Background: The adenovirus E4orf4 protein must bind protein phosphatase 2A (PP2A) for its functions.
Results: The E4orf4 binding site in PP2A was mapped to the α1,α2 helices of the B55α subunit.
Conclusion: The E4orf4 binding site in PP2A-B55α lies above the substrate binding site and does not overlap it.
Significance: A novel functional significance was assigned to the α1,α2 helices of the PP2A-B55α subunit.
Keywords: Adenovirus, Bioinformatics, PP2A, Protein-Protein Interactions, Signal Transduction
Abstract
The adenovirus E4orf4 protein regulates the progression of viral infection and when expressed outside the context of the virus it induces nonclassical, cancer cell-specific apoptosis. All E4orf4 functions known to date require an interaction between E4orf4 and protein phosphatase 2A (PP2A), which is mediated through PP2A regulatory B subunits. Specifically, an interaction with the B55α subunit is required for induction of cell death by E4orf4. To gain a better insight into the E4orf4-PP2A interaction, mapping of the E4orf4 interaction site in PP2A-B55α has been undertaken. To this end we used a combination of bioinformatics analyses of PP2A-B55α and of E4orf4, which led to the prediction of E4orf4 binding sites on the surface of PP2A-B55α. Mutation analysis, immunoprecipitation, and GST pulldown assays based on the theoretical predictions revealed that the E4orf4 binding site included the α1 and α2 helices described in the B55α structure and involved at least three residues located in these helices facing each other. Loss of E4orf4 binding was accompanied by reduced contribution of the B55α mutants to E4orf4-induced cell death. The identified E4orf4 binding domain lies above the previously described substrate binding site and does not overlap it, although its location could be consistent with direct or indirect effects on substrate binding. This work assigns for the first time a functional significance to the α1,α2 helices of B55α, and we suggest that the binding site defined by these helices could also contribute to interactions between PP2A and some of its cellular regulators.
Introduction
The adenovirus type 2 or 5 E4orf44 protein is a small protein of 14 kDa and 114 residues that contributes to the control of progression from early to late phases of viral replication. Its contribution involves down-regulation of expression of cellular and early viral genes (1–5), induction of alternative splicing of viral mRNAs (6, 7), and regulation of protein translation through an interaction with the mTOR pathway (8). When expressed outside the context of viral infection, E4orf4 induces nonclassical apoptosis that is caspase-independent in many cell lines and which is preceded by G2/M arrest (9–11). E4orf4-induced cell death is more efficient in oncogene-transformed cells (12), indicating that study of E4orf4 signaling may have implications for cancer therapy. Furthermore, at least part of the E4orf4 signaling network is highly conserved in evolution from yeast to mammalian cells (10, 13–15), underscoring its importance to cell regulation. Studies of the mechanisms underlying E4orf4 action have revealed several E4orf4 partners, including the B55/Bα (16) and B56 (17) subunits of protein phosphatase 2A (PP2A), the ATP-utilizing chromatin assembly and modifying factor (ACF) (18), Src family kinases (19, 20), the anaphase-promoting complex/cyclosome in the budding yeast (10), a subset of splicing serine/arginine-rich proteins (6), and Ynd1/Golgi uridine diphosphatase (14).
PP2A is a major E4orf4 partner, and its interaction with E4orf4 was shown to contribute to all E4orf4 functions known to date (1, 6, 8, 12, 16, 21). PP2A is composed of three subunits: the catalytic C subunit, a scaffolding A subunit, and one of several regulatory B subunits encoded by at least four unrelated gene families, PR55/B55/B, PR61/B56/B′, B″, and B‴ (22), which dictate substrate specificity of the PP2A holoenzyme. The interaction of E4orf4 with the B55α subunit of PP2A, but not the B56 subunits, contributes to E4orf4-induced cell death and cell cycle arrest in both yeast and mammalian cells (10, 17). Previous reports suggested that some E4orf4 functions required the PP2A phosphatase activity (7, 16, 23), whereas another report maintained that inhibition of PP2A contributed to E4orf4 function (24). Because physiological substrates of the E4orf4-PP2A complex have not been identified to date, it is not clear yet how E4orf4 affects PP2A activity toward them.
To gain better insight into the E4orf4-PP2A interaction, mapping of the interaction sites in E4orf4 and PP2A-B55α was undertaken. Most of the previously published work was aimed at identifying E4orf4 residues important for its association with PP2A (12, 13, 25). However, two reports described attempts to identify residues in the PP2A-B55α subunit that contributed to the interaction with E4orf4 (26, 27). Both these investigations were performed before publication of the structure of the PP2A holoenzyme that included the B55α subunit (28). They relied on random mutagenesis followed by a selection in yeast for mutations in the yeast B55 gene that conferred resistance to E4orf4-induced toxicity. This approach was powerful in that it identified biologically significant mutations in B55α that interfered with E4orf4 function. However, mutations that inhibited the association with E4orf4 could potentially be situated in the interior of the protein and could affect E4orf4 binding indirectly by changing B55α conformation or fold rather than by interfering directly with the E4orf4 binding site.
In this work we undertook a bioinformatics- and structure-oriented approach to the identification of the E4orf4 binding site in the PP2A-B55α subunit. The work was based on the solved structure of the PP2A holoenzyme and on modeled E4orf4 structures and utilized several known bioinformatics programs to predict B55α residues that interact with E4orf4. We then investigated the ability of mutations in the predicted sites to interfere with the E4orf4-PP2A-B55α interaction and function in cells. We conclude from these studies that the α1 and α2 helices, which lie above the substrate interaction domain proposed by Xu et al. (28), compose the E4orf4 binding site in B55α.
EXPERIMENTAL PROCEDURES
Plasmids, Primers, Cell Lines, and Transfections
The following plasmids have been used in this work: pHis-E4orf4 (16) and pcDNA3.1-B55α-3HA (12). The B55α is of rat origin, which shares 96% identity with human B55α. The same B55α cDNA was also fused to the glutathione S-transferase (GST) open reading frame. Various mutations were introduced into B55α cloned in the pcDNA3.1 plasmid or fused to GST by site-directed mutagenesis using the Stratagene QuikChange mutagenesis kit according to the manufacturer's instructions. All mutations were verified by sequencing. The primers used for the mutagenesis are shown in Table 1. Primers for generation of a PP2A-B55α mutant resistant to the PP2A-B55α-shRNA were previously described (1).
TABLE 1.
Primer list
fwd, forward; rev, reverse.
| Primer name | Primer sequence |
|---|---|
| B55α-L1-mut fwd | 5′-GAGAATACAATGTTTACAGCGCCTTCGCGAGCCATGAGCCAGAATTTG-3′ |
| B55α-L1-mut rev | 5′-CAAATTCTGGCTCATGGCTCGCGAAGGCGCTGTAAACATTGTATTCTC-3′ |
| B55α-L2-mut fwd | 5′-TGTTTAGGCCCATGGATCTAGCGGCGGCGGCCAGTCCGCG-3′ |
| B55α-L2-mut rev | 5′-CGCGGACTGGCCGCCGCCGCTAGATCCATGGGCCTAAACA-3′ |
| B55α-R1-mut fwd | 5′-TGAGCCAGAATTTGACTACTTGAAAGCCTTAGAAATAGAAGAGAAGATCAA C-3′ |
| B55α-R1-mut rev | 5′-GTTGATCTTCTCTTCTATTTCTAAGGCTTTCAAGTAGTCAAATTCTGGCTCA-3′ |
| B55α-R2-mut fwd | 5′-GCCTGCCAATATGGCGGAGCTCACGGAGG-3′ |
| B55α-R2-mut rev | 5′-CCTCCGTGAGCTCCGCCATATTGGCAGGC-3′ |
| B55α-R3-mut fwd | 5′-CTGCCAATATGGAGGAGGCCGCGGAGGTGATCACAGCC-3′ |
| B55α-R3-mut rev | 5′-GGCTGTGATCACCTCCGCGGCCTCCTCCATATTGGCAG-3′ |
| B55α-R4-mut fwd | 5′-GAAGATCCCAGCAACAGGTCGGCTTTTTCTGAAATCATCTCCTC-3′ |
| B55α-R4-mut rev | 5′-GAGGAGATGATTTCAGAAAAAGCCGACCTGTTGCTGGGATCTTC-3′ |
| B55α-R5-mut fwd | 5′-CAGAAGTAAACTTTGCTCACTGGCTGAAAACGACTGCATATTTGAC-3′ |
| B55α-R5-mut rev | 5′-GTCAAATATGCAGTCGTTTTCAGCCAGTGAGCAAAGTTTACTTCTG-3′ |
| B55α-R6-mut fwd | 5′-CTGTATGAAAACGACTGCATAGCTGACAAATTTGAATGTTGTTGG-3′ |
| B55α-R6-mut rev | 5′-CCAACAACATTCAAATTTGTCAGCTATGCAGTCGTTTTCATACAG-3′ |
Clone 13, a HEK293-derived cell line containing tetracycline-inducible E4orf4 (1) was propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) guaranteed to be tetracycline-free (BD Biosciences), 5 μg/ml blasticidin (Invitrogen), and 200 μg/ml zeocin (Invitrogen). E4orf4 expression was induced by the addition of 1 μg/ml doxycycline (Sigma). IRBα cells (1) were propagated in DMEM supplemented with 10% FCS and 1 μg/ml puromycin. Transfections were carried out using the jetPEITM reagent according to the manufacturer's instructions (Polyplus Transfection).
Immunoprecipitations, GST Pulldown Assays, and Western Blot Analysis
For immunoprecipitation experiments, whole cell extracts were prepared in lysis buffer (50 mm Tris-HCl (pH 7.4), 250 mm NaCl, 5 mm EDTA, 0.1% Triton X-100, 0.5% Nonidet P-40, and a 1/10 volume of Complete protease inhibitor mixture (Roche Applied Science)) and were subjected to immunoprecipitation with HA-specific antibodies (Covance). Input lysates and immune complexes were separated by SDS-PAGE and subjected to Western blot analysis with antibodies to E4orf4 (29) and to the HA tag. Protein levels in Western blots were quantified by densitometry. For GST pulldown assays, GST or GST fused to WT and mutant B55α proteins were expressed in bacteria and purified on glutathione-agarose beads. The purified bacterial proteins were incubated with bacterially expressed His-E4orf4, and the bound material was chromatographed on SDS-PAGE and analyzed by a Western blot stained with antibodies to E4orf4 and GST.
Cell Death Assays
Cells were transfected with plasmids expressing E4orf4 and HA-tagged B55α proteins. 24 h post transfection, the cells were fixed with 4% paraformaldehyde and stained with antibodies to E4orf4 and the HA tag and with DAPI (Sigma) as described previously (18, 30). Fluorescent cells were visualized by a Zeiss Axioskop microscope at a 400-fold magnification. The fraction of E4orf4-expressing cells with condensed, abnormal, or fragmented nuclei was determined in each experiment by counting 100 transfected nuclei. The average of two experiments, each containing duplicate plates, was calculated. Statistical significance of the results was determined by χ2-based statistics.
Programs and Databases Used in the Structural Prediction Analysis
The following programs were used in this work: Blast (31); ConSurf/ConSeq, which identifies functional regions or residues in a given protein (32–35); Promate, which predicts the location of protein-protein binding sites through analysis of chemical compositions of these sites, geometric properties, and crystal structure of the protein (36); the QUARK ab initio protein structure prediction program (37); Chimera, used for visualization of the molecular structures (38); ProSa, which calculates the quality score for an input protein structure (this score is compared with the solved structure of native proteins, and a local quality score plot can point at local problematic regions of the model (39, 40)); PatchDock and FireDock (Patchdock predicts molecular docking based on the rigid geometric surface of two given molecules in a native conformation, and Firedock refines Patchdock output using side-chain optimization, rigid-body optimization, and ranking based on energetic calculations (41–43)); ClusPro 2.0 predicts molecular docking. It uses its own rigid docking program (Piper), providing 1000 low energy results to the ClusPro clustering program to attempt to find the native site under the assumption that it will have a wide free-energy attractor with the largest number of results. The models coming out of ClusPro are ranked by cluster size (44–46).
RESULTS
Identification of Significant Conserved Residues of PP2A B55α
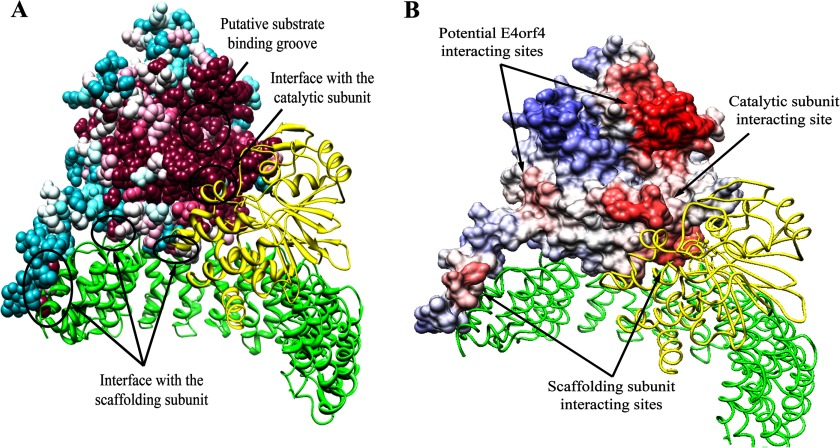
The B55α regulatory subunit of PP2A mediates an interaction between the adenovirus E4orf4 protein and the PP2A holoenzyme (12, 15, 16, 21). We set out to search for the E4orf4-binding domain in B55α using structural considerations. Predictions of the binding site were based on the solved structure of B55α (PDB ID 3dw8), containing a 7-bladed β propeller composed of 7 WD repeats (28). Because the E4orf4 signaling network is highly conserved in evolution from yeast to mammalian cells (10, 13–15), we looked for evolutionary conserved B55α residues that may interact with E4orf4. To do so, BLAST was used to compare B55α protein sequences from several organisms, and the results were subjected to multiple sequence alignment. To identify conserved residues on the surface of the B55α protein, we used the ConSurf program. This program generates a surface mapping of a protein using multiple sequence alignment to predict the functional regions of the protein surface based on the conservation of its residues. When this program was provided with the known PP2A B55α structure, it predicted which residues were conserved and were, therefore, potentially important for the function or structure of the protein (Fig. 1A). By examining Fig. 1A it can be concluded that the residues in the putative substrate binding groove that were identified based on the known PP2A structure (28), and the residues around it are very conserved. Other conserved residues are located at the interface between the B55α subunit and the scaffolding and catalytic subunits, reflecting the highly conserved interaction between them.
FIGURE 1.
Prediction of the B55α protein-protein interaction sites based on structure conservation. A, results of the analysis of the PP2A holoenzyme by the ConSurf program are shown. The scaffolding A and the catalytic C subunits are represented by ribbons colored green and yellow, respectively. B55α is represented by a space-filling model, colored by the following conservation scale: dark purple residues are the most conserved; white residues are the average on the conservation scale; cyan residues are variable. Known protein-protein interaction sites are marked by arrows. B, the surface of B55α was colored according to the ProMate prediction of protein-protein interaction sites as follows; residues with a higher probability for involvement in protein-protein interactions are marked in darker red, and residues with a low probability are in blue. In addition to the correctly predicted interfaces with the scaffolding (green) and catalytic (yellow) subunits, two more sites have been predicted to be involved in protein-protein interactions and are marked by arrows as potential E4orf4 interacting sites.
Next, it was of interest to predict the location of protein-protein interaction sites within the surface of PP2A-B55α. This was done using the ProMate program, and the results are shown in Fig. 1B. As a validation for the prediction made by this program, the B55α regions, which were reported to interact with the scaffolding and the catalytic subunits of PP2A, were indeed predicted as potential protein-protein binding sites. Interestingly, two more regions were predicted as protein-protein interacting sites: one, just above the major groove of B55α, was assigned a very high probability for protein-protein interaction, and another region was assigned a lower protein interaction probability that was still above background levels. These sites could potentially include the E4orf4-interacting site.
Modeling the Structure of E4orf4
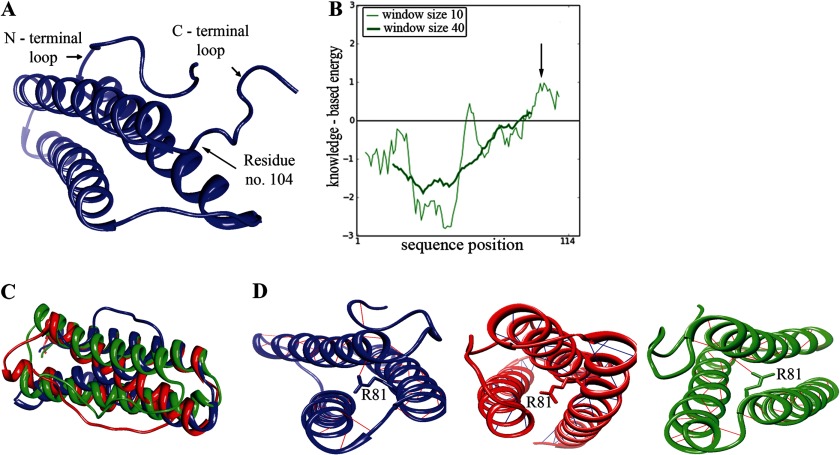
To investigate the interaction between E4orf4 and PP2A, it is essential to understand the structural properties of the viral protein. However, currently there is no solved structure of this protein, and there is no known E4orf4 homologue that could serve as a template for modeling the E4orf4 protein structure. Therefore, we performed ab initio modeling of the 114-residue E4orf4 protein using the recently published Quark computer algorithm, specifically designed to predict the structure of proteins smaller than 200 amino acids (37). This program predicted 10 structure models for E4orf4. For each model the energy based on the folding of the protein was calculated by the ProSa program. This energy calculation was translated into a quality score given to the model by the program in such a way that a lower energy of the structure indicated a better likelihood for the correctness of the model. All E4orf4 models demonstrated a C-terminal loop that caused a locally high energy structure (Fig. 2A,B). Moreover, in all the proposed models, the C-terminal loop (residues 105–114) showed great variability, indicating that the modeling of this region may be highly inaccurate. Furthermore, residues known to be of importance to the interaction between E4orf4 and PP2A were not previously found within the E4orf4 C terminus. Therefore, we omitted the C-terminal loops of the models (residues 105–114) before further analysis. Models Q1, Q3, and Q5 showed the least similarity among the different proposed models as determined by calculating the root mean square deviation between them (Fig. 2, C and D). We chose these models for further analysis as representing all proposed E4orf4 structural models.
FIGURE 2.
Modeling the E4orf4 structure. A, the first model of E4orf4 structure proposed by the Quark program is shown. The N terminus and C terminus loops are marked as well as residue 104, which is the last residue retained in the models used for further analysis. B, ProSA analysis of the first E4orf4 model offered by the Quark program is shown, demonstrating the positive energy for the C-terminal loop, marked with an arrow, and a satisfying negative energy for the rest of the protein. The analysis was done using two window sizes of 10 or 40 aa. This analysis has been repeated for all E4orf4 models with similar results. C, superposition of three Quark models of E4orf4: Q1, Q3, Q5. The structural alignment of Q1, Q3, and Q5 presents a high similarity in the α-helical structure yet shows differences in the inner structure, such as inner angels and the position of the N-terminal loop. D, E4orf4 models share the same α-helical structure with highly stabilizing H-bonds. These models (Blue, Q1; red, Q3; green, Q5) are different in the orientation of the helices, as can be seen by the Arg-81 residue, protruding out of the helix at different angels with stabilizing H-bonds in Q1 and Q5 or without them in Q3.
Prediction of PP2A B55α Residues Required for the Interaction with E4orf4
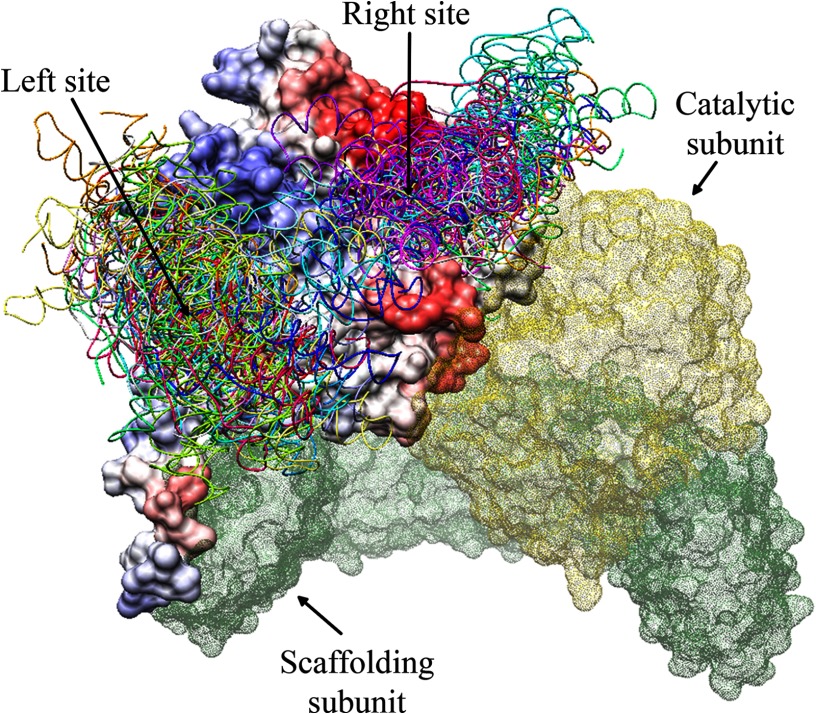
To identify the E4orf4 interaction site on the surface of B55α, each of the three structural models chosen for E4orf4 were used together with the solved structure of PP2A B55α in the PatchDock program. PatchDock is an algorithm for molecular docking of two molecules that produces a list of potential complexes sorted by shape complementarity criteria. The top 1000 docking results generated by the PatchDock program for each E4orf4 model were then delivered to the FireDock program for further refinement. The FireDock program addresses the refinement of protein-protein docking solutions by targeting the problem of flexibility and scoring solutions produced by shape-based methods. FireDock refines and scores these solutions according to its energy function. The top 10 results generated by the FireDock analysis of each E4orf4 model (adding up to 30 docking results altogether) were analyzed. All these docking models had the same order of magnitude of negative global energy, indicating their similar likelihood of correctness. As further refinement was needed, all docking results causing steric interference of E4orf4 with the PP2A subunits A and C were deleted because the A and C subunits were known components of the E4orf4-PP2A complex (15, 17). The remaining docking results were visualized together with ProMate prediction for protein-protein interaction sites in B55α. Surprisingly, all the docking predictions for E4orf4 clustered into two major regions of the B55α subunit, designated “left site” or “right site.” These clusters were consistent with the ProMate prediction of protein-protein interaction sites, which pointed at the right-site clustering pocket as a highly probable protein-protein interacting region and at the left site as a lower probability site.
To further test the reliability of the previous predictions, a different and more advanced docking program, ClusPro 2.0, was used to predict the E4orf4 binding sites in the PP2A-B55α subunit. ClusPro is a docking program that not only uses rigid geometry and energy-based solutions but also attempts to find the native site under the assumption that it will have a wide free-energy attractor with the largest number of results. This approach provides another alternative for clustering of possible docking solutions. The solutions from the ClusPro program were similar to the previous docking solutions from the PatchDock and FireDock programs, yet were better clustered to the same two sites described above (Fig. 3).
FIGURE 3.
ClusPro predicts that E4orf4 may bind the PP2A B55α subunit at one of two major sites. The B55α surface is represented in the ProMate color-coded prediction of protein-protein interacting sites, as shown in Fig. 1B, whereas the surfaces of the A and C subunits are represented in colored dots: green for A and yellow for C. The E4orf4 docking solutions, shown in various colors for the different solutions, are clustered in two sites, marked as Left and Right sites.
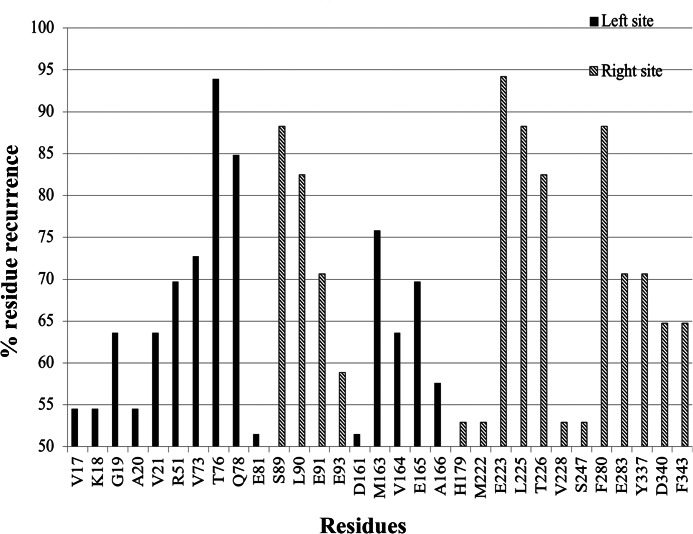
To identify specific residues involved in the E4orf4 docking sites, the Intersurf function of the Chimera program was used, selecting residues adjacent to the proposed E4orf4 docking sites in the B55α subunit. The Intersurf function was employed for each of the docking solutions, and the selected residues were noted as belonging to either the right docking site or the left docking site in B55α. Using these lists of residues, a histogram was produced. Each bar of the histogram represented the percentage of instances of finding a certain residue in an interaction with E4orf4 out of all residues noted as binding within a given site (left/right). To narrow down the number of candidate residues participating in the binding to E4orf4, a 50% threshold was set for the histogram (Fig. 4 and Table 2). Residues were considered as major candidates of crucial importance for binding E4orf4 if they received a high percentage of occurrences or if they were also previously shown to be important for E4orf4 binding in yeast (26).
FIGURE 4.
A histogram of B55α residues recurring in the E4orf4 docking predictions. The histogram summarizes the percentage of recurrence of B55α residues in ClusPro docking solutions for E4orf4 binding by PP2A-B55α. The residues shown here received percentage of appearance higher than the set 50% threshold and were divided according to their attributed site: left in black, and right in gray lines.
TABLE 2.
Summary of proposed B55α residues interacting with E4orf4
Listed are the B55α residues that have been characterized as interacting with E4orf4 according to the analysis described earlier, as shown in Fig. 4. The corresponding mutant column denotes the assigned mutant name for mutations generated based on this analysis. Several residues shown here have already been tested for their interaction with E4orf4 in yeast; however, only Tyr-344 was shown to affect E4orf4 binding.
| Residue | Percentage of occurrence | Right/Left site | Corresponding residue in yeast Cdc55 | Corresponding mutant |
|---|---|---|---|---|
| Val-17 | 54.5 | Left | Cys-13 | |
| Lys-18 | 54.5 | Left | Phe-14 | |
| Gly-19 | 63.6 | Left | Gly-15 | |
| Ala-20 | 54.5 | Left | Lys-17 | |
| Val-21 | 63.6 | Left | Ala-18 | |
| Arg-51 | 69.7 | Left | Arg-48 | |
| Val-73 | 72.7 | Left | Phe-64 | |
| Thr-76 | 93.9 | Left | Glu-67 | L1 |
| Gln-78 | 84.8 | Left | Gln-69 | |
| Glu-81 | 51.5 | Left | Asp-72 | |
| Ser-89 | 88.2 | Right | Ser-80 | R1 |
| Leu-90 | 82.4 | Right | Leu-81 | |
| Glu-91 | 70.6 | Right | Glu-82a | |
| Glu-93 | 58.8 | Right | Glu-84 | |
| Asp-161 | 51.5 | Left | Asp-168 | |
| Met-163 | 75.8 | Left | Ile-170 | L2 |
| Val-164 | 63.6 | Left | Ile-171 | |
| Glu-165 | 69.7 | Left | Ala-172 | |
| Ala-166 | 57.6 | Left | Ala-173 | |
| His-179 | 52.9 | Right | His-186 | |
| Met-222 | 52.9 | Right | Met-229 | |
| Glu-223 | 94.1 | Right | Glu-230 | R2 |
| Leu-225 | 88.2 | Right | Leu-232 | R3 |
| Thr-226 | 82.4 | Right | Thr-233a | |
| Val-228 | 52.9 | Right | Val-235 | |
| Ser-247 | 52.9 | Right | Ser-254 | |
| Phe-280 | 88.2 | Right | Phe-287 | R4 |
| Glu-283 | 70.6 | Right | Glu-290b | |
| Tyr-337 | 70.6 | Right | Tyr-344b | R5 |
| Asp-340 | 64.7 | Right | Asp-347 | |
| Phe-343 | 64.7 | Right | Phe-350 | R6 |
Examination of the Contribution of Predicted E4orf4-Binding Residues in B55α to the B55α-E4orf4 Interaction in Vivo and in Vitro
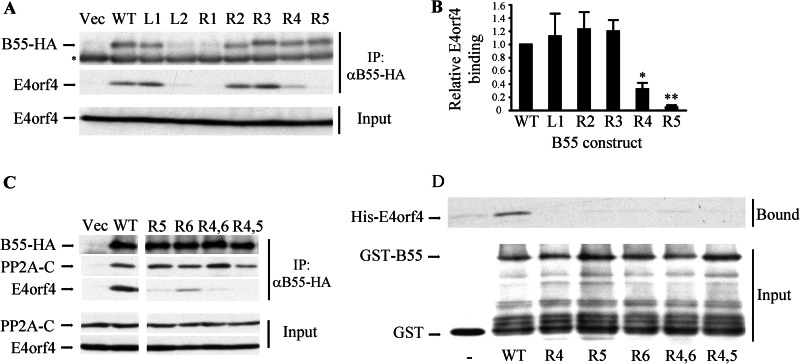
PP2A B55α residues that were considered as major candidates for binding E4orf4 had to be evaluated for their importance to E4orf4 binding. To analyze the relevance of these residues to the binding, we constructed several B55α mutants based on the previously described theoretical experiments. A plasmid expressing B55α fused to an HA tag was subjected to site-directed mutagenesis of various investigated residues described in Table 2, changing them to alanines. Plasmids expressing seven different mutants, a plasmid expressing the wild type B55α protein, and an empty vector were transfected into clone 13 cells, which stably express E4orf4 under doxycycline induction. 24 h post transfection, cells were induced for E4orf4 expression and 5 h later were harvested. Whole cell extracts were produced, and equal protein amounts were subjected to immunoprecipitation with the HA-specific antibody. Immune complexes and unprecipitated lysates were analyzed by Western blots (Fig. 5A). The Western blot analysis revealed that two of the B55α mutations, L2, targeting residues Met-163, Val-164, and Glu-165 in the predicted left E4orf4 binding site, and R1, targeting residue Ser-89 in the predicted right binding site, significantly reduced B55α protein levels, thus possibly affecting protein stability. Therefore, the ability of these mutants to bind E4orf4 was not studied further. To get a quantitative measure of the ability of the other mutants to bind E4orf4, E4orf4 levels present in the immune complexes were quantified by densitometry and normalized to expression levels of the corresponding B55α mutants. The value for the WT B55α protein was defined as 1. Fig. 5B demonstrates that three of the mutants, L1 (T76Q78A), R2 (E223A), and R3 (L225T226A), bound E4orf4 similarly to the WT B55α protein. In contrast, mutants R4 (F280A) and R5 (Y337A) bound E4orf4 at significantly reduced levels (p < 0.009). The R4 and R5 mutations affect aromatic amino acids located in two α helices identified in the B55α structure that face each other (Fig. 7, (28)). To further probe the B55α right site implicated in E4orf4 binding, we generated an additional mutation (R6) affecting another hydrophobic amino acid (Phe-343) located at the same site and facing both Phe-280 and Tyr-337 (Fig. 7C). Two double mutations including R4 and R6 (R4,6) or R4 and R5 (R4,5) were generated as well. An immunoprecipitation experiment was performed to test whether these B55α mutants could bind E4orf4. As seen in Fig. 5C, the new mutants lost the ability to bind E4orf4. Moreover, mutants that were unable to bind E4orf4 could still interact with the PP2A catalytic subunit, indicating that both single and double mutants retained the ability to incorporate into the PP2A holoenzyme.
FIGURE 5.
B55α mutants bind E4orf4 at significantly reduced levels. A, clone 13 cells were transfected with an empty vector (Vec) or with plasmids expressing HA-tagged wild type B55α (WT) or various mutant B55α proteins. Cell lysates were immunoprecipitated with HA-specific antibodies, and immune complexes as well as input lysates were separated by SDS-PAGE and subjected to Western blot analysis with E4orf4- or PP2A-B55-specific antibodies. Input protein levels represent 10% of total protein amounts used for the immunoprecipitation (IP). The asterisk marks the endogenous PP2A-B55 band, which is detected similarly in all lanes, thus serving as a loading control. B, the levels of B55α and E4orf4 proteins in the immune complexes were quantified by densitometry, and E4orf4 binding levels were normalized to B55α expression levels. E4orf4 binding by WT B55α was defined as 1. Mutants L2 and R1, which were expressed at significantly reduced levels, were not subjected to the quantitative analysis. The graph summarizes the results of three independent experiments, and error bars represent S.E. A paired one-tailed t test indicated that E4orf4 binding to R4 and R5 was reduced significantly relative to its binding to WT B55α (*, p = 0.009 for R4; **, p = 0.0005 for R5), whereas there was no statistically significant difference between the binding of the other three well expressed mutants and the WT protein. C, experiments with additional B55α mutants were carried out as in A. Western blots were stained with antibodies to the HA tag, PP2A-C, and E4orf4. Double mutants were R4,6: R4+R6; R4,5: R4+R5. D, GST fused to the WT B55α subunit of PP2A or to B55α mutants as well as GST alone and His-E4orf4 were expressed in bacteria. The GST proteins were purified on glutathione beads and incubated with His-E4orf4. Bound E4orf4 and input GST proteins were detected by a Western blot.
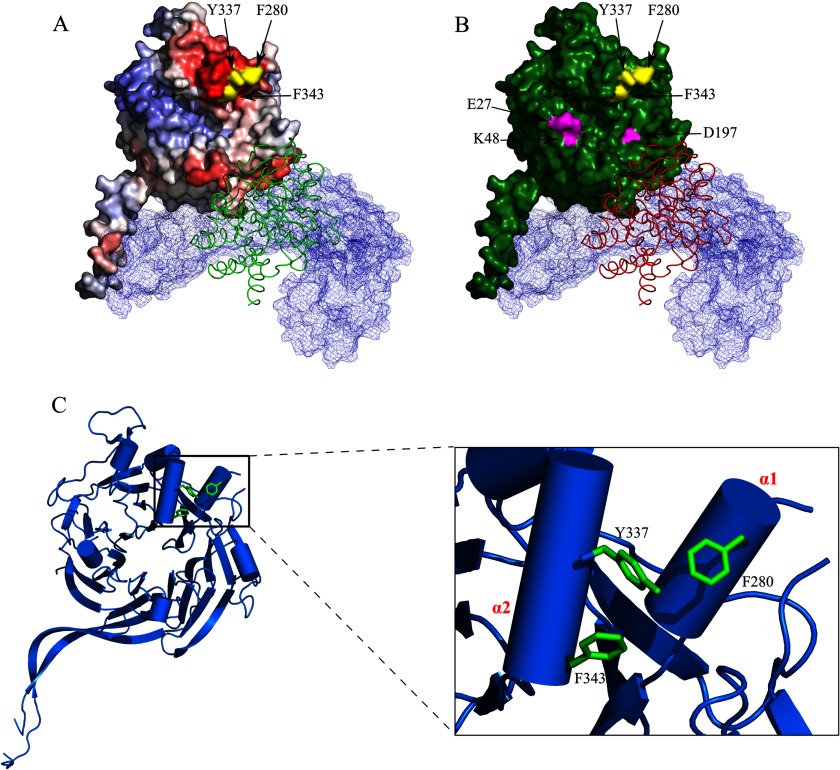
FIGURE 7.
The E4orf4 binding site in PP2A-B55α. A, the location of the B55α residues Phe-280, Tyr-337, and Phe-343 required for E4orf4 binding is shown. The B55α subunit is colored according to the Promate protein-protein interaction prediction as described in Fig. 1B, and the B55α residues Phe-280, Tyr-337, and Phe-343 are colored in yellow. The PP2A A subunit is mesh-surfaced in light blue, and the ribbon representing the C subunit is colored in green. B, the E4orf4 binding site in B55α does not overlap with the Tau binding site. B55α residues Phe-280, Tyr-337, and Phe-343 are colored in yellow, and the Tau protein binding residues Glu-27, Lys-49, and Asp-197 (28) are colored in magenta. The B55α subunit is colored in green. The PP2A A and C subunits are represented as described in A, except that the C subunit is colored red. C, the B55α residues Phe-280, Tyr-337, and Phe-343 are part of the α1 and α2 helices and face each other. The B55α structure including the core β-propeller (ribbons) and the α-helices (cylinders) is shown. Residues Phe-280, Tyr-337, and Phe-343 are colored in green. A close-up of the α1,α2 helices is shown on the right, displaying residues Phe-280, Tyr-337, and Phe-343 as they protrude from the α1 and α2 helices, facing each other.
To confirm that the mutations in the B55α right site interfered with a direct binding of E4orf4 to PP2A-B55α, an in vitro binding assay was performed using proteins expressed in bacteria. GST alone or GST fused to WT PP2A-B55α or to the PP2A-B55α mutant proteins were purified on glutathione-agarose beads and incubated with His-E4orf4. The bound proteins were analyzed by Western blots. Fig. 5D demonstrates that WT PP2A-B55α bound His-E4orf4 at much higher levels than background adsorption to the unfused GST protein, whereas none of the mutants bound His-E4orf4 above background levels. Thus, the results of both in vivo and in vitro experiments confirm the conclusion that part of the right site in B55α, which includes Phe-280, Tyr-337, and Phe-343 and which was predicted by several bioinformatics analyses with high probability to play a role in E4orf4 binding, was indeed important for this interaction.
B55α Mutants with a Diminished Ability to Bind E4orf4 Cooperate Less Efficiently with the Viral Protein to Induce Cell Death
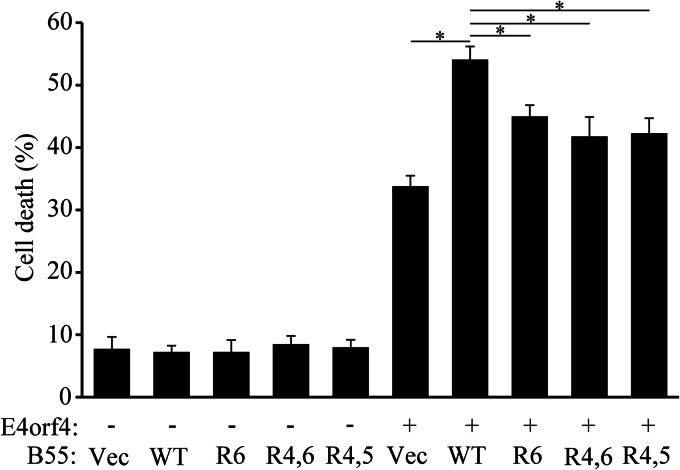
It has been previously demonstrated that overexpression of the PP2A-B55α subunit enhanced the ability of E4orf4 to induce cell death, possibly by increasing the cellular PP2A holoenzyme pools that contain the B55α subunit and are able to transduce the E4orf4 toxic signal (17). We tested, therefore, whether B55α mutants that lost the ability to bind E4orf4 were deficient in their ability to assist E4orf4 in induction of cell death. IRBα cells containing a constitutively expressed B55α-specific shRNA were utilized in this experiment, and WT and mutant B55α were rendered resistant to the shRNA by introduction of silent mutations at the appropriate site. It has been previously shown that the most typical morphologies associated with E4orf4-induced cell death include membrane blebbing, nuclear condensation, and cell detachment, whereas morphologies associated with classical apoptosis such as DNA fragmentation, caspase activation, phosphatidylserine externalization, or mitochondrial changes do not always accompany E4orf4-induced cell death (11, 47, 48). We thus measured E4orf4-induced cell death by assaying nuclear condensation and fragmentation. IRBα cells were transfected with an empty vector or a vector expressing E4orf4 together with plasmids expressing WT or mutant B55α proteins or the corresponding empty vector, and cell death was measured 24 h later. As shown in Fig. 6, E4orf4 alone induced an average of 34% cell death, whereas the addition of WT B55α increased cell death to 54%. The addition of the mutant B55α proteins R6, R4,6, and R4,5 was significantly less efficient and enhanced cell death to 42–45% (p < 0.003).
FIGURE 6.
Loss of E4orf4 binding reduces the ability of B55α mutants to enhance E4orf4-induced cell death. IRBα cells were transfected with an empty vector or a vector expressing E4orf4 together with plasmids expressing WT or mutant B55α proteins or the corresponding empty vector (Vec). Cell death was determined 24 h later by measuring the percentage of transfected cells exhibiting nuclear condensation and fragmentation. The average of two independent experiments with two duplicates each is shown. Error bars represent the S.E. *, p < 0.003.
DISCUSSION
The interaction between E4orf4 and the PP2A holoenzyme was discovered two decades ago (16). Nevertheless, little was known about the structural properties of this interaction. E4orf4 was shown to interact with the B55α and B56 regulatory subunits of PP2A, although only binding to the B55α subunit was required for E4orf4-induced cell death (10, 12, 15, 17, 21).
Several attempts have been made to figure out the importance of various residues within E4orf4 and the PP2A-B55α subunit for their interaction; however, the majority of this effort was aimed at identifying PP2A-binding residues within E4orf4 (12, 13, 15, 17, 21). More recently, the focus has shifted to the identification of E4orf4 binding domains in the B55α subunit of PP2A, with the hope that this will lead to a better understanding of the functional characteristics of this interaction. However, published research suffered from two main drawbacks; first, it was centered mainly on the E4orf4 interaction with the yeast B55 subunit orthologue, Cdc55. Cdc55 is different from human B55 in several regions of the protein, although there is a high degree of conservation in other regions, and both B55α and Cdc55 are required for E4orf4 functions in mammalian cells and yeast, respectively (10, 15). Despite the similarities between B55α and Cdc55, a recent report failed to find B55α residues required for E4orf4 binding based on analysis of the yeast orthologue Cdc55 (27). Second, the attempt to locate residues important for the binding had little structural guidance because no solved structure was available for either PP2A or E4orf4. Although a powerful genetic selection was used to identify biologically significant mutations among randomly generated mutations, such mutations could inhibit E4orf4 binding indirectly by changing global protein fold rather than affecting the interaction site directly (26, 27).
Our present attempt to establish the nature of the interaction between E4orf4 and the PP2A B55α subunit was structure-oriented using the solved structure of B55α-containing PP2A (28) and a modeled structure of E4orf4 and involving theoretical predictions as well as actual biological assays. Initially we used a bioinformatics approach to search for the B55α residues responsible for E4orf4 binding. This search comprised a series of successive bioinformatics analysis steps including identification of conserved sites of the B55α structure (Fig. 1A), prediction of protein-protein interaction sites (Fig. 1B), modeling the E4orf4 structure (Fig. 2), and predicting the E4orf4 docking sites on the surface of B55α (Figs. 3 and 4). The theoretical results suggested that the E4orf4 binding site could be located in one of two sites identified by clustering the results of docking predictions (Figs. 3 and 4 and Table 2). We did not use any assumption in our computations except for the known presence of the A and C subunits in the PP2A complex when E4orf4 was bound. One of the predicted sites, the right site, was identified by the calculations with a higher probability of being involved in protein-protein interactions (Fig. 1B). Mutations in representative residues within the proposed binding domains were then tested for their ability to prevent co-immunoprecipitation of E4orf4 with B55α (Fig. 5). This assay revealed that part of the right site predicted with high probability to be involved in protein-protein interactions was indeed required for E4orf4 binding, whereas residues in the lower probability site did not contribute to E4orf4 binding (Fig. 5). In vitro assays with proteins expressed in bacteria further confirmed that the mutations identified by co-immunoprecipitation assays to interfere with the binding of E4orf4 to PP2A-B55α indeed affected the direct binding between the two proteins (Fig. 5D). One mutation that was found to prevent E4orf4 binding, Y337A, corresponded to the Y344A mutation that prevented yeast Cdc55 binding to E4orf4 (26). Other mutations that reduced E4orf4 binding, F343A and F280A, affected amino acids that were spatially adjacent to Tyr-337 (Fig. 7). These residues are located in two α-helices (α1 and α2, see Fig. 7C) found above the putative substrate binding groove described previously to contain residues required for binding the microtubule-associated protein Tau (Fig. 7B and Ref. 28). Residues Tyr-337, Phe-343, and Phe-280 protrude from the α1 and α2 helices, facing each other (Fig. 7C). Additional mutations that were previously reported to affect E4orf4 binding included V514D in yeast (26), corresponding to mammalian Val-433, and mutations L43A/L112A in mammalian cells (27). The solved structure of PP2A-B55α revealed that these mutated residues were located in the protein core and may thus affect E4orf4 binding indirectly. Therefore, our present work supports the conclusion that a bioinformatics and structure-oriented approach for identifying protein-protein interaction sites is highly productive even in the absence of complete crystallographic information and can identify residues involved directly in the binding sites more efficiently than other methods. Although two potential E4orf4 binding sites were predicted by the theoretical experiments but only one interacting site was confirmed by biochemical experiments, the bioinformatics approach was crucial in focusing the mutagenesis analysis to few specific sites on the surface of B55α.
Loss of E4orf4 binding by the B55α mutants was shown to significantly reduce their ability to enhance E4orf4-induced cell death (Fig. 6). However, it is intriguing that they could still partially transduce the E4orf4 toxic signal. It has been previously reported that knockdown of various B55 subunits led to stress-induced Src activation (49). Src is another major E4orf4 partner that contributes to E4orf4-induced cell death, and E4orf4 was shown to affect Src activity independently of its interaction with PP2A (19, 20). However, Src and PP2A may cooperate in some activities, such as JNK activation (19). It is possible that overexpression of mutant B55 proteins affects Src activity and enhances all or part of the Src-dependent branch of E4orf4-induced cell death.
The effect of E4orf4 binding on PP2A activity is currently unresolved. It was demonstrated that E4orf4 associated with an active phosphatase (12) and that the PP2A inhibitor okadaic acid inhibited some E4orf4 functions, including down-regulation of gene expression (16) and regulation of alternative splicing (7). On the other hand it was suggested that E4orf4 inhibited PP2A activity toward some substrates, but not others, and that PP2A inhibition contributed to E4orf4-induced cell death (24). It has been recently demonstrated that E4orf4 targets PP2A to the ACF chromatin remodeling factor and to chromatin (18). This finding suggests that E4orf4 targets PP2A to new substrates, in which case it should not inhibit PP2A activity toward them. Therefore, identification of the E4orf4-PP2A substrates in the cell will be crucial to understanding how E4orf4 affects PP2A activity under physiological circumstances and in what way this effect contributes to E4orf4 functions. The identification of the E4orf4 binding site in PP2A-B55α advances our understanding of the interaction between E4orf4 and PP2A; however, it does not solve the problem of whether E4orf4 interferes with PP2A activity toward some substrates. On the one hand, the E4orf4 binding site does not appear to overlap with the proposed substrate binding site described by Xu et al. (Ref. 28 and Fig. 7B), consistent with the finding that E4orf4 associates with an active PP2A (12). On the other hand, E4orf4 may protrude from its binding site to block partial access of some substrates to the substrate binding groove. It is also possible that E4orf4 binding could cause a conformation change in PP2A that may either block substrate access, or in contrast, may facilitate access to substrates that do not usually fit in the binding groove, thus allowing targeting of PP2A to proteins that are not usually PP2A substrates. Further crystallographic and biochemical analyses are required to solve these conundrums.
This work assigned for the first time a functional significance to the α1 and α2 helices of B55α. The finding that these helices are part of the binding site for the B55α viral protein partner E4orf4 may indicate that these helices are also involved in interactions with cellular proteins that could regulate PP2A either by affecting its activity or by allowing it to be targeted to new substrates. Identification of novel cellular binding partners of the E4orf4 interaction site in B55α could add insights into novel modes of regulation of PP2A in the cell.
Acknowledgments
We are grateful to Dr. Fabian Glaser and Dr. Anna Brestovitsky (Technion) for helpful comments.
This work was supported by Israel Science Foundation Grant 769/07, by the Deutsche Forschungsgemeinschaft within the framework of The German-Israeli Project Cooperation (DIP), and by the Rappaport Family Institute for Research in the Medical Sciences.
- E4orf4
- early region 4 open reading frame 4
- PP2A
- protein phosphatase 2A.
REFERENCES
- 1. Ben-Israel H., Sharf R., Rechavi G., Kleinberger T. (2008) Adenovirus E4orf4 protein down-regulates MYC expression through interaction with the PP2A-B55 subunit. J. Virol. 82, 9381–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bondesson M., Ohman K., Manervik M., Fan S., Akusjärvi G. (1996) Adenovirus E4 open reading 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J. Virol. 70, 3844–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mannervik M., Fan S., Ström A. C., Helin K., Akusjärvi G. (1999) Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256, 313–321 [DOI] [PubMed] [Google Scholar]
- 4. Medghalchi S., Padmanabhan R., Ketner G. (1997) Early region 4 modulates adenovirus DNA replication by two genetically separable mechanisms. Virology 236, 8–17 [DOI] [PubMed] [Google Scholar]
- 5. Müller U., Kleinberger T., Shenk T. (1992) Adenovirus E4orf4 protein reduces phosphorylation of c-fos and E1A proteins while simultaneously reducing the level of AP-1. J. Virol. 66, 5867–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Estmer Nilsson C., Petersen-Mahrt S., Durot C., Shtrichman R., Krainer A. R., Kleinberger T., Akusjärvi G. (2001) The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20, 864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanopka A., Mühlemann O., Petersen-Mahrt S., Estmer C., Ohrmalm C., Akusjärvi G. (1998) Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393, 185–187 [DOI] [PubMed] [Google Scholar]
- 8. O'Shea C., Klupsch K., Choi S., Bagus B., Soria C., Shen J., McCormick F., Stokoe D. (2005) Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 24, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ben-Israel H., Kleinberger T. (2002) Adenovirus and cell cycle control. Front. Biosci. 7, d1369–d1395 [DOI] [PubMed] [Google Scholar]
- 10. Kornitzer D., Sharf R., Kleinberger T. (2001) Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in S. cerevisiae and interacts with the anaphase promoting complex/cyclosome. J. Cell Biol. 154, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li S., Szymborski A., Miron M. J., Marcellus R., Binda O., Lavoie J. N., Branton P. E. (2009) The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene 28, 390–400 [DOI] [PubMed] [Google Scholar]
- 12. Shtrichman R., Sharf R., Barr H., Dobner T., Kleinberger T. (1999) Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc. Natl. Acad. Sci. U.S.A. 96, 10080–10085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Afifi R., Sharf R., Shtrichman R., Kleinberger T. (2001) Selection of apoptosis-deficient adenovirus E4orf4 mutants in Saccharomyces cerevisiae. J. Virol. 75, 4444–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maoz T., Koren R., Ben-Ari I., Kleinberger T. (2005) YND1 interacts with CDC55 and is a novel mediator of E4orf4-induced toxicity. J. Biol. Chem. 280, 41270–41277 [DOI] [PubMed] [Google Scholar]
- 15. Roopchand D. E., Lee J. M., Shahinian S., Paquette D., Bussey H., Branton P. E. (2001) Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 20, 5279–5290 [DOI] [PubMed] [Google Scholar]
- 16. Kleinberger T., Shenk T. (1993) Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down-regulates E1A-enhanced junB transcription. J. Virol. 67, 7556–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shtrichman R., Sharf R., Kleinberger T. (2000) Adenovirus E4orf4 protein interacts with both Bα and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Bα. Oncogene 19, 3757–3765 [DOI] [PubMed] [Google Scholar]
- 18. Brestovitsky A., Sharf R., Mittelman K., Kleinberger T. (2011) The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res. 39, 6414–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Champagne C., Landry M. C., Gingras M. C., Lavoie J. N. (2004) Activation of adenovirus type 2 early region 4 ORF4 cytoplasmic death function by direct binding to Src kinase domain. J. Biol. Chem. 279, 25905–25915 [DOI] [PubMed] [Google Scholar]
- 20. Lavoie J. N., Champagne C., Gingras M.-C., Robert A. (2000) Adenovirus E4 open reading frame 4-induced apoptosis involves dysregulation of Src family kinases. J. Cell Biol. 150, 1037–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marcellus R. C., Chan H., Paquette D., Thirlwell S., Boivin D., Branton P. E. (2000) Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Bα subunit of protein phosphatase 2A. J. Virol. 74, 7869–7877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eichhorn P. J., Creyghton M. P., Bernards R. (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795, 1–15 [DOI] [PubMed] [Google Scholar]
- 23. Kleinberger T. (2004) Induction of transformed cell-specific apoptosis by the adenovirus E4orf4 protein. Prog. Mol. Subcell. Biol. 36, 245–267 [DOI] [PubMed] [Google Scholar]
- 24. Li S., Brignole C., Marcellus R., Thirlwell S., Binda O., McQuoid M. J., Ashby D., Chan H., Zhang Z., Miron M. J., Pallas D. C., Branton P. E. (2009) The adenovirus E4orf4 protein induces G2/M Arrest and cell death by blocking PP2A activity regulated by the B55 subunit. J. Virol. 83, 8340–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marcellus R. C., Lavoie J. N., Boivin D., Shore G. C., Ketner G., Branton P. E. (1998) The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J. Virol. 72, 7144–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koren R., Rainis L., Kleinberger T. (2004) The scaffolding A/Tpd3 subunit and high phosphatase activity are dispensable for Cdc55 function in the Saccharomyces cerevisiae spindle checkpoint and in cytokinesis. J. Biol. Chem. 279, 48598–48606 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Z., Mui M. Z., Chan F., Roopchand D. E., Marcellus R. C., Blanchette P., Li S., Berghuis A. M., Branton P. E. (2011) Genetic analysis of B55α/Cdc55 protein phosphatase 2A subunits. Association with the adenovirus E4orf4 protein. J. Virol. 85, 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Y., Chen Y., Zhang P., Jeffrey P. D., Shi Y. (2008) Structure of a protein phosphatase 2A holoenzyme. Insights into B55-mediated Tau dephosphorylation. Mol. Cell 31, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shtrichman R., Kleinberger T. (1998) Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 72, 2975–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brestovitsky A., Sharf R., Kleinberger T. (2012) Preparation of cell lines for conditional knockdown of gene expression and measurement of the knockdown effects on E4orf4-induced cell death. J. Vis. Exp. 68, 4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 32. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. (2004) ConSeq. The identification of functionally and structurally important residues in protein sequences. Bioinformatics 20, 1322–1324 [DOI] [PubMed] [Google Scholar]
- 34. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) ConSurf. Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]
- 35. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) ConSurf 2005. The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neuvirth H., Raz R., Schreiber G. (2004) ProMate. A structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 338, 181–199 [DOI] [PubMed] [Google Scholar]
- 37. Xu D., Zhang Y. (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 39. Sippl M. J. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17, 355–362 [DOI] [PubMed] [Google Scholar]
- 40. Wiederstein M., Sippl M. J. (2007) ProSA-web. Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andrusier N., Nussinov R., Wolfson H. J. (2007) FireDock. Fast interaction refinement in molecular docking. Proteins 69, 139–159 [DOI] [PubMed] [Google Scholar]
- 42. Mashiach E., Nussinov R., Wolfson H. J. (2010) FiberDock. A web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Res. 38, W457–W461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005) PatchDock and SymmDock. Servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro. A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro. An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 46. Kozakov D., Brenke R., Comeau S. R., Vajda S. (2006) PIPER. An FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 [DOI] [PubMed] [Google Scholar]
- 47. Lavoie J. N., Nguyen M., Marcellus R. C., Branton P. E., Shore G. C. (1998) E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140, 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Livne A., Shtrichman R., Kleinberger T. (2001) Caspase activation by adenovirus E4orf4 protein is cell line-specific and is mediated by the death receptor pathway. J. Virol. 75, 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eichhorn P. J., Creyghton M. P., Wilhelmsen K., van Dam H., Bernards R. (2007) A RNA interference screen identifies the protein phosphatase 2A subunit PR55γ as a stress-sensitive inhibitor of c-SRC. PLoS Genet. 3, e218. [DOI] [PMC free article] [PubMed] [Google Scholar]