Abstract
Iron is an essential element for the hyperthermophilic archaeon Pyrococcus furiosus, and many of its iron-containing enzymes have been characterized. How iron assimilation is regulated, however, is unknown. The genome sequence contains genes encoding two putative iron-responsive transcription factors, DtxR and Fur. Global transcriptional profiles of the dtxR deletion mutant (ΔDTXR) and the parent strain under iron-sufficient and iron-limited conditions indicated that DtxR represses the expression of the genes encoding two putative iron transporters, Ftr1 and FeoAB, under iron-sufficient conditions. Under iron limitation, DtxR represses expression of the gene encoding the iron-containing enzyme aldehyde ferredoxin oxidoreductase and a putative ABC-type transporter. Analysis of the dtxR gene sequence indicated an incorrectly predicted translation start site, and the corrected full-length DtxR protein, in contrast to the truncated version, specifically bound to the promoters of ftr1 and feoAB, confirming its role as a transcription regulator. Expression of the gene encoding Ftr1 was dramatically upregulated by iron limitation, but no phenotype was observed for the ΔFTR1 deletion mutant under iron-limited conditions. The intracellular iron concentrations of ΔFTR1 and the parent strain were similar, suggesting that under the conditions tested, Ftr1 is not an essential iron transporter despite its response to iron. In contrast to DtxR, the Fur protein appears not to be a functional regulator in P. furiosus, since it did not bind to the promoters of any of the iron-regulated genes and the deletion mutant (ΔFUR) revealed no transcriptional responses to iron availability. DtxR is therefore the key iron-responsive transcriptional regulator in P. furiosus.
INTRODUCTION
The hyperthermophilic anaerobic archaeon Pyrococcus furiosus has an optimal growth temperature of 100°C (1). It is of biotechnological interest as a source of highly thermostable enzymes and because of its ability to produce hydrogen gas (2, 3). It is well established that the organism requires iron for efficient growth, and a number of iron-containing enzymes involved in the primary metabolic pathways of P. furiosus have been characterized. These include three hydrogenases (MBH, SHI, and SHII) (4, 5), which are involved in hydrogen metabolism, superoxide reductase (SOR) and rubrerythrin, which are involved in the response to oxidative stress (6, 7), and the primary redox proteins, ferredoxin and rubredoxin, both of which contain iron (8, 9). Furthermore, ferritin and Dps of P. furiosus have been shown to be potential iron storage proteins, and the elemental-sulfur-induced protein A (SipA) was found to sequester intracellular iron and sulfide when iron and sulfur were present in the growth medium (10–12). Given the abundance of iron-containing proteins in this organism, the goal of the present study was to understand its response to iron availability.
Iron metabolism is usually tightly regulated in aerobic organisms at the transcriptional level due to potential oxidative damage. In the presence of oxygen, the solubility of ferric iron is extremely low and excess iron can react with reactive oxygen species to generate radicals via Fenton chemistry, leading to damage of DNA, proteins, and fatty acids (13). However, in an anaerobic environment, iron is expected to remain mostly in the ferrous form, which is readily soluble and accessible, and the likelihood of oxidative damage is minimized. It has been reported that the global transcription profiles of the hyperthermophilic archaeon Thermococcus kodakarensis, a close relative of P. furiosus, were similar under iron-sufficient and iron-limited conditions, suggesting a less-than-stringent response to iron availability (14). Similar nonresponsive transcriptional effects of iron were also observed with the obligately anaerobic and mesophilic bacteria Coxiella burnetii and Dichelobacter nodosus (15, 16).
To study the regulation of iron metabolism in P. furiosus, we first searched its genome for genes encoding potential iron-responsive transcriptional regulators. Homologs of the bacterial transcription factors Fur and DtxR were found, while homologs of other previously identified regulators, including RirA, Rrf2, Aft1, Aft2, and IRP, were absent (17–19). Both DtxR and Fur are known to regulate the transcription of genes involved in iron metabolism, including transporters and storage proteins, and their homologs are widespread in many bacterial species (18, 20). As might be expected, DtxR homologs can be found in all members of the Thermococcaceae. The DtxR in T. kodakarensis is a potential regulator of the production of FeoAB and ferritin according to transcriptional analysis of a deletion mutant. However, the DtxR protein does not bind to the promoters of feoAB or ferritin genes based on electrophoretic mobility shift assays (14). In contrast, and for unknown reasons, Fur is poorly conserved in the Thermococcaceae, as it is found only in P. furiosus, Thermococcus barophilus (82% similarity to P. furiosus Fur), and T. kodakarensis (86% similarity to P. furiosus Fur). The fur homolog in T. kodakarensis contains a frameshift mutation that results in the expression of a truncated protein; however, correcting the frameshift in vivo did not affect the iron response or the transcription of any genes (14).
Utilizing the genetic system recently established for P. furiosus (21), we constructed deletion mutants of the putative iron-responsive transcription factors DtxR and Fur in order to investigate their roles in iron metabolism by global transcriptional analyses. Their abilities to bind to the promoters of the iron-responsive genes were also examined, and a deletion of the gene encoding the iron permease Ftr1 was also constructed to investigate its function. The results provide the first insights into how iron assimilation is regulated in a hyperthermophilic archaeon.
MATERIALS AND METHODS
Protein expression and purification.
The vectors containing the PF1194 sequence, encoding Fur (genome coordinates 1138175 to 1138558), the PF0851 sequence, predicted by NCBI to encode the truncated DtxR (genome coordinates 824720 to 825121), and the PF0851 sequence, containing the correct translation start (genome coordinates 824684 to 825121), were constructed using pET24dBAM. The pET24dBAM vector harboring the clones was a derivative of pET24d (Novagen), which was modified to incorporate an N-terminal His6-tag on the expressed protein. The clone was under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 promoter and was transformed into BL21-CodonPlus(DE3)-RIPL cells (Stratagene) according to the manufacturer's protocol. Cells were grown in Luria-Bertani broth (Fisher Bioreagents) to an optical density at 600 nm (OD600) of 0.8 to 1.0, and expression was induced with 1 mM IPTG. Cells were harvested 4 h after induction and suspended in 20 ml binding buffer (20 mM sodium phosphate, 0.5 M NaCl, pH 7.4) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 10 mM β-mercaptoethanol (Sigma). Cells were sonicated briefly and centrifuged for 60 min at 21,000 rpm in a Beckman JA25.5 rotor. The recombinant protein in the supernatant fraction was purified using a 1-ml HisTrap FF column (GE Healthcare) on the Äkta purifier system (Amersham Bioscience) according to manufacturer's instructions. The column was washed with eluting buffer (20 mM sodium phosphate, 0.5 M NaCl, 0.5 M imidazole, pH 7.4) with a 25-ml 0 to 100% gradient. Purified fractions were pooled and buffer exchanged into 20 mM sodium phosphate–150 mM NaCl, pH 7.4. The resulting proteins were estimated to be >95% pure in SDS-PAGE (Bio-Rad). Protein concentrations were determined by the Bradford method (Bio-Rad protein assay kit).
Gel filtration.
Gel filtration was carried out using a Superdex75 HiLoad 16/60 preparative-grade column (GE Healthcare) to determine the quaternary structures of His-tagged Fur and DtxR. The column was washed with one column volume of 50 mM sodium phosphate containing 0.05 M NaCl (pH 7.3) and then equilibrated with two column volumes of 50 mM sodium phosphate containing 0.15 M NaCl (pH 7.3) at a flow rate of 1.5 ml/min. Carbonic anhydrase (29 kDa, 3 mg/ml; Sigma-Aldrich) and albumin (66 kDa, 5 mg/ml; Sigma-Aldrich) were used as protein standards to determine approximate molecular masses.
EMSA.
The putative promoter regions (position −200 to +50 from ATG) of ftr1, feoAB, fur, dtxR, and a gene unrelated to iron metabolism (PF0849) were PCR amplified from P. furiosus genomic DNA. Incubation of DNA with protein was carried out in 10 μl electrophoretic mobility shift assay (EMSA) buffer (20 mM sodium phosphate, 200 mM KCl, 5% [vol/vol] glycerol, 1 mM dithiothreitol [DTT], pH 7.3) using a 5× stock. A 100 nM concentration of DNA was used in each assay, and the protein concentration was adjusted according to the molar amount of DNA. For EMSA with divalent metals or EDTA, 10× stock solutions were made freshly using (NH4)2Fe(SO4)2 · 6H2O, MnCl2 · 4H2O, ZnCl2, or CoCl2 · 6H2O (J. T. Baker) or Na2EDTA · 2H2O (Sigma). The final concentration of the metal ion or EDTA in EMSA was 50 μM. Although the assay was carried out aerobically, it was shown that that ferrous iron added remained in the ferrous form over the duration of the EMSA using bathophenanthroline disulfonate, which specifically chelates ferrous iron. For EMSA with heparin, 10× stocks containing 2 to 20,000 ng μl−1 heparin were prepared. Reaction mixtures were incubated at 60°C for 20 min and immediately loaded onto a 5% Tris-borate-EDTA (TBE) Ready gel (Bio-Rad). Gels were stained with ethidium bromide and imaged via UV transillumination.
Media and growth conditions.
The P. furiosus strains used or constructed in this study are listed in Table 1. The maltose-based growth medium was the same as previously reported (22) except that no elemental sulfur was added and the 1,000× trace minerals solution was made without FeCl3 and used as the iron-limited medium. This medium contained <0.5 μM Fe as determined by inductively coupled plasma mass spectrometry (ICP-MS) using an Agilent 7500ce instrument. The iron-sufficient medium was made by adding 10 μM (NH4)2Fe(SO4)2 · 6H2O to the iron-limited medium. All the auxotrophic strains (COM1, ΔFUR, and ΔFTR1) were grown in medium containing 20 μM uracil. The inocula underwent three successive transfers in the iron-limited medium to eliminate intracellular iron stores. Cell growth was measured either by optical density or by protein determination. Experiments to determine the effects of iron on cell growth were carried out in biological triplicate in 125-ml serum bottles with 80 ml medium at 98°C. All glassware used in growth experiments was washed in 2% (vol/vol) nitric acid overnight and rinsed with ultrapure water.
Table 1.
P. furiosus strains used in this study
| Strain designation | Genotype | Parent strain | Genome region deleteda |
|---|---|---|---|
| DSM3638 | Wild type | ||
| COM1 | ΔpyrF | Wild type | PF1114 (1062504–1063123) |
| COM1c2 | ΔpyrF::pyrF | COM1 | None, restored |
| ΔFURb | ΔpyrF Δfur | COM1 | PF1194 (1138175–1138558) |
| ΔDTXRb | ΔpyrF ΔdtxR::pyrF | COM1 | PF0851 (824642–825058) |
| ΔFTR1b | ΔpyrF Δftr1 | COM1 | PF0723 (721320–722156) |
Locus tags of deleted genes are listed, followed by nucleotides of the deleted region in parentheses (P. furiosus DSM 3638 genome, GenBank accession number AE009950.1).
Strain constructed in this study.
RNA isolation and cDNA synthesis.
RNA was obtained from 500-ml cultures harvested at exponential phase for quantitative PCR and microarray analyses. Total RNA was extracted from P. furiosus cells using acid-phenol and further purified by a second acid-phenol extraction. Contaminating DNA was digested by Turbo DNase (Ambion) treatment for 30 min at 37°C, and the RNA was acid-phenol extracted again. cDNA synthesis for microarray analyses was prepared using the AffinityScript quantitative PCR (qPCR) cDNA synthesis kit (Agilent) according to protocol except that the cDNA synthesis reaction was carried out at 42°C for 4 h. The cDNA products were used directly for qPCR. For DNA microarray analyses, base hydrolysis of RNA was performed and cDNA was purified by phenol-chloroform extraction and ethanol precipitated before use.
Construction of gene deletions.
Deletions of fur (PF1194) and ftr1 (PF0723) were constructed with 1-kb flanking regions joined by splice-overlap extension (SOE)–PCR (23) and cloned into pGLW015 containing the PgdhpyrF cassette for prototrophic selection. The deletion of dtxR (PF0851) was constructed by SOE-PCR with 1-kb flanking regions on either side of the PgdhpyrF cassette. Plasmid deletion constructs for fur and ftr1 were transformed into P. furiosus COM1 (ΔpyrF), with selection for uracil prototrophy on solid defined medium followed by counterselection for loss of the plasmid using 5-fluoroorotic acid (5-FOA) resistance as previously described (21). The PCR product containing a deletion of dtxR was transformed directly with selection for uracil prototrophy, resulting in marker replacement. The COM1c2 strain was constructed to restore the ΔpyrF allele in the COM1 strain to the wild type as previously described (24). DNA was extracted from transformants as previously described (21) and screened for deletion by PCR amplification of the locus using primers outside the homologous flanking regions used to construct the deletions. Isolates containing the deletions were further colony purified by serial passage on solid medium. PCR products amplified from the target regions were sequenced, and quantitative PCR was used to verify the deletions.
qPCR.
Quantitative PCRs (qPCRs) were carried out in technical triplicate using an Mx3000P instrument (Agilent) and the Brilliant SYBR green qPCR master mix (Agilent). The constitutively expressed gene encoding the pyruvate ferredoxin oxidoreductase (POR) gamma subunit (PF0971) was used as an internal control. The comparative cycle threshold method was used to analyze the resulting data, which are expressed as a ratio of the change in gene expression (n-fold).
DNA microarray analyses.
The COM1c2 and ΔDTXR strains (500 ml) were grown in iron-sufficient and iron-limited media in duplicate and harvested at a cell density of ∼4 × 107 cells/ml. cDNA was labeled using the Alexa Fluor labeling kit (Invitrogen) with Alexa dye 546, 594, or 647. The labeled cDNA was purified using Micro Bio-Spin chromatography columns (Bio-Rad) according to the manufacturer's instructions. Differentially labeled cDNAs derived from the same strain grown in iron-sufficient or iron-limited medium were combined and hybridized to the microarrays. Microarray analysis was performed as previously described (25). Each log2 value represents an average from four hybridization experiments performed using cDNA derived from four different cultures, two grown under iron-limited conditions and two grown under iron-sufficient conditions.
Intracellular iron content.
The COM1 and ΔFTR1 strains were grown and transferred in the iron-limited medium three times to eliminate intracellular iron stores. These were then used to inoculate iron-sufficient and iron-limited media (500 ml), and cells were harvested in exponential phase (∼3 × 107 cells/ml) and stationary phase (∼2 × 108 cells/ml). Pellets were washed three times with base salt solution (22) containing 1 mM EDTA and once with the base salt solution to remove extracellular traces of metals. Cells were lysed using buffer containing 20 mM sodium phosphate (pH 7.4) at a ratio of 3 ml per gram of cells. Extracts were incubated at room temperature with vigorous shaking for 30 min. Lysates were then centrifuged at 72,000 × g for 1 h, and the supernatant was collected. The intracellular iron content of cells grown from various conditions was analyzed by ICP-MS in technical triplicate. The iron contents measured by ICP-MS were normalized to the protein concentration of each sample.
RESULTS
Growth of DtxR and Fur deletion mutants.
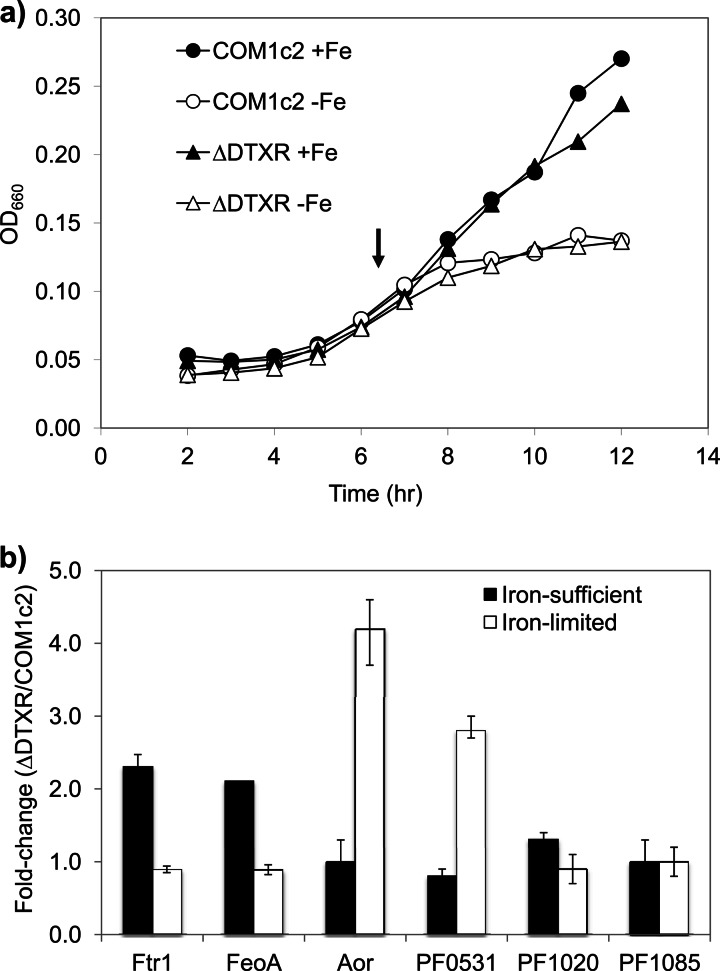
Optimal growth of P. furious in the standard maltose-containing medium required 10 μM iron, and the cell yield decreased by about half when the iron concentration was decreased to 0.5 μM (see Fig. S1 in the supplemental material). These were therefore taken to represent iron-sufficient and iron-limited conditions, respectively. To investigate the possible roles of DtxR and Fur as iron-responsive transcription factors in P. furiosus, strains containing deletions of the dtxR (ΔDTXR) and fur (ΔFUR) genes were constructed and were cultured in iron-sufficient (10 μM Fe) and iron-limited (0.5 μM Fe) media. Strain ΔFUR was compared with the parent strain COM1 (both lack the pyrF marker and require uracil for growth), while ΔDTXR (a marker-replaced deletion mutant containing pyrF) was compared with COM1c2 (the COM1 strain containing a restored pyrF locus). Growth of all strains under iron-sufficient conditions reached a significantly higher cell density than growth under iron-limited conditions, but ΔDTXR and ΔFUR did not exhibit any growth phenotypes compared to the parent strains (Fig. 1a; see Fig. S2 in the supplemental material).
Fig 1.
Characterization of the ΔDTXR strain. (a) Growth of ΔDTXR and the parent (COM1c2) strains under iron-sufficient (+Fe, 10 μM iron added) and iron-limited (−Fe, no iron added) conditions. Growth was monitored by optical density at 660 nm. Results are shown for COM1c2 (circles) and ΔDTXR (triangles) under iron-sufficient (solid symbols) and iron-limited (open symbols) conditions. The arrow indicates the culture harvest point for RNA isolation for DNA microarray analysis. (b) The effect of dtxR deletion on gene expression was measured in ΔDTXR and COM1c2 using quantitative PCR. Total RNA was prepared from ΔDTXR and COM1c2 grown under iron-sufficient and iron-limited conditions. The constitutively expressed gene encoding the pyruvate ferredoxin oxidoreductase (POR) gamma subunit (PF0971) was used as an internal control. Results are expressed as a change in gene expression, comparing ΔDTXR to COM1c2 under iron-sufficient (closed bars) and iron-limited (open bars) conditions.
Effect of iron availability on the transcription profile of the parent strain.
The parent strain COM1c2 was grown under iron-limited and iron-sufficient conditions to late log phase, and transcriptional analysis was carried out using a whole-genome DNA microarray (see Tables S1 and S2 in the supplemental material). Expression of genes encoding the putative iron transporters Ftr1 (PF0723) and FeoAB (PF0857-PF0858) was significantly upregulated by iron limitation (by 3.1- and 2.0-fold, respectively). The expression of a putative phosphate transporter encoded by PF1020-PF1021 also increased by approximately 3-fold in response to iron limitation. PF1020 is homologous to the sodium-dependent phosphate transporter Pho-4 in Neurospora crassa, the transcription of which is increased in the presence of Na+ (26). The PF1020-PF1021 transporter system therefore appears to be associated with iron uptake, although its physiological role is unclear. The genes whose expression was downregulated by iron limitation included that encoding the iron-sulfur cluster-containing enzyme aldehyde ferredoxin oxidoreductase (AOR) (by 3-fold), a known tungsten-containing protein that oxidizes aldehydes and is proposed to be involved in fermentative pathways (27). aor is one of the most highly expressed genes under iron-sufficient growth conditions, with AOR representing about 1% of the total soluble protein in P. furiosus (28). Hence, the downregulation of aor could spare iron when iron availability is limited. Expression of the gene encoding peroxiredoxin (PF1033) decreased by 4.3-fold under iron limitation. Although PF1033 is not predicted to be an iron-containing protein, the gene encoding it is also upregulated by oxidative stress (hydrogen peroxide) (29). Bacterial peroxiredoxins are peroxidases that catalyze the reduction of hydrogen peroxide, organic hydroperoxides, and peroxynitrite at their cysteine-containing active sites. The homolog in the related species Pyrococcus horikoshii (PH1217) catalyzes the reduction of cumene hydroperoxide, and its expression is also regulated by oxidative stress (30). The downregulation of PF1033 during iron limitation is likely due to a decrease in the concentration of highly reactive oxygen species generated by iron-dependent Fenton reactions, suggesting that P. furiosus downregulates its scavenging system when cells are less threatened by oxidative stress.
Iron limitation also caused decreased expression (by about 2-fold) of the genes encoding three ABC-type transporters, which are potentially involved in peptide (PF0191-PF0196), cobalt (PF0528-PF0531), and sugar (PF1936-PF1938) transport. The gene cluster PF1936-PF1938 encodes one of the two sugar-specific transporters in P. furiosus and recognizes maltodextrins with high specificity but not maltose. It has been shown previously that both the maltodextrin-specific transporter (Mal-II) and the trehalose/maltose-specific transporter (PF1739-PF1742) (Mal-I) are induced when grown on maltose, even though Mal-II does not transport maltose (31). On the other hand, expression of the Mal-II operon decreases with iron limitation, while Mal-I appears to be constitutively expressed.
Transcription analysis of a DtxR deletion mutant and regulation by DtxR.
To provide insight into the regulatory targets of DtxR, the transcriptional profiles of the ΔDTXR deletion strain were compared with those of the parent strain COM1c2. In ΔDTXR, the transcription of genes that were originally under the control of DtxR in response to iron was predicted to be independent of iron. Indeed, a more limited transcriptional response to iron was seen in the ΔDTXR strain than in COM1c2, with significantly fewer genes regulated by iron (by greater than 2-fold) in the deletion strain (see Tables S1 and S2 in the supplemental material). The remaining iron-responsive genes in ΔDTXR were all found to be similarly regulated in COM1c2. Therefore, the genes that no longer respond to iron in ΔDTXR were considered potentially regulated by DtxR.
The expression levels of genes encoding the iron transporters Ftr1 and FeoAB were similar under iron-limited and iron-sufficient conditions in ΔDTXR, indicating that these two genes are regulated by DtxR. The decrease in the expression of AOR (PF0346) in ΔDTXR in response to limiting iron is 1.8-fold, which is significantly different from that in COM1c2 (3.0-fold). The regulation of putative phosphate and cobalt transporters PF1020-PF1021 and PF0528-PF0531 was also muted in ΔDTXR. The regulation of the alpha subunit of ACSII (PF1085) by iron also differs in the two strains, even though there is no obvious relevance to iron metabolism. Several genes of unknown function, including PF1085.1 and PF1951, were also differentially regulated in ΔDTXR; however, there is no evidence to suggest that there is any direct relationship between these genes and iron metabolism in P. furiosus. Presumably, the response of these genes to iron is caused by a secondary, as-yet-unknown regulatory system.
Quantitative PCR confirmation of the genes differentially regulated in COM1c2 and ΔDTXR by microarray analysis showed a generally similar pattern of iron-responsive regulation, except for PF1085, the transcription of which was not regulated by iron as judged by qPCR (Table 2). To reveal the effect of DtxR on gene expression, transcription levels were also compared between strains. The expression levels of the genes encoding Ftr1 and FeoAB were similar under iron-limited conditions in the ΔDTXR and COM1c2 strains, while under iron-sufficient conditions, their expression in COM1c2 was decreased compared to that in ΔDTXR (Fig. 1b). This indicates that DtxR likely functions as a transcriptional repressor for the Ftr1 and FeoAB genes under iron-sufficient conditions. In contrast, the expression of AOR and PF0531 in ΔDTXR was significantly higher than that in COM1c2 under iron-limited conditions, suggesting that DtxR represses the transcription of AOR and PF0531 when iron is limited. The gene PF1020 seems not to be regulated by DtxR, since its transcription levels in COM1c2 and ΔDTXR were similar under all conditions.
Table 2.
Quantitative PCR analysis of the regulation of genes in the ΔDTXR and parent strains
| Open reading frame | Protein descriptiona | Fold change (−Fe vs +Fe)b in strain: |
|
|---|---|---|---|
| COM1c2 | ΔDTXR | ||
| PF0723 | FTR1 iron permease | 5.5 | 2.2 |
| PF0858 | Ferrous iron transporter A | 2.1 | 0.9 |
| PF1020 | Phosphate transport protein | 4.1 | 2.7 |
| PF0531 | Cobalamin biosynthesis protein | 0.5 | 1.7 |
| PF0346 | Aldehyde ferredoxin oxidoreductase | 0.1 | 0.5 |
| PF1085 | Acetyl coenzyme A synthetase Q3 alpha | 1.5 | 1.4 |
Description derived from the annotation in either the NCBI or TIGR database.
Fold changes for iron-limited (−Fe) versus iron-sufficient (+Fe) growth conditions.
Regulation of transcription by Fur.
The iron-responsive regulation of putative iron transporters was measured in ΔFUR and the parent COM1 strain using quantitative PCR. The transcription levels of ftr1, feoAB, and the other putative iron transporter genes (Table 1) were similarly regulated in the deletion and parent strains in response to iron, indicating that Fur is not likely to be an effective iron-responsive regulator in P. furiosus. It was also shown that Fur has no effect on transcription of dtxR, which encodes the other putative iron-responsive regulator (see Fig. S1 in the supplemental material).
DNA binding of recombinant P. furiosus DtxR and Fur proteins.
Sequence analysis of P. furiosus DtxR along with homologs in other members of the Thermococcaceae suggested that the translation start site predicted by the NCBI and TIGR databases was incorrect (see Fig. S3a in the supplemental material). An analysis of the upstream sequence of the P. furiosus DtxR gene revealed an alternative start codon, TTG, which is a less common start codon than ATG but is known to be functional in other genes (32). The new start site results in an additional 12 amino acids at the N terminus, and these exhibited sequence similarity with the DtxR homologs in the other members of the Thermococcaceae.
Recombinant DtxR and Fur were expressed in Escherichia coli. For DtxR, both the NCBI-predicted “truncated” protein (t-DtxR) and the full-length protein (DtxR) were produced. Gel filtration results showed that all of these proteins form dimers in solution (data not shown). The DNA-binding abilities of t-DtxR, DtxR, and Fur were examined using EMSA. The upstream regions containing the putative promoters of the iron transporter genes ftr1 and feoAB were selected as binding targets, since their transcription is regulated by iron according to microarray and qPCR data. However, while no binding was observed for t-DtxR and Fur (see Fig. S4 in the supplemental material), mobility shifts were observed for full-length DtxR (see Fig. S5, lanes 1 to 5, in the supplemental material), indicating that it is able to bind to both promoters to form specific protein-DNA complexes and confirming that the N-terminal “extension” in DtxR is essential for its DNA-binding ability. Addition of iron to the EMSA mixture did not change the binding of DtxR, indicating that the recombinant protein already contains iron. The ability of Fur to bind to its own promoter was also investigated. However, no mobility shift was observed when Fur was incubated with the upstream region of fur with and without iron, nickel, manganese, or zinc (data not shown), indicating that Fur does not appear to regulate its own expression.
To verify that the binding between (full-length) DtxR and the promoters of ftr1 and feoAB was the result of specific protein-DNA interactions, heparin, a DNA structural analog, was used in EMSA as a nonspecific competitor (33). The protein concentration was kept at 3.2 μM, which was sufficient to cause a shift in the mobility of 100 nM DNA, while heparin was added at from 0.2 to 2,000 ng μl−1. The promoter of gene PF0849 was used as a negative control. Although DtxR binds to this promoter at high protein concentrations, no specific mobility shifts were generated, and the binding did not occur with a heparin concentration of 20 ng μl−1 (see Fig. S5, lanes 6 to 10, in the supplemental material). In comparison, specific DtxR-binding complexes with the ftr1 and feoAB promoters were observed even with increasing heparin concentrations and were not prevented until the heparin concentration reached 2,000 ng μl−1, indicating a higher affinity of DtxR for these promoters.
The binding of DtxR with promoters of the AOR and PF0531 genes, the transcription of which appears to be regulated by DtxR under iron-limited conditions, was also examined. However, DtxR was not able to bind these promoters, and the addition of either EDTA or the divalent metals Fe2+, Zn2+, Mn2+, and Co2+ had no effect on binding. Binding of DtxR to its own promoter was also examined. Again, no specific binding complex was observed, suggesting that DtxR is not autoregulated (data not shown).
Role of Ftr1 in iron acquisition.
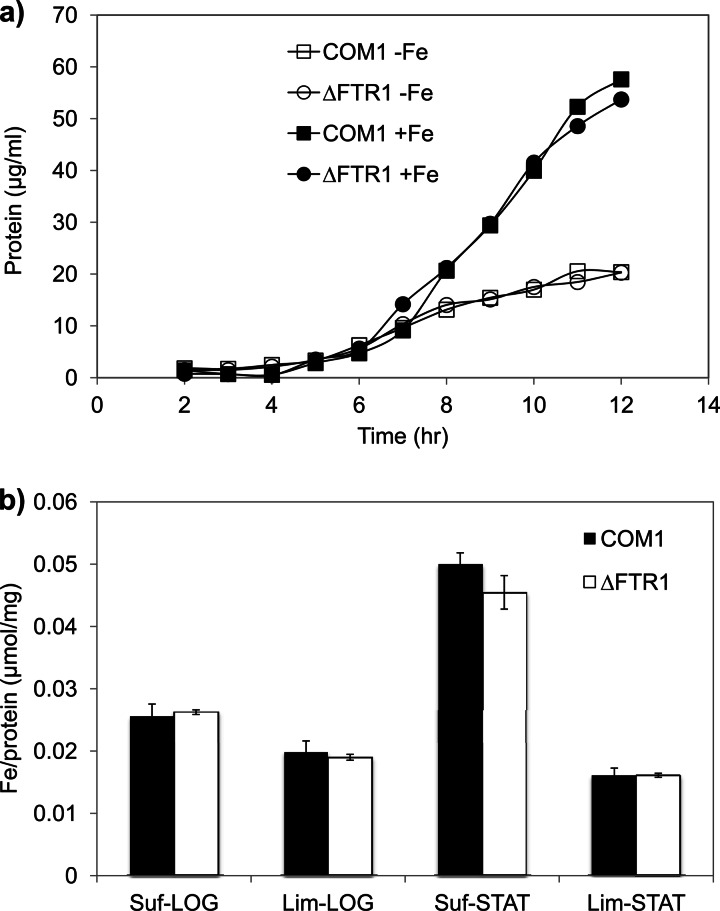
The putative iron permease Ftr1 was thought to play an important role in iron uptake in P. furiosus, since microarray data indicated that its gene was the most highly upregulated iron transporter gene in response to iron limitation. Therefore, a mutant with a deletion of the ftr1 gene was constructed (ΔFTR1) to study the role of Ftr1 in iron transport. ΔFTR1 and the parent strain COM1 were grown in iron-sufficient and iron-limited media (Fig. 2a). Surprisingly, ΔFTR1 displayed no growth phenotype associated with iron limitation. To rule out the chances of Ftr1 being involved in the transport of other metals, a metal-limited medium omitting all the trace minerals was used; however, the growth of ΔFTR1 and that of the control strains were still very similar (data not shown).
Fig 2.
Characterization of the ΔFTR1 mutant. (a) Growth of ΔFTR1 and COM1 under iron-sufficient [+Fe, 10 μM (NH4)2Fe(SO4)2 added] and iron-limited (−Fe, no iron source added) conditions. Cultures were grown in 125-ml bottles at 98°C with 5 g/liter maltose, 0.5 g/liter yeast extract, and 20 μM uracil. Cell growth was monitored by assaying total cell protein at each time point. Results are shown for COM1 (squares) and ΔFTR1 (circles) under iron-sufficient (closed symbols) and iron-limited (open symbols) conditions. (b) Cultures of ΔFTR1 (open bars) and COM1 (closed bars) grown under iron-limited (Lim) and iron-sufficient (Suf) medium were harvested at exponential (LOG) and stationary (STAT) phases. Cell pellets were washed three times with 1× base salt solution to remove extracellular metals and lysed using lysis buffer. Lysates were then centrifuged, and the supernatant was collected for the measurement of intracellular iron content using ICP-MS. The iron contents measured by ICP-MS were normalized to the protein concentration of each sample.
The intracellular iron contents of ΔFTR1 and COM1 under iron-sufficient and iron-limited growth conditions were determined by ICP-MS analyses (Fig. 2b). Cells removed from cultures in exponential phase grown under iron-sufficient and iron-limited conditions had similar intracellular iron concentrations. During stationary phase, the intracellular iron concentration in iron-sufficient medium was up to 3-fold higher, while in the iron-limited medium, the iron content remained at the same level as during log phase. Despite the differences in intracellular iron concentration observed at different growth phases for the two types of medium, the intracellular iron content of ΔFTR1 was not significantly different from that of COM1 in either case. The expression of other genes encoding putative iron transporters was not significantly different in ΔFTR1 and COM1 (see Fig. S6 in the supplemental material). Therefore, it appears that while other transporters are able to compensate for the lack of Ftr1 under the growth conditions tested, their expression levels do not change in response to the loss of Ftr1.
DISCUSSION
We conclude from the results of the transcriptomic studies of gene deletion strains and sequence-specific DNA-binding properties of the recombinant protein that DtxR is a major iron-responsive transcription factor in P. furiosus (see Fig. S7 in the supplemental material). It binds directly to the promoter regions of the genes encoding two putative iron transporters, ftr1 and feoAB. The expression of Ftr1 and FeoAB increases under iron-limited conditions only when DtxR is present. DtxR is also required for downregulating the expression of AOR and the putative ABC-type cobalt transporter PF0528-PF0531 under iron-limited conditions. However, we did not detect any DNA binding by the recombinant DtxR protein to the promoters of their genes, indicating that the regulation is indirect in response to iron, possibly caused by a secondary response triggered by DtxR.
On the other hand, Fur is not likely to be a functional iron-dependent transcription factor in P. furiosus, since none of the iron-responsive genes were differentially regulated in the fur deletion strain. The lack of an iron-responsive function for Fur in P. furiosus was not completely unexpected, given that when Fur was restored to a T. kodakarensis strain, there was no regulatory function in response to iron according to DNA microarray analyses (14). Fur is a member of a large protein family that includes regulators that are responsive to iron (Fur), manganese (Mur), zinc (Zur), nickel (Nur), and hydrogen peroxide (PerR). Sequence alignments and, particularly, comparing the metal-binding residues show that P. furiosus Fur is distinct from the Mur, Zur, and Nur proteins although not from members of the PerR subfamily (34). Accordingly, it was shown here that P. furiosus Fur did not bind to the promoter of its own gene in the presence of Fe2+, Zn2+, Mn2+, or Ni2+ ions. Although it cannot be ruled out that Fur has a PerR-like function and plays a role in oxidative stress, expression of fur was not affected by the addition of hydrogen peroxide (29). Fur is poorly conserved not only in the Thermococcaceae but in Euryarchaeota in general, and its function in only a limited number of species is not clear at present.
Homologs of the iron-responsive transcription factor DtxR found in P. furiosus are present in all members of the Thermococcaceae. P. furiosus DtxR bound to the promoters of ftr1 and feoAB and repressed the expression of these genes under iron-sufficient growth conditions. The sequence of P. furiosus DtxR has similarity to that of the manganese transport regulator MntR (60% identity) in Bacillus subtilis (see Fig. S3b in the supplemental material), and these similarities include the metal ion-binding residues (35, 36). The protein sequence of P. furiosus DtxR predicted by the databases does not contain a 12-amino-acid N-terminal sequence that is conserved in B. subtilis MntR and all other DtxR homologs in the Thermococcaceae and was shown here to be required for binding promoter DNA. Interestingly, two of the metal-binding sites characterized in B. subtilis MntR (E8 and E11) reside within the N-terminal 12 amino acids not present in the truncated DtxR protein. Mutating the metal-binding residues has been reported to modify DtxR metal selectivity and responsiveness, as the metal-binding site helps drive quaternary structural changes and mediate the coupling between the metal- and DNA-binding sites (37). Hence, it is not surprising that the N-terminal region of P. furiosus DtxR is essential for it to function as an iron-responsive regulator.
In most aerobic bacteria, a large number of genes are regulated by iron-responsive transcription factors such as Fur and DtxR. For example, in E. coli, the global transcription factor Fur regulates the transcription of over 100 genes involved in iron and energy metabolism in response to iron (38). Similarly, in Corynebacterium glutamicum, DtxR regulates the expression of 64 genes encoding iron transporters, iron-containing proteins, and transcriptional regulators (39). In the anaerobe P. furiosus, however, there seems to be a much smaller set of genes regulated by DtxR, and Fur does not appear to play any role in iron metabolism. Similar results have been reported for several other anaerobes. The pathogenic bacterium Coxiella burnetii lacks annotated iron acquisition and storage systems, and its Fur regulon consists of three genes (the iron transporter gene feoB, a gene encoding a putative cation efflux protein, and a gene predicted to encode an iron-binding protein) (15), indicating that iron-dependent transcriptional regulation plays a minor role in iron metabolism. In Dichelobacter nodosus, the Fur regulon includes genes encoding two putative iron transporters (YfeA and CbiK) and a manganese superoxide dismutase (SodA) (16). In the sulfate-reducing bacterium Desulfovibrio vulgaris, numerous genes respond to iron limitation, but only a few of them are regulated by Fur in response to iron, while the regulatory pathway for the rest remains unclear (40). Thermococcus kodakarensis DtxR regulates the transcription of the putative FeoAB systems, a ZIP family heavy metal transporter, ferritin, and a potential superoxide reductase (SOR) (14).
Besides iron transporters, the transcription of genes encoding iron storage proteins (ftnA and bfr), iron-sulfur assembly system (the suf operon), and iron-containing proteins is regulated by the iron-responsive transcription factor Fur in E. coli. Homologs of genes encoding ferritin and Suf proteins are found in P. furiosus; however, their gene transcription was not regulated by iron limitation. The upregulation of expression of the suf operon in P. furiosus is a secondary response to elemental sulfur, presumably due to a decrease in the intracellular iron concentration during sulfur reduction (31). Interestingly, a recent study of an iron-sulfur detoxification protein termed SipA in P. furiosus showed that transcription of its gene is regulated by sulfide in an iron-dependent manner (12). Our EMSA results for DtxR with the putative sipA promoter showed that a specific binding complex was formed (data not shown), indicating that DtxR is possibly involved in the regulation of sipA in response to iron. On the other hand, the indirect transcriptional regulation of the aor and PF0531 genes by DtxR suggests the presence of secondary regulators, the nature of which is not known. It is known that positive regulation by Fur is often indirectly mediated by Fur-dependent repression of an antisense regulatory small RNA (sRNA), such as RyhB. This RNA acts at the posttranscriptional level to repress expression of a large number of genes encoding iron-containing proteins, resulting in the redistribution of the intracellular iron in a variety of organisms, including E. coli (41). Other iron-responsive sRNAs include Fur-regulated NrrF in Neisseria meningitidis (42) and PrrF1 and PrrF2 from Pseudomonas aeruginosa (43). Also, recent study on the regulon of the DtxR family manganese-responsive regulator MntR in E. coli revealed a protein-encoding small gene, mntS (formerly rybA), the deletion of which perturbs manganese-dependent repression of the manganese transporter gene mntH. However, homologs of none of these iron-responsive secondary regulators are found in the P. furiosus genome, and the mechanism of indirect regulation by DtxR in P. furiosus remains to be determined.
Consistent with the less stringent regulation of iron metabolism in P. furiosus, only a small number of putative iron transporter genes and no genes for siderophore synthesis are found in its genome (see Table S3 in the supplemental material). It was confirmed by a protein-DNA binding assay and microarray analysis that DtxR and/or iron does not regulate any components of the annotated ABC-type iron transporters. Unexpectedly, the most upregulated iron permease, Ftr1, under iron limitation is not essential for iron uptake. Interestingly, a putative iron-binding protein (PF1774) of the ABC-type iron transporter is more highly expressed than other iron transporters in P. furiosus. Moreover, its expression is at a level similar to those of the most highly expressed genes in P. furiosus, including genes encoding the S-layer protein, superoxide reductase, and rubrerythrin. The last two proteins contain iron, are involved in the detoxification of peroxide, and are constitutively expressed at high levels, rather than being regulated by oxidative stress, indicating that P. furiosus is constantly armed against oxidative stress, possibly in order to respond to rapid environmental change (29). Therefore, the housekeeping protein PF1774 might also play a role in the P. furiosus iron acquisition system. Further studies of the role of PF1774 in iron assimilation are under way.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences, of the U.S. Department of Energy for funding strain construction through grant DE-FG05-95ER20175 (to M.W.W.A.) and the Office of Biological and Environmental Research of the Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy, for funding strain analyses through grant FG02-08ER64690 (to R.A.S.).
We thank Karen Stirrett and Gina Lipscomb for providing the ΔFUR and ΔFTR1 mutant strains and for critical reading of the manuscript.
Footnotes
Published ahead of print 15 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02280-12.
REFERENCES
- 1. Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100-degrees C. Arch. Microbiol. 145:56–61 [Google Scholar]
- 2. Atomi H. 2005. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 9:166–173 [DOI] [PubMed] [Google Scholar]
- 3. Verhaart MR, Bielen AA, van der Oost J, Stams AJ, Kengen SW. 2010. Hydrogen production by hyperthermophilic and extremely thermophilic bacteria and archaea: mechanisms for reductant disposal. Environ. Technol. 31:993–1003 [DOI] [PubMed] [Google Scholar]
- 4. Sapra R, Verhagen MFJM, Adams MWW. 2000. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182:3423–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma KS, Weiss R, Adams MWW. 2000. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182:1864–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jenney FE, Jr, Verhagen MF, Cui X, Adams MW. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306–309 [DOI] [PubMed] [Google Scholar]
- 7. Weinberg MV, Jenney FE, Jr, Cui X, Adams MW. 2004. Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J. Bacteriol. 186:7888–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heltzel A, Smith ET, Zhou ZH, Blamey JM, Adams MW. 1994. Cloning, expression, and molecular characterization of the gene encoding an extremely thermostable [4Fe-4S] ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 176:4790–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurihara K, Tanaka I, Chatake T, Adams MW, Jenney FE, Jr, Moiseeva N, Bau R, Niimura N. 2004. Neutron crystallographic study on rubredoxin from Pyrococcus furiosus by BIX-3, a single-crystal diffractometer for biomacromolecules. Proc. Natl. Acad. Sci. U. S. A. 101:11215–11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Honarmand Ebrahimi K, Hagedoorn PL, Hagen WR. 2010. Inhibition and stimulation of formation of the ferroxidase center and the iron core in Pyrococcus furiosus ferritin. J. Biol. Inorg. Chem. 15:1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramsay B, Wiedenheft B, Allen M, Gauss GH, Lawrence CM, Young M, Douglas T. 2006. Dps-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J. Inorg. Biochem. 100:1061–1068 [DOI] [PubMed] [Google Scholar]
- 12. Clarkson SM, Newcomer EC, Young EG, Adams MW. 2010. The elemental sulfur-responsive protein (SipA) from the hyperthermophilic archaeon Pyrococcus furiosus is regulated by sulfide in an iron-dependent manner. J. Bacteriol. 192:5841–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wardman P, Candeias LP. 1996. Fenton chemistry: an introduction. Radiat. Res. 145:523–531 [PubMed] [Google Scholar]
- 14. Louvel H, Kanai T, Atomi H, Reeve JN. 2009. The Fur iron regulator-like protein is cryptic in the hyperthermophilic archaeon Thermococcus kodakaraensis. FEMS Microbiol. Lett. 295:117–128 [DOI] [PubMed] [Google Scholar]
- 15. Briggs HL, Pul N, Seshadri R, Wilson MJ, Tersteeg C, Russell-Lodrigue KE, Andoh M, Baumler AJ, Samuel JE. 2008. Limited role for iron regulation in Coxiella burnetii pathogenesis. Infect. Immun. 76:2189–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parker D, Kennan RM, Myers GS, Paulsen IT, Rood JI. 2005. Identification of a Dichelobacter nodosus ferric uptake regulator and determination of its regulatory targets. J. Bacteriol. 187:366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rutherford JC, Bird AJ. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 3:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hantke K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172–177 [DOI] [PubMed] [Google Scholar]
- 19. Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, Rodionov DA. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals 20:501–511 [DOI] [PubMed] [Google Scholar]
- 20. Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect. Immun. 77:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE, Scott RA, Adams MWW, Westpheling J. 2011. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77:2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams MW, Holden JF, Menon AL, Schut GJ, Grunden AM, Hou C, Hutchins AM, Jenney FE, Jr, Kim C, Ma K, Pan G, Roy R, Sapra R, Story SV, Verhagen MF. 2001. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higuchi R, Krummel B, Saiki RK. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bridger SL, Clarkson SM, Stirrett K, DeBarry MB, Lipscomb GL, Schut GJ, Westpheling J, Scott RA, Adams MW. 2011. Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J. Bacteriol. 193:6498–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schut GJ, Zhou J, Adams MW. 2001. DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a new type of sulfur-reducing enzyme complex. J. Bacteriol. 183:7027–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Versaw WK, Metzenberg RL. 1995. Repressible cation-phosphate symporters in Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 92:3884–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heider J, Ma K, Adams MW. 1995. Purification, characterization, and metabolic function of tungsten-containing aldehyde ferredoxin oxidoreductase from the hyperthermophilic and proteolytic archaeon Thermococcus strain ES-1. J. Bacteriol. 177:4757–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukund S, Adams MW. 1990. Characterization of a tungsten-iron-sulfur protein exhibiting novel spectroscopic and redox properties from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Biol. Chem. 265:11508–11516 [PubMed] [Google Scholar]
- 29. Strand KR, Sun C, Li T, Jenney FE, Jr, Schut GJ, Adams MW. 2010. Oxidative stress protection and the repair response to hydrogen peroxide in the hyperthermophilic archaeon Pyrococcus furiosus and in related species. Arch. Microbiol. 192:447–459 [DOI] [PubMed] [Google Scholar]
- 30. Kawakami R, Sakuraba H, Kamohara S, Goda S, Kawarabayasi Y, Ohshima T. 2004. Oxidative stress response in an anaerobic hyperthermophilic archaeon: presence of a functional peroxiredoxin in Pyrococcus horikoshii. J. Biochemistry 136:541–547 [DOI] [PubMed] [Google Scholar]
- 31. Schut GJ, Brehm SD, Datta S, Adams MW. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 33. Trubetskoy DO, Zavalova LL, Akopov SB, Nikolaev LG. 2002. Purification of proteins specifically binding human endogenous retrovirus K long terminal repeat by affinity elution chromatography. J. Chromatogr. A 976:95–101 [DOI] [PubMed] [Google Scholar]
- 34. Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 35. Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. 2006. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry 45:15359–15372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454–1468 [DOI] [PubMed] [Google Scholar]
- 37. Guedon E, Helmann JD. 2003. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 48:495–506 [DOI] [PubMed] [Google Scholar]
- 38. McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478–29486 [DOI] [PubMed] [Google Scholar]
- 39. Wennerhold J, Bott M. 2006. The DtxR regulon of Corynebacterium glutamicum. J. Bacteriol. 188:2907–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bender KS, Yen HCB, Hemme CL, Yang Z, He Z, He Q, Zhou J, Huang KH, Alm EJ, Hazen TC, Arkin AP, Wall JD. 2007. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibfio vulgaris Hildenborough. Appl. Environ. Microbiol. 73:5389–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masse E, Salvail H, Desnoyers G, Arguin M. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10:140–145 [DOI] [PubMed] [Google Scholar]
- 42. Mellin JR, Goswarni S, Grogan S, Tjaden B, Genco CA. 2007. A novel Fur- and iron-regulated small RNA, NrrF, is required for indirect Fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189:3686–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.