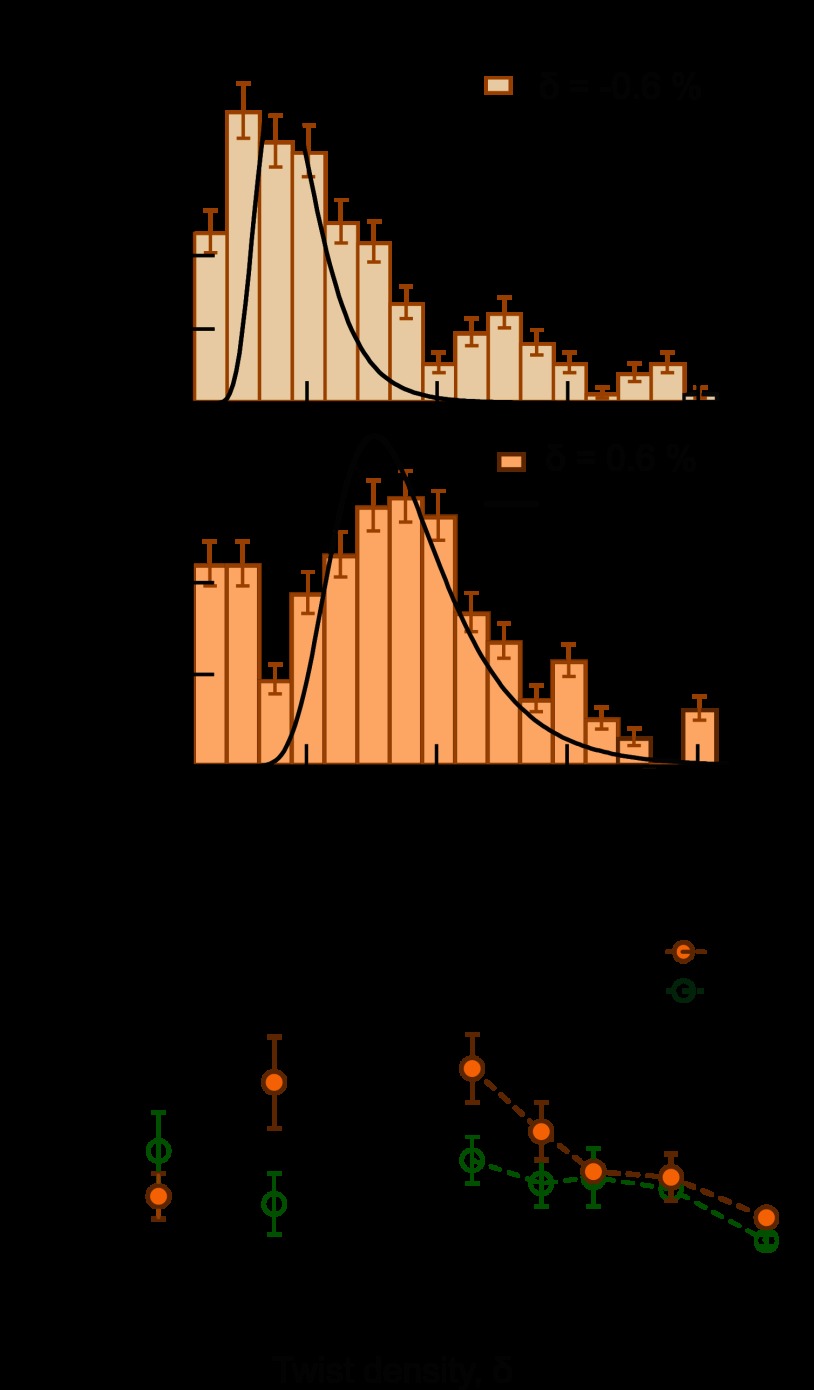
Previous ensemble studies revealed that WT htopo IIα relaxed positive supercoils faster than negative supercoils (21) and demonstrated that this chiral discrimination was entirely dictated by the CTDs (20). To elucidate the mechanistic basis of chiral discrimination by htopo IIα, we measured supercoil relaxation by WT and ΔCTD htopo IIα as a function of DNA twist density. Directly manipulating the linking number and tension of DNA with magnetic tweezers allows us to independently control the change in twist (ΔTw) and writhe (ΔWr) for a given linking number difference (ΔLk). For plasmid DNA, linking number differences are partitioned into ∼75% writhe and ∼25% twist, essentially independent of the superhelical density (36). Under tension, this partitioning depends on the superhelical density and the tension (37). As the magnetic bead is rotated, the change in the linking number of the DNA is accommodated purely by DNA twist for small values of superhelical density. At a tension-dependent critical value of twist, the DNA buckles to form a plectoneme, one manifestation of DNA writhe. Subsequent increases in linking number are entirely accommodated by increases in writhe, whereas the twist remains constant. The twist density of the DNA can therefore be accurately controlled by tension applied on the DNA. Moreover, the twist in the DNA is independent of the writhe, so supercoil relaxation experiments were carried out under conditions of fixed twist density (−0.6, 0.6, 1, 2, 2.9, and 4%). By analogy with the definition of superhelical density σ, we define twist density, δ, as the fractional change in DNA twist: δ = ΔTw/Tw0. We estimate that in ensemble measurements of chirality-dependent relaxation by htopo IIα, the maximum twist density was δ ∼ ±0.6% and that it decreased as the linking number decreased over the course of the experiment (20, 21). In the single-molecule measurements, the twist density was both higher and constant. Relaxation rates were determined by fitting the distribution of mean velocities with an inverse Gamma function (Fig. 2A, see “Experimental Procedures”). The velocity distributions are somewhat broader than expected, likely due to heterogeneous enzymatic activity. To ensure that the results are independent of this apparent heterogeneity, we computed the average strand passage rates as a function of twist density (data not shown), which are consistent with the fitting results (Fig. 2).
FIGURE 2.
Relaxation rate of positive and negative supercoils by WT and ΔCTD htopo IIα. A, two representative WT htopo IIα relaxation rate distributions. The relaxation rate distributions (bars) were fitted with inverse Gamma distributions (black line, see “Experimental Procedures”) to obtain the mean rates. B, relaxation rates of negative and positive supercoiled DNA at low twist density. WT relaxes positive supercoils ∼2-fold faster than negative supercoils. On the other hand, ΔCTD demonstrates the opposite chiral preference, relaxing negative supercoils ∼1.4-fold faster than positive supercoils. Relaxation rates (cycles × s−1 (cyc/s)) measured for WT are: 1.7 ± 0.3 (δ = −0.6%; number of events, n = 183) and 3.2 ± 0.6 (δ = 0.6%; n = 238). For ΔCTD, relaxation rates are: 2.3 ± 0.5 (δ = −0.6%; n = 228) and 1.6 ± 0.4 (δ = 0.6%; n = 295). C, relaxation rate as a function of positive DNA twist density. The relaxation rates for both WT and ΔCTD decrease as a function of DNA twist density. Relaxation rates (cycles × s−1) measured for WT are: 3.2 ± 0.6 (δ = 0.6%; n = 238), 2.1 ± 0.5 (δ = 1.0%; n = 258), 1.4 ± 0.1 (δ = 2.0%; n = 61), 1.3 ± 0.5 (δ = 2.9%; n = 31), and 0.6 ± 0.1 (δ = 4.0%; n = 24). For ΔCTD, rates are: 1.6 ± 0.4 (δ = 0.6%; n = 300), 1.2 ± 0.4 (δ = 1.0%; n = 173), 1.3 ± 0.5 (δ = 2.0%; n = 87), and 1.2 ± 0.2 (δ = 2.9%; n = 202), and 0.2 ± 0.1 (δ = 4.0%; n = 53). Measurements for all figures are reported as mean ± S.D. of the fit parameters. The average relaxation rates plotted as a function of DNA twist are qualitatively the same and support the same overall conclusions concerning chiral discrimination and the effects of twist on strand passage rates (data not shown).
At equal but opposite twist densities, δ = −0.6 and 0.6%, WT htopo IIα relaxed positive supercoils approximately twice as fast as negative supercoils (Fig. 2, A and B). Conversely, ΔCTD displayed the opposite chiral preference, relaxing negative DNA supercoils 1.4-fold faster than positive supercoils (Fig. 2B). As the twist density was increased (1–4.0%), the relaxation rate of the WT initially decreased more significantly than the ΔCTD. For twist densities of ∼2% and above, the relaxation of both enzymes was comparable and continued to decrease slightly (Fig. 2C). Twist dependence may be universal among eukaryote Topo IIA enzymes because Drosophila and yeast Topo II demonstrate comparable twist sensitivity in contrast to E. coli Topo IV, which is relatively insensitive to twist at low twist densities (38, 39).
htopo IIα Is Highly Processive on Both Positively and Negatively Supercoiled DNA
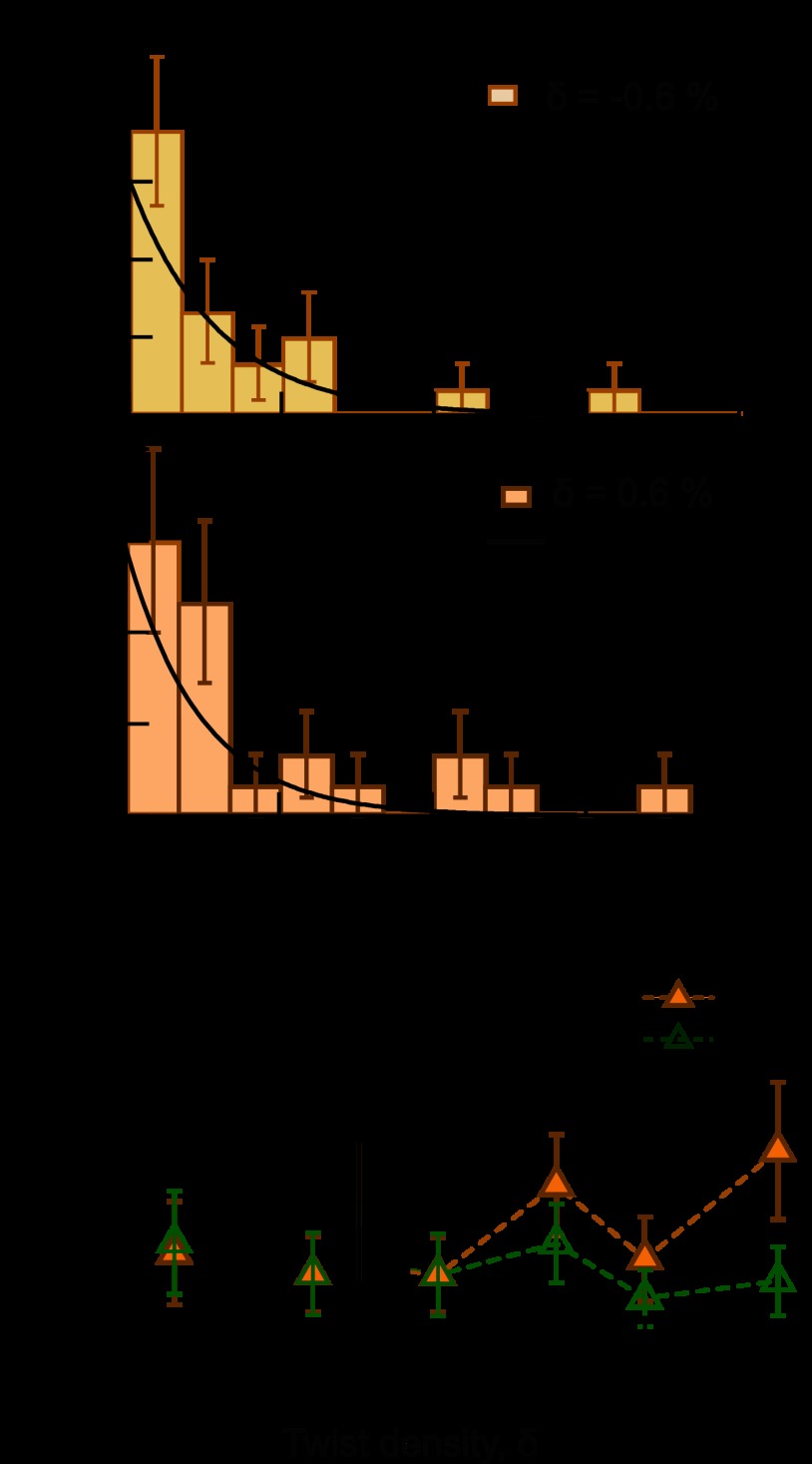
Chiral discrimination by Topo IV results from dramatic chirality-dependent differences in processivity; the enzyme completes many T-segment transfer cycles per G-segment binding on positive supercoils, whereas it can only complete one cycle on negative supercoils (29). Despite sharing chiral discrimination activity with Topo IV, htopo IIα was highly processive under all measurement conditions (Fig. 3). Processivity was directly determined by measuring the number of cycles completed in individual bursts of activity in the single-molecule supercoil relaxation records. Automatic computer-controlled supercoiling of the DNA as the enzyme relaxed supercoils created an infinite supercoiled substrate, permitting the accurate measurement of hundreds of strand passages during a single binding event (Fig. 1C). Given the low enzyme concentration and the long latency periods separating processive bursts of activity, it is reasonable to assume that each burst of activity corresponded to the action of an individual enzyme. Under conditions for which chiral discrimination was observed, WT htopo IIα relaxed both negative (δ = −0.6%) and positive (δ = 0.6%) supercoiled DNA with similar processivity (60–80 cycles; Fig. 3, A and B). ΔCTD was equally processive on both positively and negatively supercoiled DNA at these twist densities, suggesting that the core domain alone stably binds G-segment DNA at low twist density (Fig. 3B). At higher twist density (δ > 0.6%), the processivity of WT htopo IIα was slightly higher than that of the ΔCTD (Fig. 3C), suggesting a role for the CTDs in stabilizing G-segment binding at higher twist densities. Overall, these results reveal that, in contrast with Topo IV, htopo IIα processivity does not depend strongly on the supercoil chirality. Furthermore, the htopo IIα CTDs are not essential for processivity, but rather enhance DNA binding stability, particularly at elevated twist densities at which the relaxation rate decreased (Fig. 2C).
FIGURE 3.
Processivity of WT and ΔCTD htopo IIα. A, two representative processivity distributions for WT htopo IIα. Error bars correspond to the square root of the number of events in each bin. Each distribution was fitted with a single exponential (black line) to obtain the mean processivity. B, comparison of processivity at low negative and positive DNA twist density. Processivity for both htopo IIα enzymes was slightly higher for negative supercoils. WT processivity was: 84 ± 33 (δ = −0.6%; n = 22) and 71 ± 24 (δ = 0.6%; n = 23), whereas ΔCTD processivity was: 91 ± 33 (δ = −0.6%; n = 19) and 71 ± 26 (δ = 0.6%; n = 18). C, processivity plotted as a function of positive DNA twist density. The processivity of the WT enzyme increased slightly with increasing twist density, unlike the processivity of the ΔCTD enzyme that was relatively constant. WT htopo IIα processivity was: 71 ± 24 (δ = 0.6%; n = 23), 128 ± 32 (δ = 1.0%; n = 41), 81 ± 27 (δ = 2.0%; n = 16), and 150 ± 44 (δ = 2.9%; n = 14). ΔCTD processivity was: 71 ± 26 (δ = 0.6%; n = 18), 91 ± 25 (δ = 1.0%; n = 25), 56 ± 18 (δ = 2.0%; n = 16), and 67 ± 22 (δ = 2.9%; n = 18).
Chiral Discrimination by htopo IIα Depends on DNA Writhe
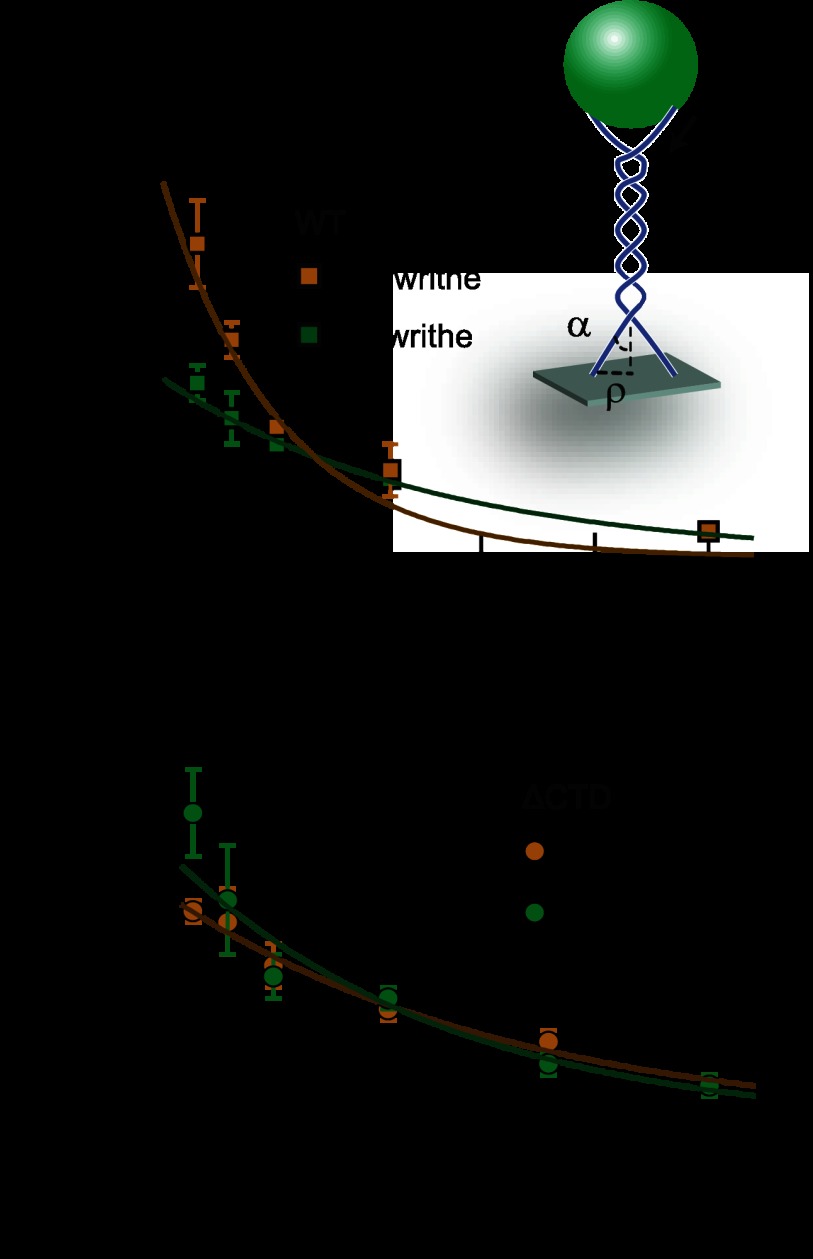
Relaxation measurements confirmed chiral discrimination by htopo IIα and demonstrated that it arises from chirality-dependent changes in the strand passage rate. However, the question remained as to whether htopo IIα distinguishes chirality based on DNA twist or writhe, which are convolved in both ensemble and single-molecule supercoil relaxation experiments. To distinguish the effects of twist and writhe, we employed a DNA braiding assay (Figs. 1 and 4, inset) in which the magnetic bead is tethered by two rotationally unconstrained DNA molecules (24, 37). Rotating the bead braids the two strands, generating an intertwined structure similar to that of a plectoneme with well defined writhe but no twist. The introduction of writhe was accompanied by a decrease in the extension of the bead, which was calibrated to obtain the relationship between writhe (number of DNA links) and extension (Fig. 1B). Rotating the bead in the clockwise direction imposes left-handed crossings (negative writhe), whereas rotating the bead in the counterclockwise direction imposes right-handed crossings (positive writhe). The geometry, i.e. the juxtaposition angle (α), of the crossings within the braid is determined by the tension on the DNA, the DNA length, and the spacing between the two DNA molecules (ρ; Fig. 4A, inset) (24, 37). The braiding assay can therefore be used to distinguish the effects of writhe and DNA tension on topoisomerase activity without the potentially confounding influence of DNA twist. Experiments were performed in a manner similar to the supercoil relaxation experiments. Rotating the magnets imposed writhe and decreased the height of the bead. The number of imposed turns was kept below the critical value at which the braided DNA buckles to form a four helix plectoneme (37). To determine whether chiral discrimination depends on writhe, we measured the rate at which WT htopo IIα relaxed positively and negatively braided 12-kb DNA molecules at five levels of tension. Positive writhe was relaxed faster than negative writhe at low tension (0.25 and 0.4 pN), indicating that chiral discrimination depends on DNA writhe rather than twist (Fig. 4A). However, as tension increased, unlinking rates for both positive and negative writhe decreased to a similar extent with the concomitant loss of chiral discrimination. The tension-dependent decrease in unlinking rate indicates a tension-sensitive rate-limiting step. The unlinking data were therefore further analyzed within the framework of a single energy barrier-crossing model in which the enzyme performs work, i.e. moves or distorts the DNA, against the DNA tension, resulting in a tension-dependent rate (24, 40).
Here v is the unlinking rate, v0 is the unlinking rate at zero applied tension, and 〈T〉 = F/(2〈cosα〉) is the mean tension along the DNA for a force, F, on the magnetic bead with mean crossing angle α (Fig. 4, inset). Δx is the distance to the transition state associated with the tension-dependent step, and kBT is the thermal energy (24). The tension-dependent unlinking rate was well fit by this model for negative writhe with fitting parameters (v0 = 2.2 ± 0.2 s−1; Δx = 3.5 ± 0.5 nm; χv2 = 1.92) that are similar to the positive and negative tension-dependent unlinking rates obtained for Drosophila Topo II (24). However, the unlinking rates of positive writhe were poorly fit with the single energy barrier-crossing model (v0 = 5.2 ± 0.9 s−1; Δx = 8.1 ± 1.4 nm; χv2 = 10.8) as there are two different tension-dependent phases (Fig. 4A).
FIGURE 4.
Braided DNA unlinking rate as a function of tension (T) fitted with an energy barrier-crossing model. A, WT htopo IIα unlinking rate for positive (gold squares) and negative (green squares) writhe as a function of applied tension. WT htopo IIα shows preferential unlinking of positive writhe at low tension (0.25 and 0.4 pN). The chiral preference disappears with increasing tension. Relaxation rates (cycles × s−1 (cyc/s)) measured for negative writhe are: 2.0 ± 0.2 (T = 0.25 pN, n = 70), 1.6 ± 0.3 (T = 0.4 pN; n = 40), 1.3 ± 0.1 (T = 0.6 pN; n = 27), 0.9 ± 0.1 (T = 1.1 pN; n = 7), and 0.3 ± 0.1 (T = 2.5 pN; n = 11). Relaxation rates for positive writhe are: 3.6 ± 0.5 (T = 0.25 pN; n = 38), 2.5 ± 0.2 (T = 0.4 pN; n = 63), 1.5 ± 0.1 (T = 0.6 pN; n = 25), 1 ± 0.3 (T = 1.1 pN; n = 5), and 0.3 ± 0.1 (T = 2.5 pN; n = 8). The relaxation rate of negative writhe as a function of tension was fit well with the energy barrier-crossing model but not the relaxation rate of positive writhe due to the chiral preference at low tension: v0positive = 5.2 ± 0.9 s−1, Δpositive = 8.1 ± 1.4 nm, and reduced χ2 = 10.8; v0negative = 2.2 ± 0.2 s−1, Δnegative = 3.5 ± 0.6 nm, and reduced χ2 = 1.9 (see “Results” for description of fit parameters). Inset: graphic (not to scale) of the braided DNA geometry. A magnetic bead (green) is tethered to the surface by two DNA molecules (blue), separated by a distance ρ. As the bead is rotated, the two DNA strands are wrapped around one another, creating writhe with an average juxtaposition angle α. No twist is introduced as rotationally unconstrained DNA molecules are used to form the interlinked braids. B, ΔCTD htopo IIα unlinking rates for positive (gold circle) and negative (green circle) writhe as a function of applied tension. ΔCTD shows a weak preferential unlinking of negative writhe at low tension (< 0.4 pN), which disappears with increasing tension similar to WT. Relaxation rates (cycles × s−1 (cyc/s)) measured for negative writhe are: 2.9 ± 0.4 (T = 0.25 pN, n = 100), 2.1 ± 0.5 (T = 0.4 pN; n = 43), 1.4 ± 0.2 (T = 0.6 pN; n = 67), 1.2 ± 0.1 (T = 1.1 pN; n = 25), 0.6 ± 0.1 (T = 2.5 pN; n = 18), and 0.4 ± 0.1 (T = 2.5 pN; n = 27). Relaxation rates for positive writhe are: 2.0 ± 0.1 (T = 0.25 pN; n = 85), 1.9 ± 0.3 (T = 0.4 pN; n = 50), 1.5 ± 0.2 (T = 0.6 pN; n = 63), 1.1 ± 0.1 (T = 1.1 pN; n = 86), 0.8 ± 0.1 (T = 1.8 pN; n = 23), and 0.4 ± 0.1 (T = 2.5 pN; n = 25). The relaxation rate of positive writhe as a function of tension was slightly better fit with the energy barrier-crossing model than the relaxation rate of negative writhe due to the chiral preference at the low tension: v0positive = 2.4 ± 0.1 s−1, Δpositive = 2.7 ± 0.3 nm, and reduced χ2 = 1.3; v0negative = 2.8 ± 0.3 s−1, Δnegative = 3.4 ± 0.4 nm, and reduced χ2 = 5.6. Error bars indicate mean ± S.D. in all panels.
Consistent with supercoil relaxation results, ΔCTD showed a slight chiral preference for negative writhe at low tension (Fig. 4B). This chiral preference diminished as the unlinking rates for both positive and negative writhe approached the same value with increasing tension. This suggests that, similar to the WT enzyme, chiral discrimination and the rate-determining step for strand passage by the core-domain are sensitive to tension. The ΔCTD unlinking rate as a function of tension for positive writhe was well fit with an energy barrier-crossing model returning similar fitting parameters as the WT unlinking rate as a function of tension for negative writhe (v0 = 2.4 ± 0.1 s−1; Δx = 2.7 ± 0.3 nm; χv2 = 1.3). On the other hand, the ΔCTD unlinking rate as a function of tension for negative writhe was less well fit with this model (v0 = 2.8 ± 0.3 s−1; Δx = 3.4 ± 0.5 nm; χv2 = 5.6).
DISCUSSION
Single-molecule supercoil relaxation measurements confirm that htopo IIα preferentially relaxes positive supercoils, distinguishing it from other eukaryote Topo IIA enzymes studied to date (20, 21). The chirality-sensing mechanism differs significantly from that of E. coli Topo IV because it results from differences in strand passage rate rather than processivity. The results from this study also indicate that both strand passage rate and chiral discrimination of htopo IIα are sensitive to DNA tension and twist density, suggesting that both phenomena are likely governed by the same mechanical step in the strand passage cycle (Figs. 2 and 4).
Although both htopo IIα and Topo IV exhibit chiral discrimination mediated by interactions between their CTDs and DNA writhe (17, 20, 21, 24, 25, 28) (Fig. 4), our results suggest that the underlying mechanisms of discrimination differ. Chiral discrimination by Topo IV results from chirality-dependent differences in processivity as the enzyme catalyzes multiple strand passage cycles per binding event on positively supercoiled DNA, whereas it catalyzes a single cycle on negatively supercoiled DNA (25). Conversely, chiral discrimination by htopo IIα results from chirality-dependent differences in the relaxation rate without significant differences in processivity. Moreover, deleting the htopo IIα CTD reverses the sign of chiral discrimination (Fig. 2), resulting in faster relaxation of negative supercoils, whereas deleting the Topo IV CTD abolishes chiral discrimination (17). Finally, the decrease in strand passage rate with increasing tension (Fig. 4) and twist (Fig. 2) is similar to that observed for other eukaryotic Topo IIA enzymes (24, 39) but distinct from that observed for Topo IV (24, 28). These results, combined with the evolutionary distance between the human and E. coli enzymes and the complete lack of sequence similarity in their CTDs (4), suggest that chiral discrimination arose from two parallel and independent evolution events.
The single-molecule results presented here are broadly consistent with ensemble results demonstrating chiral discrimination and the role of the CTDs in mediating chiral discrimination (20, 21). However, there are differences in the degree of chiral discrimination and the apparent processivity of htopo IIα. Ensemble results suggest an ∼10-fold difference in relaxation rates (21), whereas single-molecule measurements suggest a factor of ∼2 difference in relaxation rates between negatively and positively supercoiled DNA (Fig. 2). Furthermore, analysis of the relaxation products in ensemble measurements suggests that htopo IIα is much less processive on positively supercoiled DNA than negatively supercoiled DNA (21), whereas single-molecule measurements suggest that processivity is comparable for positively and negatively supercoiled DNA (Fig. 3). These apparent differences are likely due to the different topological regimes and DNA geometries employed in the two measurements. In particular, the twist density, which we have defined as: δ = ΔTw/Tw0, both is larger and remains constant in the single-molecule experiments in comparison with the ensemble measurements, where it is generally smaller and decreases to 0 over the course of the relaxation measurements. The lowest value of twist density in the single-molecule experiments (0.6%) is on par with our estimates of the initial, i.e. maximum, twist density in the supercoiled plasmid DNA used in the ensemble experiments, which diminished as the plasmid was relaxed, resulting in significantly lower average twist density (21). Given the steep dependence of the relaxation rate (Fig. 2C) and processivity (Fig. 3C) on the twist density at low values of twist, the lower processivity and more pronounced chiral discrimination observed in ensemble measurements are consistent with the single-molecule measurements performed at substantially higher levels of DNA twist. Unfortunately, a direct comparison between the single-molecule and ensemble measurements at low twist values is experimentally unfeasible due to the large decrease in resolution and increase in measurement noise (41). Direct comparison between the single-molecule and ensemble results is further complicated by the fact that the ensemble relaxation rate measurements necessarily convolve processivity and binding time with the strand passage rate, which are separately measured in the single-molecule experiments. Furthermore, the two experimental approaches employed significantly different topoisomerase concentrations: ∼50–500 pm in the single-molecule experiments and ∼1 nm in the ensemble measurements, as much as a 20-fold difference in concentration. On a final note, measuring topoisomerase activity on DNA under tension may shed light on in vivo activity when DNA is subjected to the activity of powerful molecular motors associated with replication and transcription and additional mechanical forces, particularly during mitosis. Thus, the insight we gain from investigating topoisomerase activity in the presence of tension and twist on the DNA may provide important insights into activity and the regulation of activity in vivo.
Investigating the tension dependence of the relaxation rate also allows us to probe rate-limiting steps in the strand passage cycle that involve mechanical motion or distortion of the DNA-protein complex (24, 40). The rate-limiting step in the ATP hydrolysis cycle for yeast Topo II is the release of the first inorganic phosphate (42–46). However, it is unlikely that phosphate release is the overall rate-limiting step for strand passage as single-molecule measurements of Drosophila Topo II indicate that a single tension-dependent, chirality-independent, rate-limiting step governs strand passage (24). Moreover, rapid kinetic measurements showed that ATP hydrolysis is at least 5-fold faster than the strand passage rate at saturating ATP (43). Tension-dependent unlinking measurements of htopo IIα suggest a more complex relationship between strand passage and tension than was observed with Drosophila Topo II (Fig. 4). Rates for negative writhe were similar to Drosophila Topo II and thus consistent with an energy barrier-crossing model in which the rate-limiting step corresponds to a transition over an energy barrier associated with physical motion or distortion of the DNA that is opposed by the applied tension (40) (Fig. 4). The rates for positive writhe, however, are poorly described by a single energy barrier-crossing model largely due to the dramatic increase in rate at low tension in comparison with the rates at higher tensions, which are comparable with those of negative writhe (Fig. 4A). This unusual tension-dependent behavior suggests that the rate-determining step is sensitive to both chirality and tension. At low tension, the rate-limiting step in the negative writhe strand passage cycle is much faster, and may or may not remain rate-limiting, for the positive writhe strand passage reaction. At higher tensions, it is likely that positive and negative writhe share a single, tension-dependent, rate-limiting step (Fig. 4A). We propose that the increase in strand passage rate at low tension results from weak CTD-DNA interactions that facilitate the rate-determining step. This begs the question as to which step in the strand passage cycle could both be rate-limiting and depend on CTD-DNA interactions.
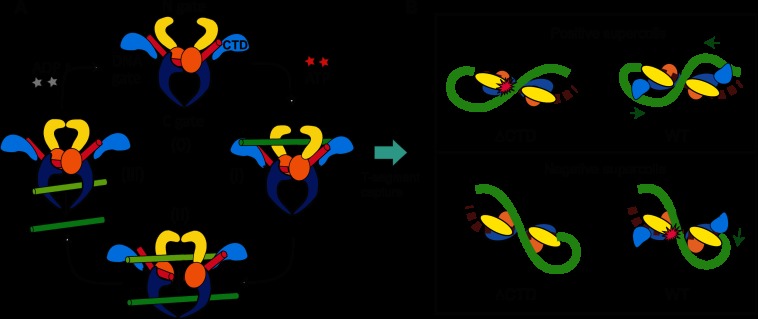
The two-gate strand passage model (5, 6, 8–10) suggests that at saturating ATP concentrations, the single-molecule relaxation rate could be determined by five potential rate-limiting steps (Fig. 5): (i) G-segment DNA cleavage; (ii) T-segment capture; (iii) T-segment transfer; (iv) G-segment DNA religation; and (v) T-segment release. G-segment binding is potentially rate-limiting; however, in our study, rates were measured within a processive burst of relaxation and thus were insensitive to G-segment binding. Because any of these five steps could be rate-limiting, further constraints are necessary to distinguish among them. The relaxation rate differences between negative and positive writhe at low tension provides this constraint. The rate difference at low tension is associated with the CTD-DNA interactions, which are the basis of chiral discrimination. However, the rate differences disappear as tension increases, indicating disruption of the CTD-DNA interactions facilitating relaxation of positive writhe. Thus, it is reasonable to assume that the step that CTD-DNA facilitates for positive writhe at low tension must be the rate-limiting step at higher tensions.
FIGURE 5.
Proposed model for CTD-dependent chiral discrimination by htopo IIα. A, graphic of the two-gate mechanism. The two-gate mechanism for strand passage by type IIA topoisomerases is well established (5, 6, 8–10). (0) G-segment DNA (red) is bound at the DNA gate (orange). (i) The binding of ATP closes the N-gate (yellow) and captures a T-segment (green). (ii) G-segment DNA is cleaved, and the T-segment is transferred through the G-segment. (iii) G-segment religation leads to opening of the C-gate and release of the T-segment. The C-terminal domains (light blue) potentially interact with T-segment DNA in a chirality-dependent manner. B, model of chirality-dependent CTD-T-segment DNA interactions (top down view). For positive supercoil relaxation (top panel), in the absence of the CTDs (top left), the T-segment DNA clashes with the N-gate (yellow-colored domain), hindering T-segment capture, whereas when the CTDs are present, the tips of the CTDs play a role in guiding the T-segment and facilitating its capture. Green arrows indicate the direction in which the CTDs reorient the T-segment DNA, facilitating T-segment capture. For negative supercoil relaxation (bottom panel), the G-segment DNA bound to the core domain is favorably oriented for T-segment capture. For WT htopo IIα, on the other hand, the CTDs pose a steric hindrance, shifting the T-segment DNA, resulting in unfavorable crossing geometry for T-segment capture. The local distortions in the DNA plectoneme structure associated with T-segment capture are energetically less favorable as the tension, or twist, in the DNA is increased, resulting in the decrease in strand passage rate.
Of the five possible rate-limiting steps, only T-segment capture is consistent with the entirety of the experimental observations as it is the only step that can reasonably be assumed to depend on both DNA writhe and interactions with the CTD, which together result in chiral discrimination. Therefore, we propose that the rate-limiting step for htopo IIα is T-segment capture. More generally, T-segment capture may be the rate-limiting step in the strand passage reaction for eukaryote Topo IIA enzymes as the tension dependence of unlinking by htopo IIα for negative writhe is similar to that of Drosophila Topo II (24). Although a scheme in which T-segment capture is rate-limiting for strand passage provides a parsimonious explanation for the observed behavior, we cannot rule out more complicated models. For example, it is possible that T-segment capture, or some other step, is in rapid tension- and writhe-dependent equilibrium with an alternative rate-limiting step. Shifting the equilibrium by changing the tension or writhe chirality could influence the overall strand passage rate by altering the occupancy of the state preceding the rate-limiting transition rather than the rate of the transition.
We propose that the steep tension dependence of the positive writhe relaxation rate results from relatively weak CTD-DNA interactions (left-handed crossings; Fig. 5B). This interaction appears to be primarily governed by a small number of residues in the CTD because deleting as few as 10 amino acids from the terminal end of the CTD results in a 30% decrease in chiral discrimination, whereas deleting 30 amino acids abolishes discrimination (20). This finding suggests a mechanism for the loss of chiral discrimination with increased tension. The critical interactions between the DNA and the terminal residues of the CTDs required for chiral discrimination are likely perturbed as the tension on the DNA increases. This scenario is supported by recent single-molecule measurements of DNA unlinking by htopo IIα that found no effect of the braid handedness on the unlinking rate (27). In this experiment, braids were created by mechanically winding two optically trapped beads tethered to the surface of a slide by long λ-DNA molecules around one another, and the extent of the braid was directly measured through fluorescence visualization of the DNA. The relaxation rate of 1.4 ± 1 cycle × s−1 measured in this experiment at a mean tension of 0.95 ± 0.26 pN (27) agrees quite well with our unlinking measurements at high tension, which at a tension of 1.1 pN were 1.0 ± 0.3 and 0.9 ± 0.1 cycle × s−1 for positive and negative writhe, respectively (Fig. 4).
Within the context of a model in which T-segment capture is the rate-limiting step (Fig. 5), the chirality-independent decrease in strand passage rate with increasing tension (Fig. 4A) suggests that the distortions of the DNA associated with productive T-segment capture become increasingly infrequent with applied tension. This scenario is further supported by the fact that the effect of tension on the strand passage rate is similar to the effect of twist for htopo IIα (Figs. 2 and 4). Remarkably, Drosophila Topo II and Topo IV also exhibit a high degree of correlation between the effect of twist and tension on relaxation rate, although the relationships are quite different for the two enzymes (38). Rather than assuming two steps in the reaction cycle that are each sensitive to twist or tension, we suggest the more parsimonious solution that tension and twist affect the same step in the cycle, namely the DNA distortions associated with productive T-segment capture. As the tension or twist increases, distortions of the plectonemes become energetically less favorable and thus less likely, resulting in the decrease in strand passage rate. This hypothesis is qualitatively consistent with the available data for htopo IIα, Drosophila Topo II, and Topo IV, but remains to be tested quantitatively.
Results obtained with the ΔCTD construct revealed two interesting aspects of the htopo IIα enzyme. First, deletion of the CTDs switches the chiral discrimination from positive to negative writhe rather than abolishing chiral discrimination as observed for Topo IV. This behavior suggests that the core domain has an inherent preference for negative writhe as ΔCTD showed a higher strand passage rate for negative than for positive writhe geometry at low twist or tension (Figs. 2C and 4B). Secondly, the CTDs appear to reduce the chiral preference by the core domain as the WT enzyme exhibited slower relaxation rates for negative writhe than the ΔCTD at the same twist or tension (Figs. 2C and 4B). Within the context of the rate-limiting T-segment capture model, we speculate that the orientation of negative supercoils favors T-segment capture at low twist density in the absence of the CTDs. This implies that the core htopo IIα enzyme, i.e. the ΔCTD construct, preferentially captures T-segments in negative writhe and that the CTDs disrupt this innate selection, whereas favoring T-segment capture in positive writhe (Fig. 5).
Overall, the interaction between the CTDs and DNA enhances positive supercoil relaxation and stabilizes the htopo IIα-DNA complex, increasing processivity. Considering its sensitivity to twist and tension as shown in our study, it is interesting that htopo IIα maintains catalytic activity in the cellular environment where significant DNA twist and tension are probably prevalent, particularly during DNA replication, transcription, and in particular mitosis, during which the separating chromosomes are likely subjected to significant mechanical stress (47). The observed tension dependence of the relaxation and unlinking rates suggests that in vivo, the activity of htopo IIα significantly decreases in response to mechanical stress on the DNA, such as during mitosis, when efficient DNA decatenation is perhaps most important. One intriguing possibility is that htopo IIα requires additional enzymes to operate efficiently under conditions of elevated DNA twist and tension. Interestingly, a recent in vitro study demonstrated that RECQL5, a human member of the RecQ family of helicases, binds to DNA and enhances the decatenation activity of htopo IIα (48).
Acknowledgments
We thank Marie-Paule Strub, (NHLBI, National Institutes of Health), Grzegorz Piszczek, (Biophysics core facility, NHLBI, National Institutes of Health), Duck-Yeon Lee, (Biochemistry core facility, NHLBI, National Institutes of Health), and Jonathan Silver for help and advice. We thank Richard Neuman for editing the manuscript.
This research was supported, in whole or in part, by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health (to K. C. N.) and by extramural National Institutes of Health Grant GM033944 (to N. O.)
This article was selected as a Paper of the Week.
- Topo IIA
- type IIA topoisomerase
- Topo IV
- E. coli topoisomerase IV
- htopo IIα
- topoisomerase IIα
- T-segment
- transfer segment
- G-segment
- gate segment
- CTD
- C-terminal domain
- WB
- wash buffer
- pN
- piconewtons.
REFERENCES
- 1. Liu L. F., Wang J. C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 3. Deweese J. E., Osheroff N. (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 37, 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forterre P., Gribaldo S., Gadelle D., Serre M.-C. (2007) Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446 [DOI] [PubMed] [Google Scholar]
- 5. Schoeffler A. J., Berger J. M. (2008) DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 [DOI] [PubMed] [Google Scholar]
- 6. Vos S. M., Tretter E. M., Schmidt B. H., Berger J. M. (2011) All tangled up: how cells direct, manage, and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 8. Roca J. (2009) Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 37, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Timsit Y. (2011) Local sensing of global DNA topology: from crossover geometry to type II topoisomerase processivity. Nucleic Acids Res. 39, 8665–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J. C. (2009) Untangling the Double Helix, Cold Spring Harbor Laboratory Press [Google Scholar]
- 11. Adachi N., Miyaike M., Kato S., Kanamaru R., Koyama H., Kikuchi A. (1997) Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nucleic Acids Res. 25, 3135–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cardenas M. E., Dang Q., Glover C. V., Gasser S. M. (1992) Casein kinase II phosphorylates the eukaryote-specific C-terminal domain of topoisomerase II in vivo. EMBO J. 11, 1785–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crenshaw D. G., Hsieh T. (1993) Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster. I. In vitro studies. J. Biol. Chem. 268, 21328–21334 [PubMed] [Google Scholar]
- 14. Jensen S., Andersen A. H., Kjeldsen E., Biersack H., Olsen E. H., Andersen T. B., Westergaard O., Jakobsen B. K. (1996) Analysis of functional domain organization in DNA topoisomerase II from humans and Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 3866–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mirski S. E., Gerlach J. H., Cummings H. J., Zirngibl R., Greer P. A., Cole S. P. (1997) Bipartite nuclear localization signals in the C terminus of human topoisomerase IIα. Exp. Cell Res. 237, 452–455 [DOI] [PubMed] [Google Scholar]
- 16. Wells N. J., Addison C. M., Fry A. M., Ganapathi R., Hickson I. D. (1994) Serine 1524 is a major site of phosphorylation on human topoisomerase IIα protein in vivo and is a substrate for casein kinase II in vitro. J. Biol. Chem. 269, 29746–29751 [PubMed] [Google Scholar]
- 17. Corbett K. D., Schoeffler A. J., Thomsen N. D., Berger J. M. (2005) The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351, 545–561 [DOI] [PubMed] [Google Scholar]
- 18. Corbett K. D., Shultzaberger R. K., Berger J. M. (2004) The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 101, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh T.-J., Yen T.-J., Lin T.-S., Chang H.-T., Huang S.-Y., Hsu C.-H., Farh L., Chan N.-L. (2010) Twisting of the DNA-binding surface by a β-strand-bearing proline modulates DNA gyrase activity. Nucleic Acids Res. 38, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McClendon A. K., Gentry A. C., Dickey J. S., Brinch M., Bendsen S., Andersen A. H., Osheroff N. (2008) Bimodal recognition of DNA geometry by human topoisomerase IIα: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain. Biochemistry 47, 13169–13178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McClendon A. K., Rodriguez A. C., Osheroff N. (2005) Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 280, 39337–39345 [DOI] [PubMed] [Google Scholar]
- 22. Ruthenburg A. J., Graybosch D. M., Huetsch J. C., Verdine G. L. (2005) A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J. Biol. Chem. 280, 26177–26184 [DOI] [PubMed] [Google Scholar]
- 23. Tretter E. M., Lerman J. C., Berger J. M. (2010) A naturally chimeric type IIA topoisomerase in Aquifex aeolicus highlights an evolutionary path for the emergence of functional paralogs. Proc. Natl. Acad. Sci. U.S.A. 107, 22055–22059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Charvin G., Bensimon D., Croquette V. (2003) Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 100, 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neuman K. C., Charvin G., Bensimon D., Croquette V. (2009) Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl. Acad. Sci. U.S.A. 106, 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roca J., Wang J. C. (1996) The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells 1, 17–27 [DOI] [PubMed] [Google Scholar]
- 27. Yogo K., Ogawa T., Hayashi M., Harada Y., Nishizaka T., Kinosita K., Jr. (2012) Direct observation of strand passage by DNA-topoisomerase and its limited processivity. PLoS One 7, e34920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stone M. D., Bryant Z., Crisona N. J., Smith S. B., Vologodskii A., Bustamante C., Cozzarelli N. R. (2003) Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 100, 8654–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neuman K. C. (2010) Single-molecule measurements of DNA topology and topoisomerases. J. Biol. Chem. 285, 18967–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seol Y., Neuman K. C. (2011) Single-Molecule Measurements of Topoisomerase Activity with Magnetic Tweezers, in Single Molecule Enzymology (Mashanov G. I., Batters C., eds) pp. 229–241, Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- 31. Seol Y., Neuman K. C. (2011) Magnetic Tweezers for Single-Molecule Manipulation, in Single Molecule Enzymology (Peterman E., J., Wuite G., eds) pp. 265–293, Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- 32. Dickey J. S., Osheroff N. (2005) Impact of the C-terminal domain of topoisomerase IIα on the DNA cleavage activity of the human enzyme. Biochemistry 44, 11546–11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kingma P. S., Greider C. A., Osheroff N. (1997) Spontaneous DNA lesions poison human topoisomerase IIalpha and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry 36, 5934–5939 [DOI] [PubMed] [Google Scholar]
- 34. Koster D. A., Palle K., Bot E. S. M., Bjornsti M.-A., Dekker N. H. (2007) Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448, 213–217 [DOI] [PubMed] [Google Scholar]
- 35. Neuman K. C., Saleh O. A., Lionnet T., Lia G., Allemand J. F., Bensimon D., Croquette V. (2005) Statistical determination of the step size of molecular motors. J. Phys. Condens. Matter 17, S3811–S3820 [DOI] [PubMed] [Google Scholar]
- 36. Bates A. D., Maxwell A. (2005) DNA Topology, 2nd Ed., Oxford University Press, New York [Google Scholar]
- 37. Charvin G., Vologodskii A., Bensimon D., Croquette V. (2005) Braiding DNA: experiments, simulations, and models. Biophys. J. 88, 4124–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Charvin G., Strick T. R., Bensimon D., Croquette V. (2005) Tracking topoisomerase activity at the single-molecule level. Annu. Rev. Biophys. Biomol. Struct. 34, 201–219 [DOI] [PubMed] [Google Scholar]
- 39. Strick T. R., Croquette V., Bensimon D. (2000) Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 404, 901–904 [DOI] [PubMed] [Google Scholar]
- 40. Tinoco I., Jr., Bustamante C. (2002) The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem. 101–102, 513–533 [DOI] [PubMed] [Google Scholar]
- 41. Neuman K. C., Nagy A. (2008) Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baird C. L., Gordon M. S., Andrenyak D. M., Marecek J. F., Lindsley J. E. (2001) The ATPase reaction cycle of yeast DNA topoisomerase II: slow rates of ATP resynthesis and Pi release. J. Biol. Chem. 276, 27893–27898 [DOI] [PubMed] [Google Scholar]
- 43. Baird C. L., Harkins T. T., Morris S. K., Lindsley J. E. (1999) Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. U.S.A. 96, 13685–13690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harkins T. T., Lewis T. J., Lindsley J. E. (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry 37, 7299–7312 [DOI] [PubMed] [Google Scholar]
- 45. Mueller-Planitz F., Herschlag D. (2006) Interdomain communication in DNA topoisomerase II: DNA binding and enzyme activation. J. Biol. Chem. 281, 23395–23404 [DOI] [PubMed] [Google Scholar]
- 46. Roca J., Wang J. C. (1992) The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71, 833–840 [DOI] [PubMed] [Google Scholar]
- 47. Roca J. (2011) The torsional state of DNA within the chromosome. Chromosoma 120, 323–334 [DOI] [PubMed] [Google Scholar]
- 48. Ramamoorthy M., Tadokoro T., Rybanska I., Ghosh A. K., Wersto R., May A., Kulikowicz T., Sykora P., Croteau D. L., Bohr V. A. (2012) RECQL5 cooperates with topoisomerase IIα in DNA decatenation and cell cycle progression. Nucleic Acids Res. 40, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strick T. R., Allemand JF, Bensimon D, Bensimon A, Croquette V. (1996) The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 [DOI] [PubMed] [Google Scholar]
- 50. Marko J. F. (2007) Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys. Rev. E Stat. Nonlin. Soft Matter. Phys. 76, 021926 [DOI] [PubMed] [Google Scholar]