Abstract
Vitamin B6 is an essential cofactor for a large number of enzymes in both prokaryotes and eukaryotes. In this study, we characterized the pyridoxal 5′-phosphate (PLP) biosynthesis pathway in Streptococcus pneumoniae. Our results revealed that S. pneumoniae possesses a de novo vitamin B6 biosynthesis pathway encoded by the pdxST genes. Purified PdxS functionally displayed as PLP synthase, whereas PdxT exhibited glutaminase activity in vitro. Deletion of pdxS, but not pdxT, resulted in a vitamin B6 auxotrophic mutant. The defective growth of the ΔpdxS mutant in a vitamin B6-depleted medium could be chemically restored in the presence of the B6 vitamers at optimal concentrations. By analyzing PdxS expression levels, we demonstrated that the expression of pdxS was repressed by PLP and activated by a transcription factor, PdxR. A pneumococcal ΔpdxR mutant also exhibited as a vitamin B6 auxotroph. In addition, we found that disruption of the vitamin B6 biosynthesis pathway in S. pneumoniae caused a significant attenuation in a chinchilla middle ear infection model and a minor attenuation in a mouse pneumonia model, indicating that the impact of vitamin B6 synthesis on virulence depends upon the bacterial infection niche.
INTRODUCTION
Vitamin B6 represents the most versatile organic cofactor of enzymes in all organisms. It refers to six biologically interconvertible compounds or vitamers: pyridoxine (PN), pyridoxamine (PM), pyridoxal (PL), and their respective phosphorylated forms, including pyridoxine 5′-phosphate (PNP), pyridoxamine 5′-phosphate (PMP), and pyridoxal 5′-phosphate (PLP) (1). PLP is considered as the only active form of vitamin B6. In addition to its role in several enzymatic processes, including amino acid and fatty acid metabolism (2, 3), vitamin B6 is also involved in oxidative stress response and acts to quench reactive oxygen species (4).
Bacteria synthesize PLP via two major pathways: a de novo pathway and a salvage pathway (5–7). There are two distinct de novo pathways, either deoxyxylulose 5-phosphate (DXP)-dependent or DXP-independent, in different organisms (5–7). The genes of the DXP-dependent pathway have been identified mostly in the γ subdivision of proteobacteria, which form a tight cluster around pdxA and pdxJ of Escherichia coli (5, 6). In this pathway, the biosynthetic product PNP is oxidized by PdxH to produce PLP (8). The DXP-independent pathway (or alternative pathway) requires two enzymes, PdxS (also named as Pdx1, SnzP, or YaaD) and PdxT (also referred to Pdx2, SnoP, or YaaE), which are found in all archaea, fungi, plants, and most bacteria (5). PdxS and PdxT directly produce PLP from a pent(μl)ose (ribose 5-phosphate or ribulose 5-phosphate) and a triose (glyceraldehyde 3-phosphate or dihydroxyacetone phosphate) in the presence of glutamine. Biochemical and structural studies revealed that PdxS and PdxT of Bacillus subtilis form a hetero-oligomeric complex (9–11).
In addition to the de novo pathway, bacteria are also able to synthesize PLP by a salvage pathway (12), in which PLP is synthesized from other B6 vitamers. In this pathway, the vitamers PL, PN, and PM are phosphorylated by a kinase, such as PdxK or PdxY (13). PMP and PNP are then oxidized to PLP by PdxH (14, 15).
The DXP-dependent and the DXP-independent biosynthesis pathways have been mainly established in E. coli and B. subtilis, respectively. However, these pathways have not been efficiently explored in pathogenic bacteria. In Helicobacter pylori, it was reported that the DXP-dependent vitamin B6 biosynthesis pathway plays a role in optimal bacterial growth, chronic colonization of mice, glycosylated flagellum synthesis, and flagellum-based motility (16). The DXP-dependent vitamin B6 biosynthesis pathway of Mycobacterium tuberculosis is essential for its survival and virulence (17).
Little is known about how the vitamin B6 biosynthesis pathways are regulated. A recent study displayed that expression of pdxST in Corynebacterium glutamicum is significantly reduced in the presence of PL and PLP (18). In this bacterium, a MocR-type transcription factor, PdxR, positively controls the transcription of PLP synthase, whereas the in vitro binding of PdxR to the pdxR-pdxST intergenic DNA is not significantly affected by the addition of B6 vitamers (18).
The vitamin B6 biosynthesis pathway has not been characterized in Streptococcus pneumoniae, a Gram-positive bacterium responsible for several human diseases, including pneumonia, septicemia, otitis media (OM), bacteremia, sinusitis, and meningitis. In the present study, we characterized the genes encoding the vitamin B6 biosynthesis pathway in S. pneumoniae using both genetic and biochemical approaches.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the bacterial strains used in the present study are listed in Table 1. S. pneumoniae D39 (serotype 2, ATCC) and its derivatives were initially grown in Todd Hewitt broth containing 0.5% yeast extract (THY) (BD Biosciences), and all of the working stocks for the present study were grown in a chemically defined medium (CDM; purchased from JRH Bioscience) supplemented with 0.1% choline, 0.25% sodium bicarbonate, and 0.073% cysteine, except as specified. A formulated CDM (complete CDM) was prepared as reported (19), which contains 1 mg of both PM and PL/liter. When necessary, both PM and PL were dropped out (designated as depleted CDM), and the depleted CDM was then supplemented with various concentrations of B6 vitamers, as specified (supplemented CDM). Competence THY medium was prepared by adding 0.2% glucose, 0.2% CaCl2, and 0.02% bovine serum albumin to THY medium (pH 7.2 to 7.4), which was used for transformation of pneumococcal strains (20). If necessary, 200 μg of kanamycin/ml, 2 μg of erythromycin/ml, or 150 μg of streptomycin/ml was used for selection. All pneumococcal strains were routinely grown at 37°C with 5% CO2. E. coli strains were grown in either Luria-Bertani (LB) or M9 minimal medium at 37°C. If necessary, 25 μg of kanamycin/ml, 100 μg of ampicillin/ml or 200 μg of erythromycin/ml was used for selection.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. pneumoniae | ||
| D39 | S. pneumoniae serotype 2 | 40 |
| ST588 | D39 ΔcbpA::Janus cassette; Kanr | 22 |
| ST606 | OM strain of S. pneumoniae; Strepr | 21 |
| ST1809 | ST606 ΔarcT::Janus cassette; Kanr | This study |
| ST2675 | D39 ΔpdxT::Janus cassette; Kanr | This study |
| ST2676 | D39 ΔpdxS::Janus cassette; Kanr | This study |
| ST2677 | D39 ΔpdxST::Janus cassette; Kanr | This study |
| ST2726 | D39 ΔpdxR::Janus cassette; Kanr | This study |
| ST2784 | ST2676(pST2782); Kanr Ermr | This study |
| ST2785 | ST2676(pVA838); Kanr Ermr | This study |
| ST2786 | ST2726(pST2783); Kanr Ermr | This study |
| ST2787 | ST2726(pVA838); Kanr Ermr | This study |
| ST2790 | ST606 ΔpdxS::Janus cassette; Kanr | This study |
| ST2792 | ST2790(pST2782); Kanr Ermr | This study |
| ST2793 | ST2790(pVA838); Kanr Ermr | This study |
| E. coli | ||
| DH5α | E. coli strain used for cloning | Lab stock |
| BL21(DE3) | E. coli strain used for expression | Novagen |
| ST2699 | E. coli BL21(DE3)(pST2697); Kanr | This study |
| ST2700 | E. coli BL21(DE3)(pST2698); Kanr | This study |
| ST2728 | E. coli BL21(DE3)(pST2727); Kanr | This study |
| ec022 | E. coli BW25113 (parent strain for the Keio Collection) | 25, 29 |
| ec048 | JW0051-1(pdxA764:kan); Kanr | 25 |
| ec053 | JW2548-1(pdxJ736:kan); Kanr | 25 |
| ec061 | ec048(pGB104); Apr | This study |
| ec065 | ec053(pGB104); Apr | This study |
| ST2683 | E. coli ec048(pST2679); Apr | This study |
| ST2684 | E. coli ec053(pST2679); Apr | This study |
| ST2688 | E. coli ec048(pST2681); Apr | This study |
| ST2689 | E. coli ec053(pST2681); Apr | This study |
| Plasmids | ||
| pET28a(+) | His tag expression vector; Kanr | Novagen |
| pST2697 | pET28a(+) carrying S. pneumoniae pdxS ORF; Kanr | This study |
| pST2698 | pET28a(+) carrying S. pneumoniae pdxT ORF; Kanr | This study |
| pST2727 | pET28a(+) carrying S. pneumoniae SPD_1225 ORF; Kanr | This study |
| pGB104 | pBR322 with E. coli crp promoter; Apr | This study |
| pST2679 | pGB104 carrying S. pneumoniae pdxS ORF; Apr | This study |
| pST2681 | pGB104 carrying S. pneumoniae pdxT ORF; Apr | This study |
| pVA838 | S. pneumoniae expression vector; Ermr | 23 |
| pST2782 | pVA838 carrying S. pneumoniae pdxS promoter and ORF; Ermr | This study |
| pST2783 | pVA838 carrying S. pneumoniae pdxR promoter and ORF; Ermr | This study |
Ermr, erythromycin resistance; Apr, ampicillin resistance; Strepr, streptomycin resistance; Kanr, kanamycin resistance.
Construction of mutants and complementation in S. pneumoniae.
All of the primers used in the present study are listed in Table S1 in the supplemental material. pdxS, pdxT, pdxST, and pdxR mutants were generated by homologous recombination similarly, as reported earlier (21, 22). Briefly, the upstream and the downstream flanking regions for each target gene were PCR amplified from either D39 or ST606 genomic DNA. The products were digested and ligated with a Janus cassette, which contains a kanamycin resistance gene and was amplified from ST588 (22). The ligated fragments were then used to transform D39 or ST606. The mutants were selected with kanamycin and verified by PCR. For complementation of the ΔpdxS and the ΔpdxR mutants, the putative promoter and the open reading frame (ORF) of the respective gene was cloned in the pVA838 plasmid (23) and was transformed into the mutant. As a control, pVA838 was also transformed into both the mutants. The transformants were selected with erythromycin and verified by PCR.
Morphological study.
S. pneumoniae D39 wild-type (WT) and its derivatives were grown in complete CDM to an optical density at 620 nm (OD620) of 0.6. Bacteria were thoroughly washed with depleted CDM and then incubated in either supplemented CDM with 10 μM PLP or depleted CDM for 4 h. The bacterial morphologies were observed under a light microscope after Gram staining (Fisher Diagnostics).
Heterogeneous expression of S. pneumoniae pdxS and pdxT in E. coli ΔpdxA and ΔpdxJ, respectively.
An E. coli crp promoter was amplified by PCR from DH5α genomic DNA and was cloned into pBR322 to generate pGB104. The flanking region is similar to that in pMBC664 (24), except that different restriction sites were introduced. ORFs of S. pneumoniae pdxS and pdxT were then PCR amplified and cloned into pGB104. These constructs, along with pGB104, were individually transformed into a ΔpdxA strain JW0051-1 (CGSC8361) and a ΔpdxJ strain JW2548-1 (CGSC10027) of E. coli (25). The recombinant strains were grown in M9 medium in the presence or absence of 0.1 mM PLP.
Protein expression and purification.
ORFs of S. pneumoniae pdxS, pdxT, and pdxR were PCR amplified from D39 genomic DNA. The PCR products were cloned into pET28a(+) (Novagen) at the same restriction sites as indicated in the respective primers (see Table S1 in the supplemental material). The recombinant plasmids were verified by sequencing and used to transform E. coli BL21(DE3). The expression of each recombinant strain was induced for 3 h at 22°C with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in LB broth. Another aliquot of recombinant bacteria that expresses PdxR was washed with phosphate-buffered saline (PBS) and then resuspended in M9 broth for induction with 0.5 mM IPTG. Protein purification was performed as described previously using Ni-NTA resin (Qiagen) (26, 27). The purified proteins were dialyzed against PBS with 10% glycerol and stored at −80°C until use.
Glutaminase activity.
Glutaminase activity was determined using a procedure as described previously (10). Briefly, 10 μM PdxT, 10 μM PdxS, or a 10 μM mixture of both proteins was incubated with 1 U of l-glutamic dehydrogenase from bovine liver (Sigma), 10 mM glutamine, 0.6 mM 3-acetylpyridine adenine dinucleotide (APAD; Sigma). After 1 h of preincubation at 37°C, the OD was measured every 10 min at 363 nm (OD363) over an 80-min period.
PLP formation assay.
PLP formation was analyzed according to an earlier report (10). Briefly, 10 μM PdxS, 10 μM PdxT, or a 10 μM mixture of both proteins was incubated in the presence of 10 mM glutamine or 10 mM NH4Cl, as specified, 1 mM d-ribose 5-phosphate, 2 mM dl-glyceraldehyde 3-phosphate, and 50 mM Tris-HCl (pH 8.0) in a final volume of 500 μl. After 30 min of preincubation at 37°C, the OD at 414 nm (OD414) was measured every 10 min for a total of 3 h.
Electrophoretic mobility shift assay (EMSA).
A 32P-end-labeled pdxS probe (0.01 pmol) was used in each 10-μl binding reaction mixture as described previously (26, 28), with modifications. Briefly, purified His-PdxR (at specified concentrations) and DNA probes were incubated for 30 min at room temperature in DNA binding buffer, followed by electrophoresis on a nondenaturing 8% polyacrylamide gel for 2 to 3 h at 14 V/cm in 0.5× Tris-borate-EDTA (TBE) buffer. A 500-fold excess of unlabeled DNA fragments was used for competition experiments. Gels were vacuum dried, exposed on a phosphor screen, scanned with a Storm 860 PhosphorImager (Molecular Dynamics), and analyzed with ImageQuant software.
RNA preparation and Northern blot analysis.
Wild-type (WT) D39 was grown in 10 ml of complete CDM to an OD620 of 0.6 and then split into two 5-ml aliquots. The bacteria were washed three times with depleted CDM and then resuspended in 5 ml of either depleted CDM or supplemented CDM with 10 μM PLP. After 4 h of incubation at 37°C with 5% CO2, RNA samples were prepared from these bacteria using an RNeasy minikit (Qiagen). Then, 10 μg of total RNA of each sample was separated on a 1% agarose gel with 1× morpholinepropanesulfonic acid buffer and 2% formaldehyde. Subsequently, the RNA was transferred overnight by capillarity to a Hybond membrane using 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as the transfer buffer.
Hybridization with a [γ-32P]ATP (MP Biomedicals)-labeled oligonucleotide specific to pdxS was performed in hybridization buffer (GE Healthcare) for 5 h at 42°C. The membrane was washed twice with a washing buffer (2× SSC, 0.1% sodium dodecyl sulfate [SDS]), exposed overnight to a phosphor screen, and visualized using a PhosphorImager (Molecular Dynamics). Hybridization with a probe specific to S. pneumoniae 16S rRNA was used as a loading control.
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee of Albany Medical College (permits 11-12002 for mice and 11-12001 for chinchillas). All efforts were made to minimize suffering.
Preparation of polyclonal antibodies and Western blot analyses.
Female BALB/c mice (Taconic) were immunized subcutaneously to generate polyclonal antibodies against purified PdxS and PdxT, respectively (n = 5). The procedure of immunization and antisera preparation were similar to those described earlier (27).
For Western blot analyses, bacteria were grown in complete CDM to an OD620 of 0.6 and treated as described in the “morphological study” section above. After 2 to 4 h of incubation, bacteria were broken by sonication, and equal amounts of proteins from each sample were loaded onto a SDS–12.5% PAGE gel. Separated proteins were transferred to a polyvinylidene difluoride membrane and probed with the antisera to PdxT or PdxS. The analysis with a polyclonal antibody against SPD_1063 was used as a loading control. After blotting with a peroxidase-conjugated goat anti-mouse IgG secondary antibody (Thermo Scientific), peroxidase detection was carried out with an ECL Western blotting detection reagents and analysis system (Thermo Scientific).
Mouse infection.
We infected 6- to 8-week-old female BALB/c mice (Taconic) with WT, ST2675, and ST2676 strains (n = 8). Briefly, bacteria were grown to an OD620 of 0.4 in complete CDM, washed four times in PBS, diluted with PBS to ∼1 × 108 CFU/ml, and then inoculated intranasally in a volume of 50 μl (∼5 × 106 CFU). The inocula were determined by serial dilution and plate counting. Mice were subsequently monitored for 9 days.
Coinfection of chinchilla middle ears.
Coinfection experiments were carried out as described previously (21) with slight modifications. Three 1-year-old female chinchillas (Ryerson Chinchilla Ranch) were used in each group. Broth cultures of WT (ST606) and its isogenic mutants were separately incubated to an OD620 of 0.4 and stocked with 25% glycerol in THY. The CFU levels of the frozen stocks were enumerated for WT in the presence of streptomycin or for mutant in the presence of kanamycin. The bacteria were diluted and mixed in PBS at a 1:1 ratio of CFU. Aliquots of 100-μl mixtures were used to infect the middle ears of chinchillas (∼1 × 104 CFU/ear, n = 6). The level of attenuation is expressed as the competitive index (CI), which is defined as the output CFU ratio (mutant/WT) divided by the input CFU ratio (mutant/WT). In addition, ST1809, a ΔarcT mutant that did not affect the bacterial virulence in our previous experiment (unpublished) was used as a control in this experiment.
Statistical analysis.
Comparisons of growth were analyzed using a two-tailed t test. Kaplan-Meier survival curves were compared using Gehan-Breslow-Wilcoxon test. All of the analyses were performed using Prism 5 (GraphPad Software), and P values of < 0.05 were considered to be statistically significant.
RESULTS
S. pneumoniae PdxS and PdxT are the enzymes for PLP synthesis.
We used the S. pneumoniae D39 strain as a model organism to study the biosynthesis pathway of vitamin B6. In this strain, SPD_1297 (pdxS) has been annotated as a putative pyridoxal biosynthesis lyase, and SPD_1296 (pdxT) encodes a putative glutamine amidotransferase subunit (30). The two genes are clustered in a putative operon (Fig. 1A). Both PdxS and PdxT share high identity (64 and 44%, respectively) in the amino acid sequences with their orthologs in B. subtilis. In the present study, S. pneumoniae pdxS and pdxT were overexpressed in E. coli, and the recombinant proteins were purified to high homogeneity using Ni-NTA chromatography. These proteins were then used to determine their activities in vitamin B6 formation in vitro.
Fig 1.
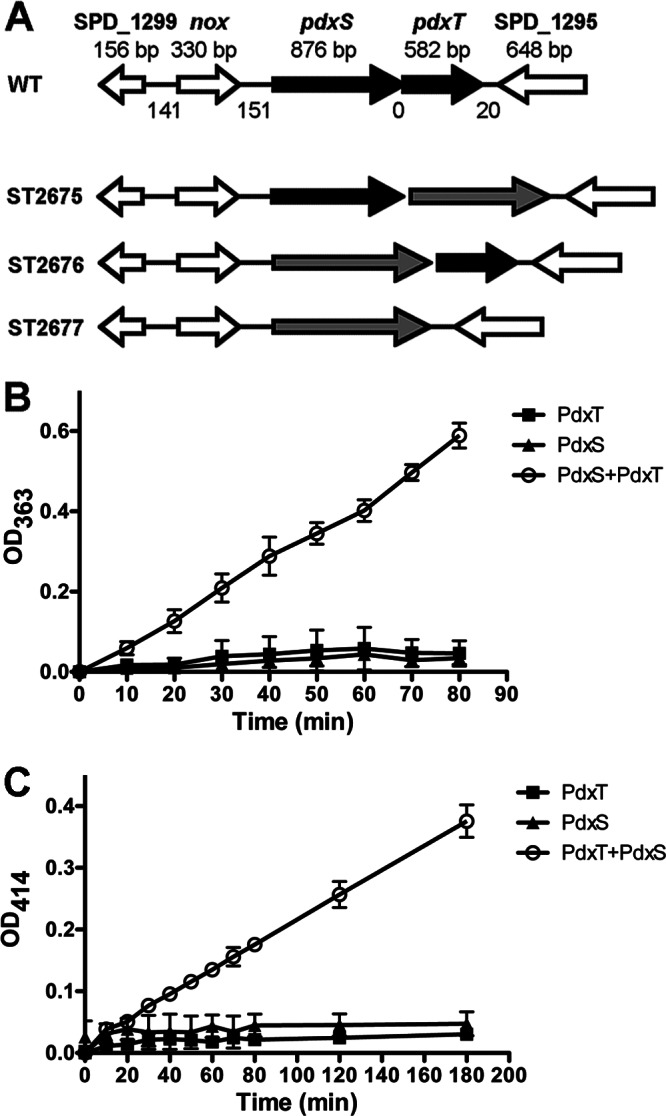
Enzymatic activity assays of PdxS and PdxT. (A) Genetic organization of pdxS and pdxT in S. pneumoniae. The size (in base pairs) of each ORF is indicated above the gene. The size of each intergenic region is also noted between adjacent genes. Each target gene was replaced with a Janus cassette (gray arrow) by homologous recombination. (B and C) Glutaminase activity (B) or PLP synthase activity (C) was determined in the presence of PdxS, PdxT, or a mixture of the two proteins at a molar ratio of 1:1. The data shown are the means of three independent assays. Error bars denote the standard error of the mean (SEM).
To test the glutaminase activity, pneumococcal PdxT was incubated individually or together with PdxS using glutamine as a substrate. The activity was exclusively detected when both proteins were present in the reaction but not with either PdxT or PdxS alone (Fig. 1B). This result indicates that the glutaminase activity detected with PdxT is dependent on the presence of PdxS. This finding coincides with previous observations in other organisms (17, 31–35).
Glutamine, glyceraldehyde 3-phosphate, and ribose 5-phosphate were used as substrates to determine the PLP synthesis activity displayed by pneumococcal PdxS, PdxT, or a mixture of the two proteins. As a result, the PLP synthase activity was detected only when both PdxS and PdxT were concomitantly present in the reaction, whereas no PLP was detectable with either individual protein (Fig. 1C). This result indicates that either the interaction between PdxS and PdxT is required for the PLP formation or the formation of PLP is a sequential reaction in which the product of one enzyme is used as a substrate for the other enzyme.
In B. subtilis, PdxT and PdxS form an active complex, which is required for PLP biosynthesis (9–11, 31). In our study, the glutaminase and the PLP synthase activities were exclusively detected in the presence of both proteins. Therefore, we examined the interaction between S. pneumoniae PdxS and PdxT in vitro, either under the same conditions as described by others (10, 31) or using conditions in which glutaminase activity or PLP synthase activity was detected. The proteins were then separated on a native gel and detected either by staining with Coomassie brilliant blue or by Western blotting analyses with both anti-PdxS and anti-PdxT antibodies. Our result showed that the mobility of PdxS and PdxT when mixed was identical to that of the two proteins analyzed singularly (data not shown). Thus, S. pneumoniae PdxS and PdxT do not appear to form a complex under these conditions.
S. pneumoniae ΔpdxS is auxotrophic for vitamin B6.
In S. pneumoniae, pdxS and pdxT genes are immediately adjacent to each other (Fig. 1A). To study the pneumococcal vitamin B6 biosynthesis pathway in vivo, we replaced pdxS, pdxT, and pdxST with a Janus cassette and designated these strains ST2676, ST2675, and ST2677, respectively (Fig. 1A). These mutants were verified by Western blotting with specific antibodies (see Fig. S1 in the supplemental material). Interestingly, PdxT was still detected in ST2676, although pdxS and pdxT are clustered in a putative operon (Fig. 1A). The expression of pdxT in ST2676 is likely controlled by a promoter from the inserted Janus cassette.
The growth of these mutants was determined in complete CDM, which contains PM and PN, and in depleted CDM which does not have any B6 vitamer. The growth rates of all of the mutants were indistinguishable from WT grown in complete CDM (data not shown). In depleted CDM, ST2675 was able to grow, but its growth rate was lower than that of the WT. In contrast, ST2676 and ST2677 were unable to grow in depleted CDM (Fig. 2A), suggesting that the ΔpdxS mutant is auxotrophic for vitamin B6. Since PdxT was detected in ST2676 (see Fig. S1 in the supplemental material), the vitamin B6 auxotrophy of this strain is mainly caused by the absence of PdxS, which is consistent with the observation that expression of pdxS alone in both ST2676 and ST2677 partially rescued the bacterial growth in depleted CDM (data not shown).
Fig 2.
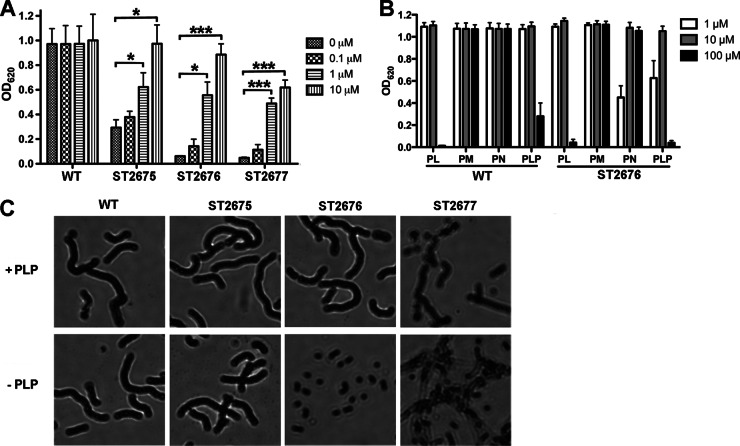
Growth phenotypes of the S. pneumoniae mutants associated with the vitamin B6 biosynthesis pathway. (A) Growth of WT (D39) and its derivatives, including ΔpdxT (ST2675), ΔpdxS (ST2676), and ΔpdxST (ST2677), in depleted CDM (0 μM) or supplemented CDM with indicated concentrations of PLP. Bacteria were grown for 8 h at 37°C with 5% CO2. The data shown are the means of three independent experiments. Error bars denote the SEM. *, P < 0.05; ***, P < 0.001. (B) Growth of WT (D39) and ST2676 in supplemented CDM with various concentrations of indicated vitamers for 8 h at 37°C with 5% CO2. The data shown are the means of three independent experiments. Error bars denote the SEM. (C) Gram staining result of WT (D39) and its derivatives after incubation in supplemented CDM with 10 μM PLP (+ PLP) or in depleted CDM (− PLP).
The defective growth of the ΔpdxS, ΔpdxT, and ΔpdxST mutants in depleted CDM was restored by exogenous PLP in a concentration-dependent manner (Fig. 2A). In contrast, addition of up to 10 μM PLP exhibited no effect on the growth of WT (Fig. 2A). Surprisingly, 100 μM PLP was toxic to both WT and ST2676 (Fig. 2B). Therefore, PLP in S. pneumoniae needs to be maintained at precise levels.
S. pneumoniae has a functional salvage pathway.
Most bacteria have a salvage pathway for vitamin B6 biosynthesis that enables them to utilize other B6 vitamers for PLP synthesis. However, PdxH and PdxK, which catalyze the conversions between different B6 vitamers, could not be found in the genome of D39 (30). To determine whether S. pneumoniae has a functional salvage pathway, WT and ST2676 were grown in supplemented CDM which was provided with different concentrations of PL, PM, or PN. In the presence of 1 or 10 μM each tested vitamer, the growth of the WT was not affected, but the growth of ST2676 was partially (at 1 μM PN) or fully (at 1 μM PL or PM or 10 μM any of the vitamers) restored to the WT levels (Fig. 2B). This result suggests that a functional savage pathway exists in S. pneumoniae.
Morphologies of the pneumococcal pdx mutants.
To analyze the effects of vitamin B6 biosynthesis on the bacterial morphology, we performed Gram-staining to compare ST2675, ST2676, and ST2677 with the WT strain. These bacterial strains were first grown in complete CDM and then incubated in either depleted CDM or supplemented CDM with 10 μM PLP. Our result showed that the chain sizes of all of the strains were similar in the presence of PLP. In the absence of PLP, both WT and ST2675 exhibited similar chain sizes comparable to that in the presence of PLP, whereas ST2676 and ST2677 appeared as mono- or diplococci (Fig. 2C). The morphological changes of ST2676 and ST2677 in the absence of PLP are consistent with their growth phenotypes in depleted CDM.
The activity of S. pneumoniae PdxS is independent of PdxT in the presence of ammonium.
It has been shown that the addition of exogenous ammonium in the growth medium compensates the defective growth rate of a ΔpdxT mutant in B. subtilis (31), suggesting that the primary role of PdxT in vitamin B6 synthesis is providing ammonium. In the present study, we grew WT, ST2675, ST2676, and ST2677 in depleted CDM, but supplemented with various concentrations of ammonium. The result showed that the defective growth of ST2675 was partially (with 0.1 or 1 mM ammonium) or fully (with 10 mM ammonium) restored to the WT levels with additional ammonium (Fig. 3A). In contrast, the growth of neither WT nor ST2676 or ST2677 was changed in the presence of 0.1 to 10 mM ammonium. These results indicate that the primary function of pneumococcal PdxT is similar to its ortholog in B. subtilis.
Fig 3.
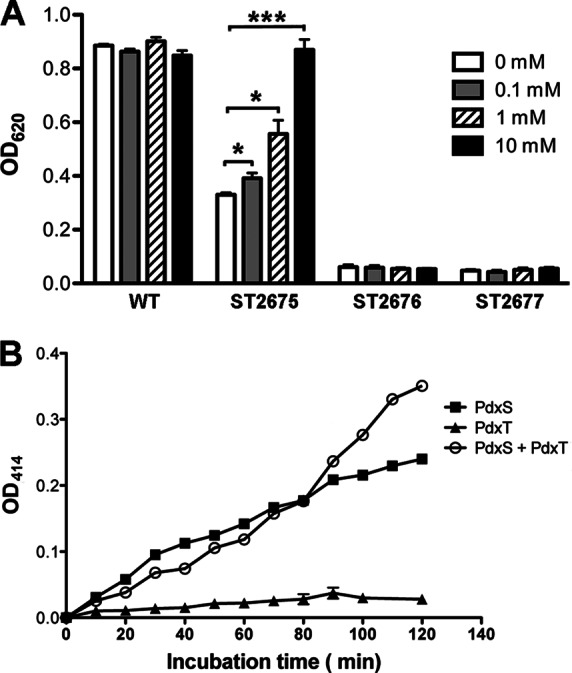
PLP synthase activity of S. pneumoniae PdxS. (A) Growth of WT (D39) and its derivatives, including ΔpdxT (ST2675), ΔpdxS (ST2676), and ΔpdxST (ST2677) strains, in depleted CDM (0 mM) or supplemented CDM with various concentrations of NH4Cl for 8 h at 37°C with 5% CO2. The data shown are the means of three independent experiments. Error bars denote SEM. *, P < 0.05; ***, P < 0.001. (B) PLP formation assay in the presence of PdxS, PdxT, or a mixture of the two proteins at a molar ratio of 1:1. The data shown are the means of triplicate samples. Error bars denote the SEM.
We showed that both PdxS and PdxT were required to synthesize PLP in vitro (Fig. 1C). Based upon the ammonium-dependent growth phenotype of ST2675, we hypothesized that the function of PdxT in this reaction was producing ammonium from glutamine. Therefore, replacing glutamine with ammonium in the reaction might allow PdxS to produce PLP in the absence of PdxT. We tested this hypothesis in vitro, and the result showed that PdxS alone could form PLP after replacing glutamine with ammonium in the reaction (Fig. 3B). These results are consistent with a two-step model for PLP formation, in which PdxT and PdxS together convert glutamine into ammonium, and PdxS utilizes ammonium, glyceraldehyde 3-phosphate (or dihydroxyacetone phosphate), and ribose 5-phosphate (or ribose 5-phosphate) to produce PLP (9, 11).
It has been reported that Cercospora nicotianae pdx1 complements an E. coli pdxJ mutation (36). We also investigated whether heterogeneous expression of S. pneumoniae pdxS or pdxT could complement E. coli mutants defective in vitamin B6 biosynthesis. The ORFs of both pdxS and pdxT were cloned into plasmid pGB104 to express these genes under the control of the E. coli crp promoter. These constructs and the control plasmid pGB104 were then individually transformed into E. coli ΔpdxA (ec048) or ΔpdxJ (ec053). These mutants and their derivatives were compared to the WT strain (ec022) for growth either in M9 broth (Fig. 4A and B) or on M9 plates (see Fig. S2 in the supplemental material), in the presence or absence of 0.1 mM PLP or PN. The results showed that neither ec048 nor ec053 grew with the minimal media in the absence of B6 vitamers, but both the mutants grew similarly to their parental strains by the addition of exogenous PN or PLP. In the absence of B6 vitamers, the growth defect of both the mutants could be restored by the expression of pneumococcal pdxS, but not by the control plasmid (Fig. 4A and B) or expression of pdxT (data not shown). This result also suggests that pneumococcal PdxS is capable of producing PLP independent of PdxT in the presence of ammonium (Fig. 4C).
Fig 4.
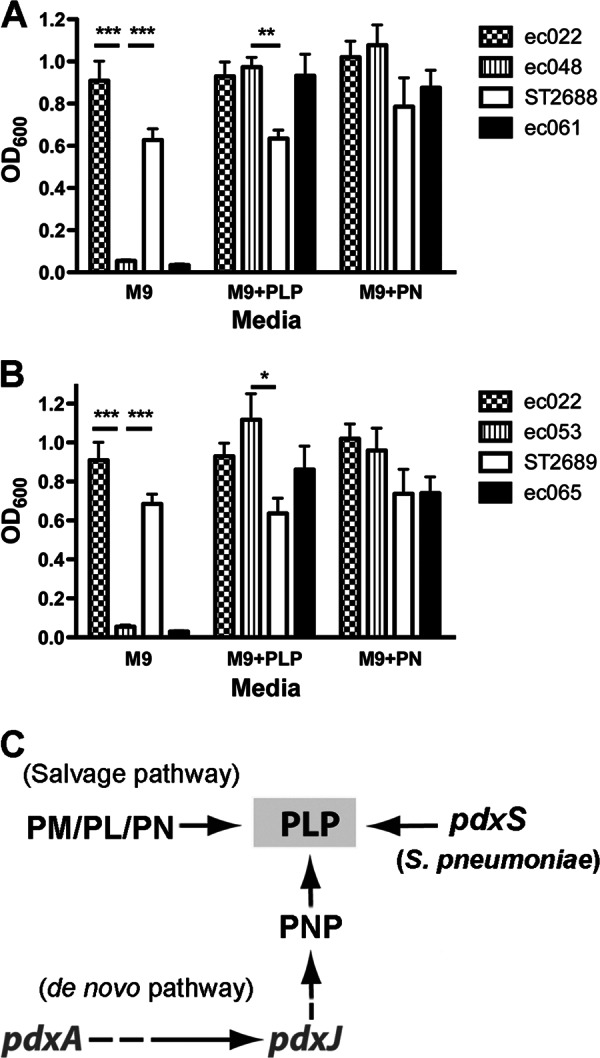
Heterogeneous expression of S. pneumoniae pdxS in the E. coli ΔpdxA and ΔpdxJ mutants. (A and B) Complementation of E. coli ΔpdxA (A) or ΔpdxJ (B) mutants by expression of S. pneumoniae pdxS. ec022 is the E. coli WT strain. ec048 and ec053 are E. coli ΔpdxA and ΔpdxJ, respectively. ST2688 and ST2689 express pneumococcal pdxS in ΔpdxA and ΔpdxJ, respectively. Strains ec061 and ec065 contain the control vector in ΔpdxA and ΔpdxJ, respectively. Bacteria were grown in M9 broth in the absence (M9) or presence of either 0.1 mM PLP (M9 + PLP) or 0.1 mM PN (M9 + PN) for 8 h shaking at 37°C. The data shown are the means of three independent experiments. Error bars denote the SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Schematic representation of E. coli vitamin B6 biosynthesis pathways and the heterogeneous expression of S. pneumoniae pdxS in the E. coli mutants.
The expression levels of PdxS are feedback inhibited by PLP.
The vitamin B6 biosynthesis pathways have been studied in several bacteria, but the knowledge regarding regulation of the PLP levels remains very limited. We grew S. pneumoniae WT in complete CDM and then incubated the bacteria in either depleted CDM or supplemented CDM with 10 μM PLP. The expression levels of PdxS were then determined by Western blotting with antibody against pneumococcal PdxS. Our result showed that PdxS was weakly expressed in CDM supplemented with PLP, but it was abundant after incubation for 2 to 4 h in the absence of PLP (Fig. 5A), suggesting a feedback inhibition of vitamin B6 biosynthesis by PLP.
Fig 5.
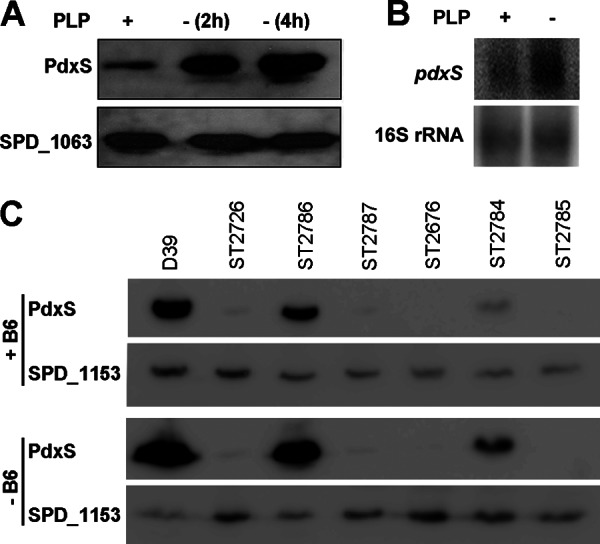
Regulation of S. pneumoniae pdxS. (A) Western blot analysis of WT (D39) incubated in either supplemented CDM with 10 μM PLP (+) or depleted CDM (−) for 2 or 4 h. The protein samples were analyzed with the indicated antibodies (right side). Blotting with antibody against SPD_1063 was used as a loading control. (B) Northern blot of total RNA samples isolated from WT (D39) incubated in either supplemented CDM with 10 μM PLP (+) or depleted CDM (−) for 4 h. The blot with the16S rRNA probe was used as a loading control. (C) Expression of PdxS in strains as indicated, including WT (D39), ST2726 (ΔpdxR), ST2786 (ΔpdxR/pdxR+), ST2787 (ΔpdxR/vector control), ST2676 (ΔpdxS), ST2784 (ΔpdxS/pdxS+), and ST2785 (ΔpdxS/vector control). Bacteria were grown in CDM with vitamin B6 (+B6) and an aliquot of each strain was washed and incubated with depleted CDM (−B6). Bacteria were subsequently disrupted by sonication, and protein samples were detected by Western blotting analyses with the anti-PdxS antibody. An antibody against SPD_1153 was used as a loading control.
We further determined the expression of pdxS using Northern blotting. The result showed that pdxS mRNA levels were enhanced in the absence PLP compared to that in the presence of PLP (Fig. 5B), which is consistent with the result of Western blot analysis. Taken together, it is likely that the expression of pdxS is transcriptionally regulated.
Expression of pdxS is directly activated by PdxR.
In C. glutamicum, it has been reported that the pdxST operon is transcriptionally regulated by a transcription factor PdxR (18), which belongs to the MocR subfamily of the GntR-type proteins. PdxR carries a winged helix-turn-helix (wHTH) domain at the N-terminal portion and an aspartate aminotransferase-like (AAT) domain at the C-terminal portion with potential PLP binding sites (18). In S. pneumoniae, we identified five genes each having a wHTH domain of the GntR superfamily (see Fig. S3A in the supplemental material). However, only SPD_1225 possesses an AAT domain and shares all of the characteristics with C. glutamicum PdxR (see Fig. S3A in the supplemental material), and the two proteins only share 21% identity in amino acid sequences. In addition, C. glutamicum pdxR is adjacent to pdxS, whereas S. pneumoniae SPD_1225 is over 70 genes away from pdxS. We designated the SPD_1225 gene pdxR and the encoded protein PdxR hereafter unless specified. To investigate the role of S. pneumoniae PdxR in regulation of pdxS, we generated a ΔpdxR mutant strain (ST2726) and complemented ST2726 with a native pdxR under its own promoter (ST2786). The ΔpdxS mutant was also complemented with a native pdxS (ST2784). The deletion and the complementation of both pdxS and pdxR were verified by PCR (Fig. 3B). We then explored the expression of PdxS in response to vitamin B6 with these mutants. Our result showed that the expression of PdxS was almost completely abolished in ST2726 either in the presence or absence of vitamin B6 (Fig. 5C), indicating that PdxR is an activator of pdxS. This phenotype can be corrected by the expression of pdxR, but not by the vector control (Fig. 5C), indicating specificity.
We grew ST2676, ST2726, and their complemented strains in either complete CDM or depleted CDM. Our result showed that both the mutants grew similarly to the WT in the presence of vitamin B6 (see Fig. S4 in the supplemental material) but were fully auxotrophic for vitamin B6 (Fig. 6). The growth phenotypes of both mutants in the absence of vitamin B6 can be corrected by complementation with their respective genes but not by the vector control (Fig. 6). These results are consistent with the Western blot analysis data (Fig. 5C).
Fig 6.
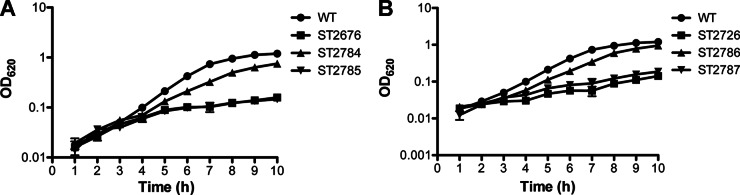
Growth of S. pneumoniae ΔpdxS (A) and ΔpdxR (B) in depleted CDM. The bacterial strains tested include WT (D39), ST2676 (ΔpdxS), ST2784 (ΔpdxS/pdxS+), ST2785 (ΔpdxS/vector control), ST2726 (ΔpdxR), ST2786 (ΔpdxR/pdxR+), and ST2787 (ΔpdxR/vector control). Bacterial growth was monitored hourly at OD620. The data shown are the means of three repeat experiments. Error bars denote the SEM. Note that ST2676 and ST2785 are indistinguishable in panel A.
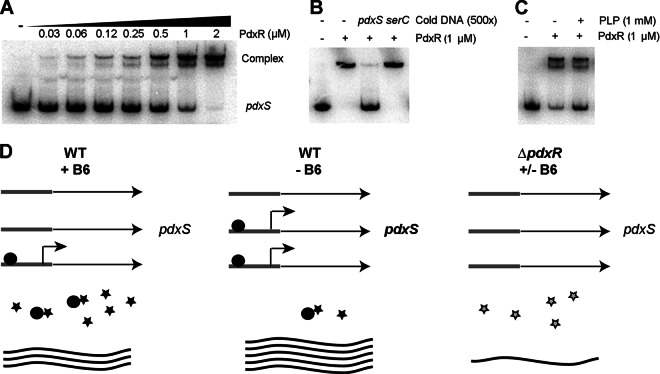
In order to determine whether the regulation of pdxS by PdxR is direct, we purified His-tagged PdxR from recombinant E. coli and then performed EMSA with this protein. Our result showed that the mobility of the labeled pdxS promoter DNA was retarded in the presence of PdxR (Fig. 7A). The free probe was detected when 500-fold excess of unlabeled pdxS DNA was present in the binding reaction (Fig. 7B). These results indicate that the interaction between the PdxR protein and the pdxS promoter DNA is specific. In addition, more unbound probe was detected when 1 mM PLP was added in the binding reaction (Fig. 7C), indicating that the affinity between PdxR-pdxS is reduced in the presence of PLP. We hypothesized that the PdxR protein we applied in the EMSA experiments had bound, at least partially, with PLP when it was purified. To test this hypothesis, we washed the recombinant E. coli thoroughly and resuspended the bacteria in M9 broth during induction for expressing PdxR. The protein purified from this procedure enhanced its affinity with pdxS promoter DNA by ∼2-fold more than that induced in LB broth (data not shown). Based on our data (Figs. 5 to 7), we propose a model for the regulation of pdxS by PLP and PdxR (Fig. 7D). At high concentrations of PLP, more PdxR binds PLP. The PLP-bound PdxR has a low affinity with the pdxS promoter DNA, thus the expression of pdxS is relatively low. However, when environmental PLP is reduced, PdxR-pdxS affinity is enhanced, which in turn activates the expression of pdxS. In the ΔpdxR mutant, pdxS is expressed at baseline levels either in the presence of absence of vitamin B6.
Fig 7.
Regulation of pdxS expression by PdxR. (A) EMSA experiment showing binding of the purified PdxR protein to the labeled putative pdxS promoter DNA. (B) Binding of the pdxS probe by PdxR in the presence of the 500-fold excess unlabeled (Cold) pdxS promoter DNA. The unlabeled M. tuberculosis serC promoter DNA is served as a nonspecific control. (C) Interaction of PdxR with the pdxS probe in the presence or absence of 1 mM PLP. This result is a representative of three repeat experiments. (D) Regulation of pdxS by PdxR and PLP under different growth conditions. A mechanistic model showing that pdxS expression requires the activator PdxR, and the expression levels of pdxS in the WT are higher when less PLP is available. In the ΔpdxR mutant, pdxS was expressed at the baseline levels either in the presence or absence of vitamin B6. Black ball, PdxR protein; black star, PLP molecule; gray star, with or without PLP; curved line, pdxS transcript.
S. pneumoniae PdxS impacts on infection in a chinchilla OM model, but not in a mouse pneumonia model.
It has been shown that the DXP-independent vitamin B6 biosynthesis pathway in M. tuberculosis plays an essential role in virulence (17). In the present study, we first determined the role of the pneumococcal vitamin B6 de novo biosynthesis pathway in virulence using a mouse pneumonia model. As a result, only 12% of the mice infected with either WT or ST2675 survived until the end of the experiment with a median survival time of 3 days. However, 50% of the ST2676-infected mice survived for the duration of the experiment with a median survival time of 6.5 days (Fig. 8A). Therefore, ST2676 was slightly attenuated compared to WT, although this difference appears to be statistically insignificant (P = 0.12).
Fig 8.
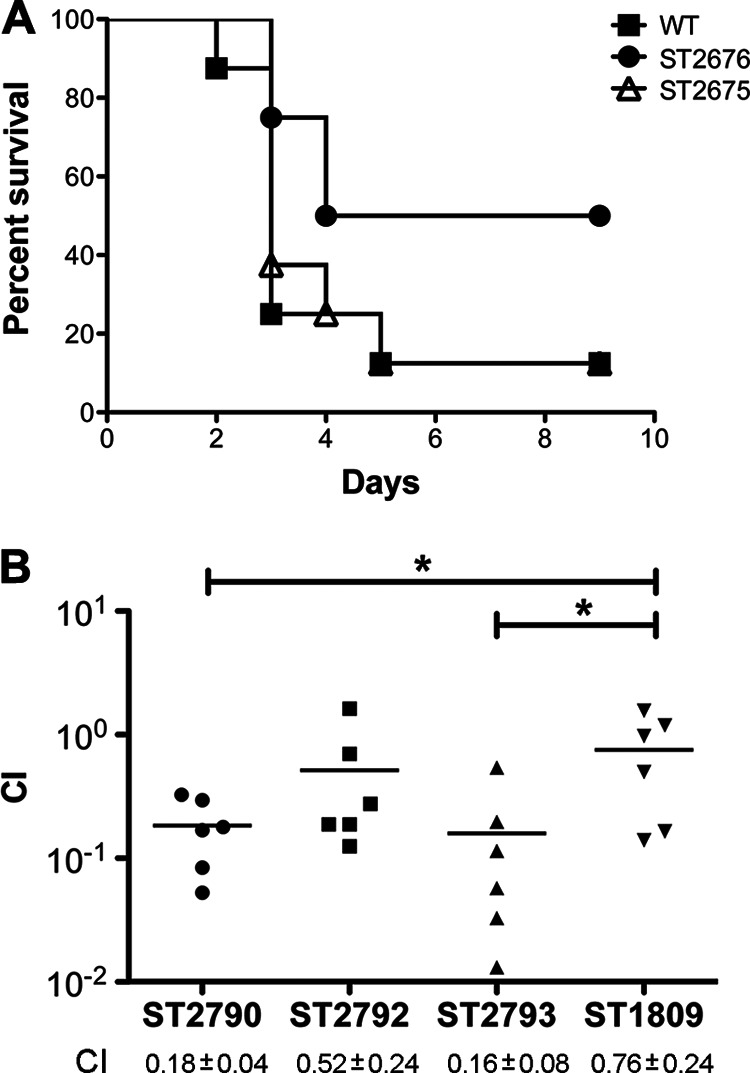
Infection of animals with S. pneumoniae pdxS mutants. (A) Survival of infected mice (n = 8). Mice were infected intranasally with WT (D39) and its derivatives, including ST2675 (ΔpdxT) and ST2676 (ΔpdxS), and the survival was monitored for up to 9 days postinoculation. (B) Coinfection of chinchilla middle ears (n = 6). Each ear was coinfected with 1:1 mixture of WT (ST606) and its derivatives, including ST2790 (ΔpdxS), ST2792 (ΔpdxS/pdxS+), ST2793 (ΔpdxS/vector control), and ST1809 (ΔarcT). The bacteria were washed out 3 days postinfection, and the CFU were counted to calculate the competitive index (CI). *, P < 0.05.
We argued that the pneumococcus might encounter different levels of vitamin B6 at various infection niches. Thus, we also infected chinchilla middle ears using a coinfection model with a ΔpdxS mutant (ST2790) generated from a clinical isolate (21). Our result showed that ST2790 was significantly attenuated (Fig. 8B). The attenuation could be partially recovered by the expression of pdxS (ST2792) but not by transformation with a vector control (ST2793) (Fig. 8B). The attenuations of both ST2790 and ST2793 are significant compared to the ST1809 control (P < 0.05).
DISCUSSION
A paradigm of DXP-independent vitamin B6 biosynthesis pathway includes two enzymes: PdxS and PdxT. This pathway has been well described in B. subtilis. In this bacterium, PdxS and PdxT physically form a complex as PLP synthase (9–11, 31, 37), which is also structurally stabilized by glutamine (9–11). The PdxS and PdxT complex is required for glutaminase activity, which converts glutamine into ammonium (9, 11). In addition, a ΔpdxT mutant of this bacterium grows well in the presence of sufficient ammonium in the medium (31), supporting that PdxT is not required for PLP synthase if ammonium is available.x
In S. pneumoniae, the glutaminase activity of PdxT is also dependent upon the availability of PdxS. However, we failed in detecting a PdxS-PdxT complex when we separated the protein mixture with a native gel. It is possible that the PdxS and PdxT proteins in S. pneumoniae differ from their orthologs in B. subtilis. We demonstrated that the S. pneumoniae ΔpdxT strain grew similarly to the WT when sufficient ammonium is available. In addition, the defective growth of the ΔpdxST mutant could be partially corrected by complementation with pdxS alone (not shown). These observations suggest that PdxS alone is functional to synthesize PLP in the presence ammonium, similar to its ortholog in B. subtilis.
In addition to a de novo vitamin B6 biosynthesis pathway, bacteria also have a salvage pathway that converts the B6 vitamers taken from the environment into PLP. It is interesting that various bacteria differ in usage of these vitamers. Our results showed that S. pneumoniae could utilize all of the vitamers we tested including PM, PN, PL, and PLP. However, 100 μM PLP is toxic to this bacterium, whereas E. coli grew well with up to 0.5 mM PLP in our study (not shown). When low concentration of a vitamer was supplemented for the growth of the S. pneumoniae ΔpdxS mutant, bacteria grew preferentially with the vitamers other than PN. This result coincides with what was observed in B. subtilis (31, 38). Mutants of the pdxS orthologs in M. tuberculosis complex bacteria grew well with PN or PL but very poorly with PM or PLP (17; G. Bai, unpublished data). It is likely that these bacteria differ in the uptake of B6 vitamers or that they are unable to convert certain vitamers to PLP, which warrants further investigation.
PLP has been extensively studied as a cofactor for various enzymes, but its regulatory role is still inadequately explored. A recent study revealed that the DXP-independent pathway in C. glutamicum is transcriptionally activated by PdxR and inhibited by PLP (18). However, a link between PdxR and PLP has not been established. In the present study, we demonstrated the same regulatory effects by PdxR and PLP in S. pneumoniae. Furthermore, with the addition of PLP in the in vitro reaction, we noticed that the affinity between the PdxR protein and the pdxS promoter DNA was reduced, even though it was not dramatic. More interestingly, the recombinant PdxR we induced in M9 broth, free of exogenous B6 vitamer, enhanced ∼2-fold in interaction with the DNA ligand, compared to the protein induced in LB broth (data not shown). These findings are consistent with the observation that a PLP-dependent transcriptional regulator GabR in B. subtilis reduces ∼2-fold in binding with its DNA ligand by addition of PLP and γ-amino-butyric acid (GABA) (39). Both PdxR and GabR have an AAT domain with putative PLP binding sites. Mutations of the PLP-binding sites in PdxR warrant further investigation to evaluate the role of PLP in the interaction between PdxR and its DNA ligands.
The knowledge pertaining to the vitamin B6 pathway in pathogenic bacteria is very limited. Recently, it has been shown that the vitamin B6 biosynthesis is essential for virulence of M. tuberculosis (17) and H. pylori (16). However, in our mouse infection experiment, the ΔpdxS mutant only showed a moderate attenuation and is not statistically significant. It is likely that the levels of vitamin B6 in the lungs are high enough to compensate the vitamin B6 defect of ST2676, as an extracellular pathogen. In contrast, the infection niche of M. tuberculosis differs from S. pneumoniae. M. tuberculosis encounters a relatively low vitamin B6 environment during intracellular replication. This hypothesis was supported by the coinfection experiment that vitamin B6 synthase gene affects the infection of S. pneumoniae in an OM model. Thus, the contribution of vitamin B6 in bacterial pathogenesis largely depends upon the particular infection niche. Understanding how bacterial metabolism pathways impact on infection will provide a molecular basis antimicrobial therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Jacobs for providing S. pneumoniae isolates from OM patients, Paul Kolenbrander for providing the pVA838 plasmid, the E. coli Genetic Center for providing the E. coli JW0051-1 (ΔpdxA) and JW2548-1 (ΔpdxJ) mutants, and Adam Underwood for critical reading of the manuscript.
This study was supported by National Institutes of Health grant DC006917 to D.W.M.
Footnotes
Published ahead of print 8 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00041-13.
REFERENCES
- 1. Eliot AC, Kirsch JF. 2004. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73:383–415 [DOI] [PubMed] [Google Scholar]
- 2. Grogan DW. 1988. Temperature-sensitive murein synthesis in an Escherichia coli pdx mutant and the role of alanine racemase. Arch. Microbiol. 150:363–367 [DOI] [PubMed] [Google Scholar]
- 3. Horrobin DF. 1993. Fatty acid metabolism in health and disease: the role of delta-6-desaturase. Am. J. Clin. Nutr. 57:732S–737S [DOI] [PubMed] [Google Scholar]
- 4. Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF. 2000. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 71:129–134 [DOI] [PubMed] [Google Scholar]
- 5. Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T. 2007. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem. J. 407:1–13 [DOI] [PubMed] [Google Scholar]
- 6. Mittenhuber G. 2001. Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J. Mol. Microbiol. Biotechnol. 3:1–20 [PubMed] [Google Scholar]
- 7. Mukherjee T, Hanes J, Tews I, Ealick SE, Begley TP. 2011. Pyridoxal phosphate: biosynthesis and catabolism. Biochim. Biophys. Acta 1814:1585–1596 [DOI] [PubMed] [Google Scholar]
- 8. Lam HM, Winkler ME. 1992. Characterization of the complex pdxH-tyrS operon of Escherichia coli K-12 and pleiotropic phenotypes caused by pdxH insertion mutations. J. Bacteriol. 174:6033–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neuwirth M, Flicker K, Strohmeier M, Tews I, Macheroux P. 2007. Thermodynamic characterization of the protein-protein interaction in the heteromeric Bacillus subtilis pyridoxalphosphate synthase. Biochemistry 46:5131–5139 [DOI] [PubMed] [Google Scholar]
- 10. Strohmeier M, Raschle T, Mazurkiewicz J, Rippe K, Sinning I, Fitzpatrick TB, Tews I. 2006. Structure of a bacterial pyridoxal 5′-phosphate synthase complex. Proc. Natl. Acad. Sci. U. S. A. 103:19284–19289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wallner S, Neuwirth M, Flicker K, Tews I, Macheroux P. 2009. Dissection of contributions from invariant amino acids to complex formation and catalysis in the heteromeric pyridoxal 5-phosphate synthase complex from Bacillus subtilis. Biochemistry 48:1928–1935 [DOI] [PubMed] [Google Scholar]
- 12. Yang Y, Tsui HC, Man TK, Winkler ME. 1998. Identification and function of the pdxY gene, which encodes a novel pyridoxal kinase involved in the salvage pathway of pyridoxal 5′-phosphate biosynthesis in Escherichia coli K-12. J. Bacteriol. 180:1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Safo MK, Musayev FN, di Salvo ML, Hunt S, Claude JB, Schirch V. 2006. Crystal structure of pyridoxal kinase from the Escherichia coli pdxK gene: implications for the classification of pyridoxal kinases. J. Bacteriol. 188:4542–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. di Salvo ML, Safo MK, Musayev FN, Bossa F, Schirch V. 2003. Structure and mechanism of Escherichia coli pyridoxine 5′-phosphate oxidase. Biochim. Biophys. Acta 1647:76–82 [DOI] [PubMed] [Google Scholar]
- 15. Zhao G, Winkler ME. 1995. Kinetic limitation and cellular amount of pyridoxine (pyridoxamine) 5′-phosphate oxidase of Escherichia coli K-12. J. Bacteriol. 177:883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grubman A, Phillips A, Thibonnier M, Kaparakis-Liaskos M, Johnson C, Thiberge JM, Radcliff FJ, Ecobichon C, Labigne A, de Reuse H, Mendz GL, Ferrero RL. 2010. Vitamin B6 is required for full motility and virulence in Helicobacter pylori. mBio 1:e00112 doi:10.1128/mBio.00112-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dick T, Manjunatha U, Kappes B, Gengenbacher M. 2010. Vitamin B6 biosynthesis is essential for survival and virulence of Mycobacterium tuberculosis. Mol. Microbiol. 78:980–988 [DOI] [PubMed] [Google Scholar]
- 18. Jochmann N, Gotker S, Tauch A. 2011. Positive transcriptional control of the pyridoxal phosphate biosynthesis genes pdxST by the MocR-type regulator PdxR of Corynebacterium glutamicum ATCC 13032. Microbiology 157:77–88 [DOI] [PubMed] [Google Scholar]
- 19. van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein L, Piccoli L, Simon D, Morrison D. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen H, Ma Y, Yang J, O'Brien CJ, Lee SL, Mazurkiewicz JE, Haataja S, Yan JH, Gao GF, Zhang JR. 2008. Genetic requirement for pneumococcal ear infection. PLoS One 3:e2950 doi:10.1371/journal.pone.0002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu L, Ma Y, Zhang JR. 2006. Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J. Biol. Chem. 281:15464–15474 [DOI] [PubMed] [Google Scholar]
- 23. Macrina FL, Evans RP, Tobian JA, Hartley DL, Clewell DB, Jones KR. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145–150 [DOI] [PubMed] [Google Scholar]
- 24. Bai G, Schaak DD, Smith EA, McDonough KA. 2011. Dysregulation of serine biosynthesis contributes to the growth defect of a Mycobacterium tuberculosis crp mutant. Mol. Microbiol. 82:180–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bai G, McCue LA, McDonough KA. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187:7795–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. 2012. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One 7:e35206 doi:10.1371/journal.pone.0035206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bai G, Gazdik MA, Schaak DD, McDonough KA. 2007. The Mycobacterium bovis BCG cyclic AMP receptor-like protein is a functional DNA binding protein in vitro and in vivo, but its activity differs from that of its M. tuberculosis ortholog, Rv3676. Infect. Immun. 75:5509–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belitsky BR. 2004. Physical and enzymological interaction of Bacillus subtilis proteins required for de novo pyridoxal 5′-phosphate biosynthesis. J. Bacteriol. 186:1191–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong YX, Sueda S, Nikawa J, Kondo H. 2004. Characterization of the products of the genes SNO1 and SNZ1 involved in pyridoxine synthesis in Saccharomyces cerevisiae. Eur. J. Biochem. 271:745–752 [DOI] [PubMed] [Google Scholar]
- 33. Gengenbacher M, Fitzpatrick TB, Raschle T, Flicker K, Sinning I, Muller S, Macheroux P, Tews I, Kappes B. 2006. Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: biochemical and structural insights. J. Biol. Chem. 281:3633–3641 [DOI] [PubMed] [Google Scholar]
- 34. Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB. 2005. Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. U. S. A. 102:13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wrenger C, Eschbach ML, Muller IB, Warnecke D, Walter RD. 2005. Analysis of the vitamin B6 biosynthesis pathway in the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 280:5242–5248 [DOI] [PubMed] [Google Scholar]
- 36. Wetzel DK, Ehrenshaft M, Denslow SA, Daub ME. 2004. Functional complementation between the PDX1 vitamin B6 biosynthetic gene of Cercospora nicotianae and pdxJ of Escherichia coli. FEBS Lett. 564:143–146 [DOI] [PubMed] [Google Scholar]
- 37. Raschle T, Speziga D, Kress W, Moccand C, Gehrig P, Amrhein N, Weber-Ban E, Fitzpatrick TB. 2009. Intersubunit cross-talk in pyridoxal 5′-phosphate synthase, coordinated by the C terminus of the synthase subunit. J. Biol. Chem. 284:7706–7718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakai A, Kita M, Katsuragi T, Ogasawara N, Tani Y. 2002. yaaD and yaaE are involved in vitamin B6 biosynthesis in Bacillus subtilis. J. Biosci. Bioeng. 93:309–312 [DOI] [PubMed] [Google Scholar]
- 39. Belitsky BR. 2004. Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator. J. Mol. Biol. 340:655–664 [DOI] [PubMed] [Google Scholar]
- 40. Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.